Abstract
Abstract We evaluated the biological basis of reduced fat gain by oleoylethanolamide (OEA) in high-fat-fed mice and sought to determine how degradation of OEA affected its efficiency by comparing its effects to those of KDS-5104, a nonhydrolyzable lipid OEA analog. Mice were given OEA or KDS-5104 by the oral route (100 mg/kg body weight). Sixty-eight variables per mouse, describing six biological processes (lipid transport, lipogenesis, energy intake, energy expenditure, endocannabinoid signaling, and glucose metabolism), spanning gene expression of biochemical and physiological parameters were examined to determine the primary target whereby OEA reduces fat gain. Although KDS-5104 but not OEA was resistant to fatty acid amide hydrolase hydrolysis, OEA was degraded by an unidentified hydrolysis system in the liver. Nevertheless, both compounds equally decreased body fat pads after 5 weeks (20%; P < 0.05). The six biological functions constructed from the 68 initial variables predicted up to 58% of adipose fat variations. Lipid transport appeared central to the explanation for body fat deposition (16%; P < 0.0001), in which decreased expression of the FAT/CD36 gene was the component most related to adipose depots. Lipid transport appears to be a determinant player in the OEA fat-lowering response, with adipose tissue FAT/CD36 expression being the most relevant bioindicator of OEA action.
Keywords: endocannabinoid, indirect calorimetry, nutrigenomics, PLS
Fatty acid ethanolamides (FAEs), or N-acylethanolamines, are amides of saturated or unsaturated fatty acids with an ethanolamine moiety. FAEs are formed in vivo from N-acetylated phosphatidylethanolamine derivatives and broken down by two different enzymes, fatty acid amide hydrolase (FAAH) (1) and N-acylethanolamine-hydrolyzing acid amidase (NAAA) (2–4). The most abundant FAEs, found in biological tissues such as brain and neuronal cells, are anandamide and oleoylethanolamide (OEA). They might also occur in trace amounts from edible sources (5). Biological effects and metabolism of intraperitoneally administered OEA have been recently reviewed (5). OEA was found to regulate feeding (6) and body weight gain (7, 8) partly through the activation of peroxisome proliferator-activated receptor-α (PPAR-α) (9), a ligand-activated transcription factor that regulates several pathways of lipid metabolism (10). Similarly, KDS-5104, a reportedly hydrolysis-resistant OEA analog, was observed to reduce food intake through a PPAR-α mechanism (see supplementary Fig. I for chemical structures). In addition to its effect on food intake, OEA has been shown to activate lipid metabolism through a decrease in neutral lipid content in hepatocytes, as well as through a decrease in serum cholesterol and triglyceride levels (9). In addition, increased lipid β oxidation in muscle and hepatic cells has been described (11), suggesting that OEA induces fat utilization as an energy substrate. Nonetheless, a direct evaluation of OEA on whole-body energy expenditure (EE) has remained unexplored.
Similar to results shown with intraperitoneal administration, previous rodent studies have shown that orally administered (8) or diet-supplemented OEA (e.g., see Ref. 14) is biologically effective, although the extent of its breakdown and, thus, its bioefficiency is not known. The only clinical study performed did not find an anorectic effect of OEA, but the dietary supplement used was phosphatidylethanolamine given as a parent OEA metabolic precursor (12).
From the above work, data regarding the various possible OEA modes of action to reduce body fat accretion appear only partial and fragmented. Furthermore, most of the reports make the assumption that the primary OEA response can be explained by the modulated expression of one or a few gene products such as PPAR-α, FAT/CD36, UCP2 (8), whereas the body fat modulation by OEA seems highly multifactorial (13). Examining together a large number of representative molecular indicators of biological functions assumed to be targeted by the treatment appears more likely to determine the complex mechanisms involved (14). However, with increasing complexity, it is difficult to interpret biological information if the numerous biomodulators are examined independently. The picture can be simplified and more easily interpreted through the use of a biological modules model (15). This perspective can be achieved by grouping and modeling the individual molecular players that belong to the same biological string into one composite bioindicator that integrates and summarizes the information (16).
By comparing the effects of long-term oral OEA supplementation with those of its nonhydrolyzable analog KDS-5104 and using an integrated modeling approach embedding molecular, biochemical, and physiological assessments, our goal was also to evaluate the influence of enzymatic breakdown of OEA on its ability to reduce body fat gain. Our primary focus was to identify the possible molecular targets of OEA that are relevant to body fat mass, as well as the underlying metabolic functions most likely affected in this process.
MATERIALS AND METHODS
Chemical synthesis
OEA was synthesized from oleic acid and ethanolamine (Sigma-Aldrich, St. Louis, MO) according to the method of Roe et al. (see Ref. 21) . Purity was checked by GC analysis (90% for OEA). KDS-5104 was synthesized according to the method of Astarita et al. (see Ref. 22) from oleoyl chloride (Sigma-Aldrich) and R-(−)-2-aminopropanol (Sigma-Aldrich) under stoichiometric conditions. Acyl chloride was preferred to the triglyceride form for oleic acid in order to induce a total reaction and to avoid heating conditions. Then, the synthesized molecule was washed three times using warm saline solution to eliminate secondary products. KDS-5104 purity was checked using a 1200 series Agilent Rapid Resolution Liquid Chromatography (RRLC) (Waldbronn, Germany) coupled through an ESI interface to a Micro Quadrupole Time of Light (microQTOFTM, Bruker Daltonik, Bremen, Germany) (99% purity).
Animals, diet, and experimental design
All experiments were conducted according to the French Regulations for Animal Experimentation (Art 19. Oct 1987, Ministry of Agriculture) and in conformity with the Public Health Service Policy after approval by our institutional Animal Care and Use Committee. Male C57Bl6j mice purchased at 8 weeks of age from Janvier Elevage (Le Genest-St.-Isle, France) were individually housed and fed ad libitum on a high-fat diet (50% of energy intake as lipids) for 2 weeks. For each kilogram, the composition of the high-fat diet was: 235 g of casein, 201 g of sucrose, 3.5 g of l-cysteine, 85 g of starch, 116 g of maltodextrin, 60 g of cellulose, 12 g of vitamins mix, 51.5 g of mineral mix, 18.4 g of canola oil, and 236 g of lard [Unité de Préparation des Aliments Expérimentaux (UPAE) INRA Jouy en Josas, France]. Subsequently, 30 mice were blocked by body weight (BW) and randomly allocated to three groups (n = 10 per group) to be fed the same high-fat diet for the subsequent 5 weeks’ experimental period. Two groups of mice were given OEA (100 mg/kg BW) or KDS-5104 (100 mg/kg BW) by addition to the diet, while the third group received the basal diet alone. Daily food intake was monitored, and mice were weighed three times per week. A second group of 24 mice was used to determine fat absorption. These mice were fed the same high-fat diet but with either 0.0, 0.1, or 1.0 g of OEA per kilogram of BW by addition to the diet for 5 weeks. During the fourth week of feeding, the mice were kept for 5 days in metabolic cages to collect feces for evaluation of fat absorption (see supplementary material).
Sampling
At the end of the experimental period, mice were euthanized by exsanguination via cardiac puncture after anesthesia with isoflurane (Abbot France, Rungis, France). Plasma was obtained by centrifugation (1,000 g for 10 min at 4°C). Mesenteric, epididymal, inguinal, and peritoneal adipose depots, as well as liver, stomach, small intestine mucosa, pancreas, and gastrocnemius muscle were removed and frozen in liquid nitrogen. Plasma, feces, and organ samples were kept at −80°C until analysis.
Biological analysis
All biological parameters and methods related to biochemical, physiological (including indirect calorimetry), gene expression, in vitro and ex vivo assays, and statistical validations are fully detailed in the supplementary material.
Statistical analysis
Univariate analysis.
Results are presented as means ± standard error of the means (SEM). Treatment effects on physiological parameters and gene expression data were tested by one-way ANOVA. A mixed-effects model for repeated measures with a first-order autoregressive variance covariance matrix was used, with each daily food intake, lipid oxidation (LOX), carbohydrate oxidation (GOX), and EE as dependent variables and group (OEA or KDS-5104) as independent variable. The effect of the interaction between Group and Time variables was tested with and without total activity adjustment (except for food intake). Statistical significance was set at a P level of <0.05. All analyses were performed with Statview and SAS version 9.1 software (SAS institute, Cary, NC).
Multivariate analysis.
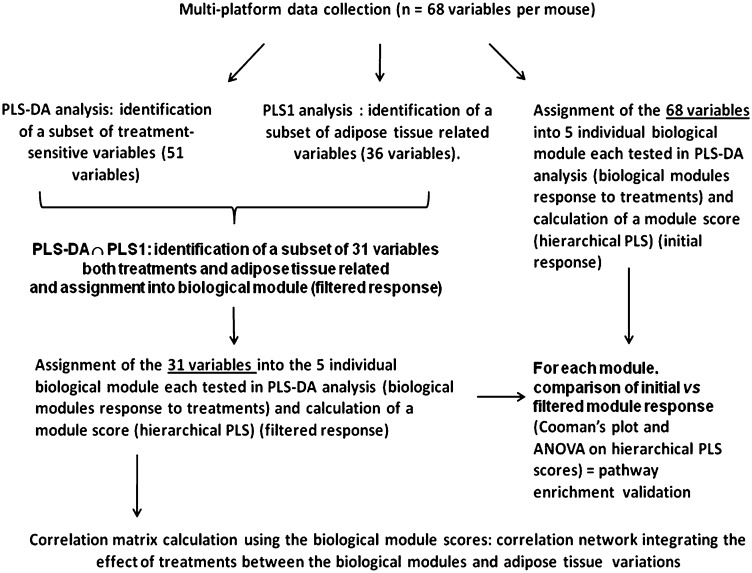
We used a multivariate statistical approach to obtain an integrated view of the biological impact of the treatments. The flowchart of multivariate statistical analysis is shown in Fig. 1. The objective was to find among selected biological processes (Table 1) those processes affected mostly by both OEA and KDS administration and also associated with the reduction of body fat gain (see a detailed description below). This included linking of gene expression with the biochemical and physiological data that belong to the same string of biological functions across various systems and specifically grouped as lipid transport, EE, energy intake, endocannabinoid signaling, lipogenesis, and glucose metabolism (Table 1). Such a statistical approach (17) allows a semiquantitation of the activity of each biological function under our nutritional challenge.
Fig.1.
Flow-chart of the statistical pipeline is shown. Validation of all models can be found in the supplementary material.
TABLE 1.
Assignment of genes, metabolites, and physiological parameters to biological processes
| Biological Cluster | Endocannabinoid Signaling | Lipid Transport | Lipogenesis | Energy Expenditure | Energy Intake | Glucose Metabolism |
|---|---|---|---|---|---|---|
| Genes | Adipose tissues CB1, upper intestine CB1 and GPR119, upper intestine and adipose tissues FAAH and NAAA, upper intestine OEAs | Adipose tissues, liver and upper intestine CD36, liver PCSK9, upper intestine and adipose tissues FIAF | Liver FAS, SREBP1c, SREBP2, liver and adipose tissues SCD1, adipose tissue SREBP1c, and SREBP2 | Liver ACO, adipose tissue epid. Adiponectin, liver, upper intestine and adipose tissues CPT1, muscle UCP2, upper intestine PPAR-α | Upper intestine CCK, stomach leptin and ghrelin, adipose tissues leptin | Muscle GLUT4, liver G6P and PEPCK, adipose tissues visfatin |
| Metabolites | Plasma cholesterol, plasma FFA, HDL-C, non-HDL-C, plasma TG | Liver TG, liver FA and glycerol | Plasma β-hydroxybutyrate | Plasma glucose | ||
| Physiological parameters | FAAH activity in intestine | Plasma insulin | Adiponectin, indirect calorimetry LOX, GOX, RQ, and EE, spontaneous activity | Plasma ghrelin, plasma leptin, cumulative food intake | Plasma insulin |
RQ, respiratory quotient; TG, triglyceride. Biological variables in boldface type indicate those variables modified by treatments (with 95% confidence interval, as determined from the PLS analysis) and used to calculate the biological module scores (hierarchical PLS).
Due to the large number of biological variables compared with the low number of mice, partial least squares discriminant analysis (PLS-DA) and partial least-square regressions (PLS) were used to identify factors that significantly contributed to the treatment effect. PLS-DA was used to select among the original 68 variables those that were treatment-responsive according to their variable importance index (VIP) (51 of 68 variables) (Fig. 1). Concurrently, PLS was used to identify the variables that were related to adipose fat gain (36 of 68 variables). The variables in common (i.e., both treatment-sensitive and adipose fat-related variables determined independently) were retained (31 of 68 variables). These were reassigned to their respective biological modules. For each biological module, significance of pathway enrichment was tested by comparing the PLS-DA model consisting of all the variables of a biological module to that calculated from the variables retained after the PLS-DA and PLS filtering (Fig. 1). Significant differences between the nonfiltered and filtered models were performed using both Cooman's plot critical distance and ANOVA for the hierarchical PLS scores from nonfiltered and filtered models (see supplementary material).
Calculations were performed using Simcap+ version 12.0 software (Umetrics, Umea, Sweden) using unit-of-variance scaling of the data. In each instance, model validation was performed using an R2 value (total variance explained) over 0.5, and a Q2 value (variance predicted after cross validation) at least equal to 2/3 of R2. Additional permutation tests (at least 100 permutations) allowed evaluation of model robustness. Variable selection was performed based on the significant (95% jackknife confidence interval) VIP with values over 0.8. All model validation indexes (Q2 after cross-validation; P values after cross-validation, ANOVA) are shown in supplementary Table II.
Hierarchical clustering of significant variables was performed using Permutmatrix software (http://www.lirmm.fr/∼caraux/PermutMatrix/PermutMatrixHome.htm) with Pearson correlation as distance and the Ward method as clustering condition. In addition, a correlation network displaying the strongest associations among the biological modules computed from the hierarchical PLS was built using Cytoscape open source freeware (18). The number of edges of the network, which represent the Pearson correlation coefficient between the biological module variables, was defined from the simplest network to display the entire biological modules. Thus, of the 21 possible relationships theoretically possible, only the 7 strongest are shown (correlation coefficient among the variables ranged from 0.703 to 0.85), validated by the false discovery rate for multiple testing.
RESULTS
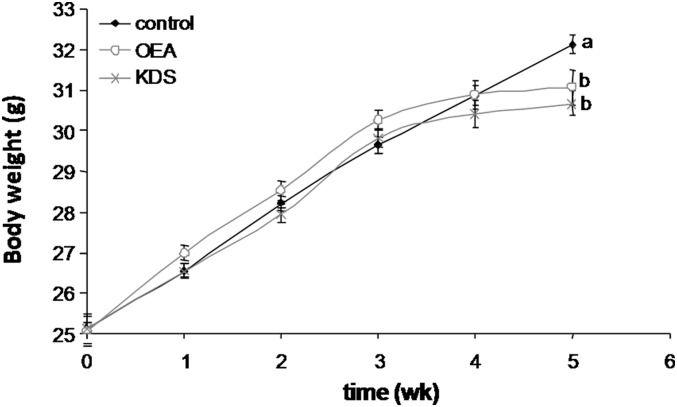
Body weight gain started to shift from control mice from 3 weeks onward after supplementation with either OEA or KDS-5104 (Fig. 2), resulting in a significant and similar decrease in body weight gain after 5 weeks’ administration of OEA (−6%, P = 0.0029) and KDS-5104 (−7%, P = 0.010) (Table 2). After 5 weeks, all fat depots were reduced following KDS-5104 administration, whereas OEA did not affect epididymal or inguinal adipose tissues (Table 2). However, total fat pads were decreased similarly by OEA and KDS-5104 (−16% for OEA and −20% for KDS-5104; P = 0.0364 and 0.0126, respectively).
Fig. 2.
Body weight changes are shown. Results are means ± SEM (N = 10 mice per group). Plots not sharing the same letter (a, b) are statistically different (P ≤ 0.05). The bold line with black squares corresponds to the control group; the continuous gray line with open circles corresponds to the 0.1 g/kg BW OEA group; and the gray line with x's corresponds to the 0.1 g/kg BW KDS-5104 group.
TABLE 2.
Basal physiological parameters of mice fed on experimental diets for 5 weeks
| Physiological parameter | Controlb | OEA (0.1 g/kg BW)b | KDS-5104 (0.1 g/kg BW)b |
|---|---|---|---|
| Cumulative food intake (g) | 103.12 ± 1.5a | 97.56 ± 0.89b | 96.69 ± 0.88b |
| Final body weight (g) | 32.6 ± 0.2a | 30.6 ± 0.6b | 30.3 ± 0.6 |
| Peritoneal adipose tissue (g) | 0.53 ± 0.02a | 0.42 ± 0.03b | 0.44 ± 0.03 |
| Mesenteric adipose tissue (g) | 0.48 ± 0.05 | 0.38 ± 0.02 | 0.72 ± 0.03 |
| Epididymal adipose tissue (g) | 1.19 ± 0.06a | 1.021 ± 0.07 | 0.936 ± 0.07b |
| Inguinal adipose tissue (g) | 0.6 ± 0.03a | 0.52 ± 0.05 | 0.49 ± 0.03b |
| Total adipose tissue (g)a | 2.8 ± 0.09a | 2.35 ± 0.16b | 2.24 ± 0.16b |
| Liver weight (g) | 1.34 ± 0.03 | 1.31 ± 0.04 | 1.27 ± 0.03 |
Corresponds to the sum of peritoneal, mesenteric, epididymal, and inguinal adipose tissues. Results are means ± SEM (N = 10 mice per group).
Values in the same row and not sharing the same superscripted letter (a, b) are statistically different (P ≤ 0.05).
Oral OEA and KDS-5104 both induced a significant decrease in daily food intake throughout the experimental period (P = 0.001). Consequently, final cumulative food intake was significantly decreased after OEA (−5.4%, P = 0.0033) and KDS-5104 (−6.3%, P = 0.0004) administration (Table 2). Indeed, correction of adipose fat weight gain for cumulative food intake by a generalized linear modeling abrogated the statistical differences among the treatment groups.
Based on respiratory quotient, OEA and KDS-5104 administration induced a significant increase in LOX relative to GOX , resulting in a 13% increase in LOX ratio over time (LOX/GOX + LOX; 2-way ANOVA; P<0.0001 and P = 0.0001, respectively) (Table 3). Analysis of EE in different treatment groups, calculated from the volume of oxygen consumed (VO2) and the volume of carbon dioxide produced (VCO2), revealed a significant increase in EE over the 24-h period (+6%, P < 0.0001) and during the postprandial period (+7%, P < 0.0001) for the OEA group but not for the KDS-5104 group, compared with that of the control group (Table 3). Total 24-h spontaneous activity was increased in groups treated with OEA (+216%, P = 0.0008) and KDS-5104 (+226%, P = 0.0002) compared with controls (Table 3). The relative lipid oxidative consumption levels and the EE were strongly correlated with the total spontaneous activity in every group (P < 0.0001 for LOX and EE, data not shown). Consequently, our statistical model was adjusted to account for the total activity level. Results obtained with the adjusted model revealed that OEA and KDS-5104 similarly, significantly, and independently increased LOX (P = 0.0071) but did not affect GOX or EE compared with control.
TABLE 3.
Indirect calorimetry results over 24 h
| Parameter | Controlb | OEA (0.1 g/kg BW)b | KDS-5104 (0.1 g/kg BW)b |
|---|---|---|---|
| ANOVA on repeated measure over 24 h | |||
| LOX (watt/kg mouse) raw | 11.26 ± 0.24a | 13.92 ± 0.26b | 12.43 ± 0.23b |
| Adjusted for activity | 11.18 ± 0.1a | 13.88 ± 0.1b | 12.31 ± 0.1b |
| GOX (watt/kg mouse) raw | 5.401 ± 0.12a | 4.01 ± 0.12b | 4.474 ± 0.10b |
| Adjusted for activity | 5.57 ± 0.03a | 3.98 ± 0.03a | 4.53 ± 0.03a |
| LOX ratio raw | 0.63 ± 0.01a | 0.72 ± 0.01b | 0.69 ± 0.01b |
| Adjusted for activity | 0.63 ± 0.01a | 0.67 ± 0.01b | 0.70 ± 0.01b |
| EE (watt/kg mouse) raw | 16.58 ± 0.1a | 17.81 ± 0.1b | 16.85 ± 0.1a |
| Adjusted for activity | 16.7 ± 0.01a | 17.7 ± 0.01b | 16.7 ± 0.01a |
| Total activity (AU)a | 198 ± 10a | 429 ± 64b | 449 ± 54b |
| ANOVA of postprandial period | |||
| LOX ratio rawc | 0.51 ± 0.04a | 0.66 ± 0.06a,b | 0.69 ± 0.05b |
| GOX ratio raw | 0.49 ± 0.04a | 0.34 ± 0.06a,b | 0.31 ± 0.05b |
Results are means ± SEM (N = 10 mice per group). Postprandial period lasting from feeding completion to 600 min afterward (from 396 to 1,008 min after the introduction of the mouse in the calorimetric system).
For total activity, the P value for time effect was 0.0192, and the P value for treatment effect was 0.0039; time × treatment is nonsignificant. Total activity corresponds to the sum of rearing and spontaneous activity, normalized to the control mice mean value.
Values in the same row and not sharing the same superscripted letter (a, b) are statistically different (P ≤ 0.05). The values and statistical indications after adjustment on total spontaneous activity are indicated.
cthinsp: LOX ratio = LOX/(LOX + GOX).
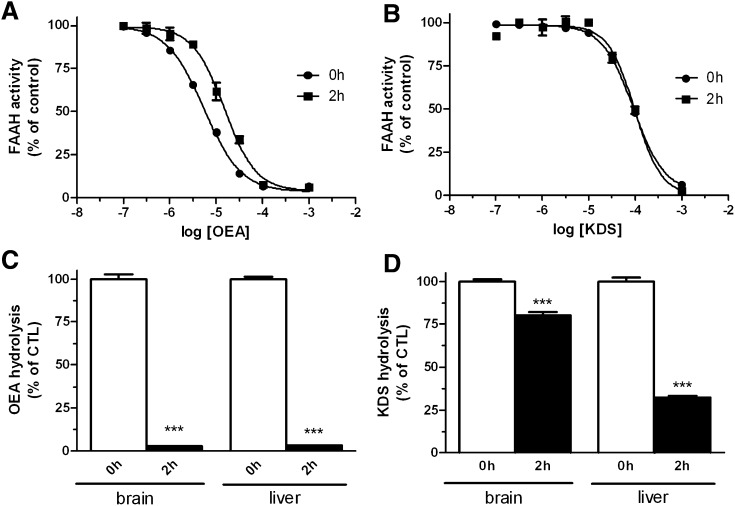
We compared the extent of OEA biological degradation with that of its reportedly nonhydrolyzable analog KDS-5104 by using both in vitro and ex vivo assays. Using recombinant FAAH, we confirmed that KDS-5104 is indeed stable under FAAH hydrolysis conditions, as its inhibition potential with FAAH activity remains unchanged after 2 h of preincubation in the presence of the enzyme. By comparison, for OEA, being a substrate of FAAH, 2 h of preincubation resulted in decreased inhibition of the enzyme (Fig. 3A, B). However, we found that the KDS-5104 inhibition potential was reduced after 2 h of preincubation with mouse tissue, suggesting it is hydrolyzed (data not shown). To confirm this finding, we incubated KDS-5104 in the presence of mouse brain or liver homogenates for 2 h and found that 25% and 70% of the KDS-5104 was hydrolyzed by the tissue preparations, respectively (Fig. 2D). A similar experiment with OEA resulted in almost complete degradation of OEA (Fig. 3C).
Fig.3.
Effects of increasing concentrations of OEA (A) and KDS-5104-5104 (B) on [3H]anandamide (AEA) hydrolysis by recombinant FAAH following no preincubation or 2 h or preincubation of the enzyme with OEA or KDS-5104, respectively. The IC50 for OEA at 0 h was 5.46 ± 0.3 µM and 15.0 ± 1.5 µM at 2 h; the IC50 for KDS-5104 at 0 h was 92.2 ± 4.6 µM and 98.8 ± 11.6 µM at 2 h. Hydrolysis of OEA (C) and KDS-5104-5104 (D) is shown, as measured by HPLC-MS after 0 h or 2 h of incubation with brain or liver mice homogenate.
Of note, when we looked at the intestinal FAAH activity of the treated mice, we noted that KDS-5104 supplementation reduced the FAAH activity by 57% compared with that of the vehicle-treated mice, whereas in OEA-fed mice, FAAH activity was decreased by 20% (P = 0.0003 and 0.0422, respectively; see supplementary Table I).
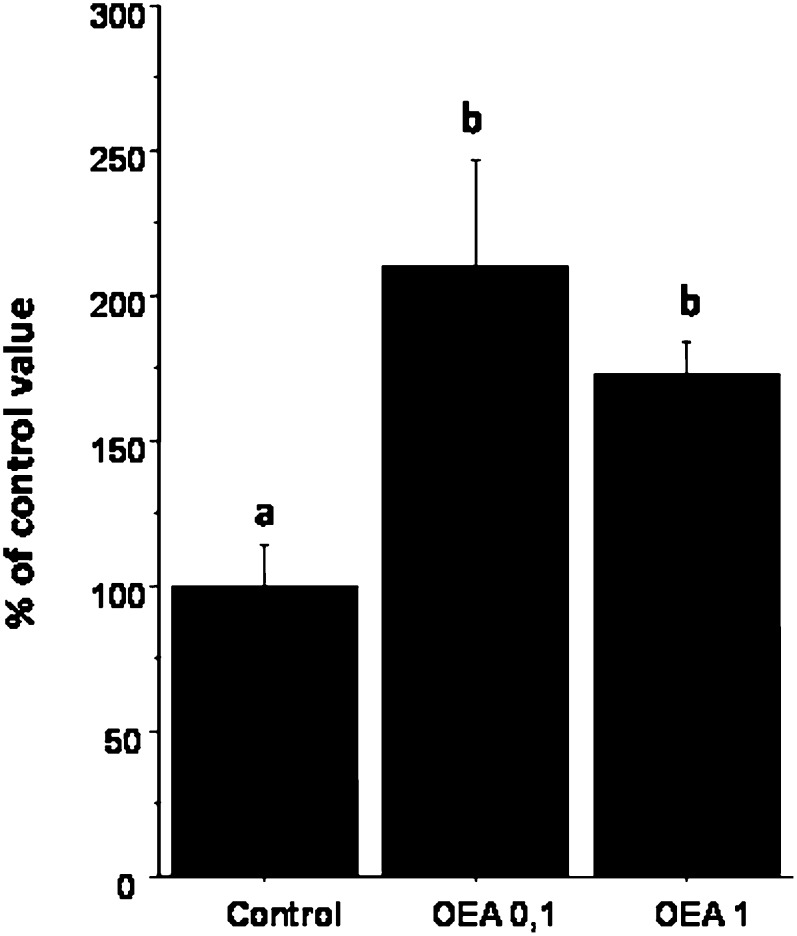
A complementary separate experiment specifically examined fecal fat excretion. The level of fecal lipids was significantly increased by OEA administration compared with that of control (a similar effect was observed for both the 0.1 and the 1 g/kg BW doses, P = 0.0042 and 0.0442, respectively) (Fig. 4). An in vitro assay of pancreatic lipase activity showed a significant decrease of the lipase hydrolytic activity after the addition of two different concentrations of OEA compared with that of control (−21.6% for 300 µmol of OEA/liter and −25.3% for 600 µmol of OEA/liter) (P = 0.02, not shown). No significant difference was observed between the two OEA concentrations.
Fig.4.
Fecal fat excretion of mice fed either 0, 0.1, or 1 g OEA/kg BW for 4 weeks (n = 8 mice per group) is shown. Results are expressed as means ± SEM, normalized to control values.
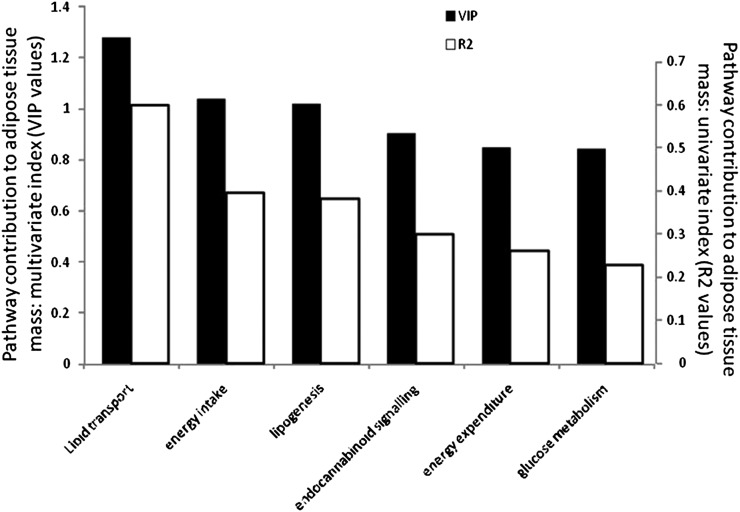
The combined contribution of the treatment-sensitive and adipose fat-related biological processes (Table 1) explained 58% of the fat mass changes (PLS model, adipose tissue variance explained R2Y = 0.58; adipose tissue variance predicted after cross validation Q2Y = 0.52; decrease of R2Y and Q2Y after 100 permutations to 0.008 and −0.14 at the intercept, respectively; P = 0.00021 after cross-validation ANOVA). Although the contribution of each function cannot be statistically compared to adipose fat mass, the R2 and VIP values (multivariate index of the pathway contribution to adipose fat mass) allowed us to rank their relationship to body fat mass variations (Fig. 5). This contribution was, in decreasing order: regulation of lipid transport, then both energy intake and lipogenesis, and similarly endocannabinoid signaling, EE, and glucose metabolism.
Fig.5.
Contribution of each biological function to adipose fat pad mass variations is shown as a multivariate coefficient (variable importance in projection [VIP]), or the determination coefficient R2.
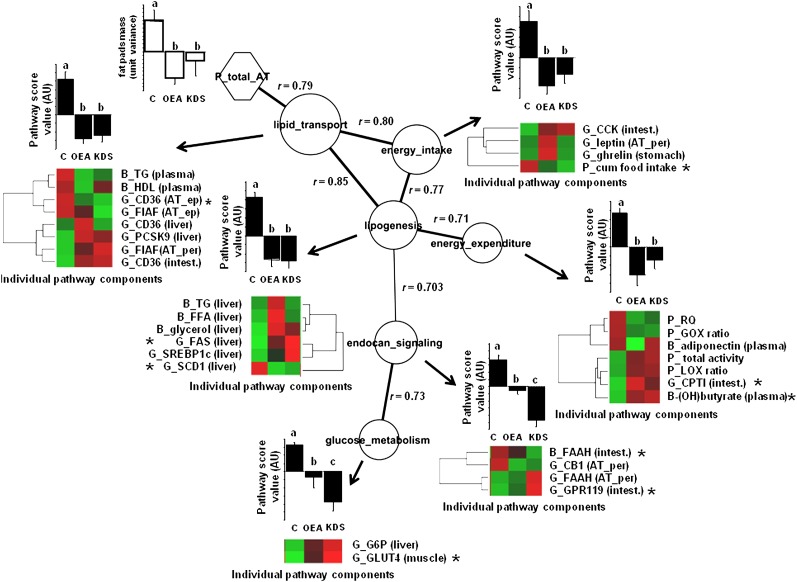
We also performed a pair-wise correlation analysis to show the relationships among the biological events associated with body fat. The pathway correlation analysis (Fig. 6) indicates that the lipid transport function was the most closely associated to adipose fat content. The comparison of the individual scores of each function indicated that both OEA and KDS-5104 modified the regulation of the biological functions in the same direction, but the strength of the effect differed: the effect was equal for lipid transport and lipogenesis; there was a tendency to be higher (but not statistically different) in OEA fed-mice for energy intake and EE; and the effect was greater in KDS-5104 fed-mice for endocannabinoid signaling and glucose metabolism. Finally, the individual PLS VIP values (Fig. 6) showed which individual components of the biological functions studied were more likely affected by both OEA and KDS-5104 in relationship with adipose tissue expansion: downregulation of the epididymal adipose tissue FAT/CD36 gene expression in the lipid transport function, respectively; down- and upregulation of both liver SCD1 and FAS gene expression in the lipogenesis pathway; reduced cumulative food intake in the energy intake process; upregulation of the intestine CPT-I gene, along with increased blood β-hydroxybutyrate and liver free glycerol; upregulation of the liver G6P gene expression in the glucose metabolism function; and upregulation of intestine FAAH enzyme activity and the GPR119 gene expression in the endocannabinoid-signaling module.
Fig.6.
Correlation network among processes with activity modified by both OEA and KDS-5104 treatments is shown. Each node depicts individual biological activity. Histograms represent means ± SEM of semiquantitative activity of biological functions according to treatments, based on the hierarchical PLS computation performed from the individual components, with letters a, b, and c the ANOVA significance threshold (P < 0.05). Node size represents the importance of the process to predict adipose tissue mass (based on VIP values of the PLS analysis). The edges represent the absolute Pearson coefficient ρ values among the nodes. Edge thickness represents the strength of the correlation among connected nodes. The correlation network was built on the maximum stringency calculated from the minimum structure displaying all nodes. The significance of the Pearson correlation coefficient was checked by false discovery rate with a q value of <0.05. This threshold corresponded to Pearson correlation ρ values of >0.701. By convention, pathway score values of the control mice were always positive, thus emphasizing the contrasting effect of treatments. Individual pathway components are displayed as a heat map sorted according to hierarchical clustering (red box, relative increase; green box, relative decrease [see supplementary material for significant levels across treatments). The asterisk indicates which individual components in the biological module weighs more in the biological score process and is more likely to predict adipose tissue fat pads mass (greatest VIP index of the PLS analysis). Adipose tissue mass is shown as means ± SEM of unit of variance. AT, adipose tissue; G, gene; B, biochemical; P, physiological; ep, epididymal; per, peritoneal; intest., upper intestine; muscle, gastrocnemius muscle; ES, endocannabinoid signaling; GM, glucose metabolism; LT, lipid transport; EI, energy intake; LG, lipogenesis.
DISCUSSION
We investigated which molecular and physiological processes were involved with the reduced body fat gain from oral OEA administration in a mouse model of diet-induced obesity. We also sought to determine the extent to which OEA could be broken down and how this would affect its bioefficiency by comparing its effects to those of OEA's more stable analog KDS-5104. KDS-5104 has a structure similar to OEA, and like OEA, efficiently activates PPAR-α and induces a reduction in food intake (19).
Here, both OEA and KDS-5104 were similarly effective in significantly reducing body fat gain in diet-induced obesity of C57Bl/6J male mice (16% to 20%). The biological processes involved in this outcome were also similar, although they occurred with some specificity between OEA and KDS-5104. One may have predicted a somewhat higher efficiency in the decrease of adipose fat for KDS-5104, based on its in vivo resistance to hydrolysis compared with that of OEA (19). However, although we found that KDS-5104 is resistant to FAAH degradation, we showed here that it is extensively broken down ex vivo (up to 70%) by using mouse liver homogenates. This implies that a system other than FAAH is involved in catabolism of OEA. The important observation from this study is that the amount of OEA fed (100 mg/kg BW) is high enough to exert a biological effect despite a high degradation rate compared with that of KDS-5104. This is substantiated by our published results using two oral doses of OEA (10 mg and 100 mg/kg BW) and showing that both doses exerted the same effect on fat storage, despite a 10-fold intake difference (13).
A dose extrapolation to the human, taking into account the metabolic rate difference with the mouse (20), would correspond to roughly 1 g/70 kg adult/day. If we also consider that OEA is efficiently catabolized in vivo, this would suggest that it is active at very low levels in cells and that its level in cells is tightly controlled, thus emphasizing its biological importance.
Our study is the first to investigate the impact of OEA on EE and its various components by indirect calorimetry. It revealed that OEA and KDS-5104 shifted the energy balance in favor of lipid oxidation. In our diagram of the EE pathway (Fig. 6), the LOX component clustered with the intestine CPTI gene, a gene committed in fatty acid oxidation but also with metabolic indicators of a greater lipid substrate utilization (blood β-hydroxybutyrate occurring from LOX [21]). Thus, the channeling of fat toward the oxidation pathway contributes to lower body fat expansion upon high-fat feeding. Lipid oxidation also clustered with spontaneous activity that is dramatically increased by OEA, thus OEA can contribute to nonexercise activity thermogenesis, a factor that also contributes to obesity resistance (22).
To obtain a simple picture of the biological effects associated with body fat gain, the various biological indicators were grouped as biological modules from which a response to treatment could be calculated (17). We determined that all of the biological modules evaluated were affected by the treatments and could explain up to 58% (PLS regression) of body fat variation. The part not captured by our model could occur for other reasons, as we measured only part of the biological components of the functions investigated. Instead of attempting to measure all functions, we chose to focus on dedicated processes that we wanted to assess as targets of either OEA or KDS-5104 (Table 1). Other biologically relevant events could also be involved in body fat lowering, such as fat excretion. This was evaluated in a complementary study that confirmed that intestinal fat absorption is decreased by OEA feeding. Although our in vitro results indicated a direct inhibitory effect of OEA on pancreatic lipase, it is not known if this mechanism is important under the conditions prevailing in vivo in the intestinal lumen. In our study, measured adiposity reflected the cumulative exposure to either OEA or KDS-5104 over 5 weeks, whereas our snapshot of biological evaluations can be subject to time variations. Such nonlinearity can weaken our predictions. Nevertheless, we identified important processes and some of their individual components targeted by either or both OEA and KDS-5104, which, in combination, are associated with decreased fat mass. For instance, from our model (R2 values in Fig. 5), we identified the fact that the lipid transport function can predict most of the body fat variation (R2 = 0.60, e.g., 16% of adipose mass variation, P < 0.0001); followed by both energy intake and lipogenesis (R2 = 0.4, 11% of adipose mass variations, P = 0.0006; and R2 = 0.38, 10% of adipose mass variation, P = 0.0008, respectively); then endocannabinoid-signaling regulation (R2 = 0.30, 8% of adipose mass variation, P = 0.0039); EE (R2 = 0.26, 7.0% of adipose mass variation, P = 0.0075); and glucose metabolism and (R2 = 0.23, 6% of adipose mass variation, P = 0.0078). Additionally, the pathways analyses (Fig. 6) focusing on pair-wise relationships indicated that the lipid transport process is the cornerstone function most strongly associating the other functions to adipose fat. This emphasizes its functional importance in the inhibition of body fat gain by OEA intake.
Although fecal fat cannot be modeled by our equation, as it was determined in a complementary study, its excretion also appears as an indirect expression of lipid transport in the intestine. This again underlines its importance in the control of body fatness by oral OEA. Paradoxically, in contradiction to greater fecal fat excretion, the upper-intestine fatty acid transporter FAT/CD36 gene expression was upregulated by the treatments, as observed by others with OEA (2, 13). It would be interesting to study whether this acts to signal the brain to inhibit overfeeding and trigger satiety. Finally, in the lipid transport cluster, we identified the fact that the adipose tissue FAT/CD36 gene expression was the indicator most strongly associated with reduced body fat gain in response to OEA (and KDS-5104). This makes it a highly putative target for oral OEA action in the control of body fat.
Among the other most relevant targets found here, food intake control by OEA was also identified as important to the control of body fatness. In fact, after correction for food intake, the differences in adipose fat were no longer significant among treatment groups, thus emphasizing the important role of this factor in the antiobesity action of OEA and confirming other similar findings (6, 13, 23, 24). In addition, downregulation of the stearoyl-CoA desaturase 1 (SCD1) gene activity in the liver was observed in both the OEA and KDS-5104 groups. SCD1 is a key enzyme that coordinates fat storage and fat oxidation (25). Decreased SCD1 expression has been demonstrated to be strongly associated with obesity resistance and increased lipid β-oxidation (25, 26) and also lowers food intake upon stearoylethanolamine administration (27). A similar finding was found in Zucker rats but only when 5 mg/kg injected OEA was combined with 1 mg/kg SR141716A (rimonabant) (28). We also found that downregulation of the SCD1 gene was one of the strongest factors (along with FAT/CD36 in adipose tissue) affecting cumulative food intake in a PLS regression model (not shown). The relationship found among the lipogenesis functions, the regulation of energy intake, and adipose tissue variations, as shown in Fig. 6, is also consistent with the above-described findings and makes SCD1 a possible important target of OEA antiobesity action. In contrast to downregulation of SCD1, FAS expression, which is another de novo lipogenic enzyme strongly linked to body fat, was upregulated by both OEA and KDS-5104 in our study. Decreased SCD1 activity produces lower oleic acid, which is the building block of stored triglycerides (29), thus, decreased SCD1 activity would direct de novo synthesized fatty acids from FAS to oxidation instead of storage, provided lipid oxidation is efficient (30). This is also substantiated by a decreased concentration of postprandial plasma triglyceride in OEA- (and KDS-5104-) treated mice, accompanied by higher LOX, plasma β-hydroxybutyrate, and CPT-1 gene transcription in the intestine, and lower SCD1 gene transcription. Finally, our present findings confirmed that endocannabinoid signaling, mainly through both intestinal and adipose tissue FAAH, appears to be a significant player in modulating OEA effects (13).
Last, it is interesting that the reported nonhydrolyzable OEA analog KDS-5104 did not exert more pronounced effects on fat storage in our study. The differences in responses between KDS-5104 and OEA were mainly related to glucose metabolism and endocannabinoid signaling for which the effect of KDS-5104 was greater. The impact on adipose weight gain was relatively weak, as both glucose metabolism and endocannabinoid signaling were not the primary factors explaining body fatness in our model. In contrast, OEA and KDS-5104 exerted the same effects on lipid transport, energy intake, and lipogenesis, the three main modules related to body fat deposition. The similar effects of both KDS-5104 and OEA on these three main modules are likely to explain the identical outcome on adipose fat gain.
In conclusion, we confirmed that OEA feeding decreased body fat gain upon high-fat feeding in the mouse. We found that at least six different pathways (lipid transport, energy intake, regulation of EE, endocannabinoid signaling, lipogenesis, and glucose metabolism) can be involved in this effect and explained up to 58% of the body fat variation. Other biologically relevant effects such as fecal fat excretion and other as-yet unidentified mechanisms also contributed to reduce body fat gain upon OEA feeding. Our study also pointed out that the physiological level of OEA degradation does not compromise its bioefficiency at the oral dose used. Seemingly, a degradation system other than the fatty acyl anandamide hydrolyze is able to process both OEA and its nonhydrolyzable analog KDS-5104.
Finally, our results highlight the multifactorial aspect of body fat changes that occur following OEA nutritional intervention and that take place at several levels of biological systems. Although we are aware that we captured only part of these changes, our multilevel integrated biological analysis brings new mechanical hypotheses to explain the effect of OEA on body fat control in the diet-induced obesity mouse model and allow ranking their effects in importance with regard to adipose tissue weight gain. With this perspective, we conclude that lipid transport appears as a determinant player of the OEA biological response, in which adipose tissue FAT/CD36 expression was found to be the most relevant bioindicator/effector of OEA action.
Supplementary Material
Acknowledgments
The authors are grateful to Dorothy Laflamme for excellent manuscript review. We also thank Jean-François Landrier, Myriam Moussa, and Erwan Gouranton from UMR INRA 1260 for technical support.
Footnotes
Abbreviations:
- BW
- body weight
- EE
- energy expenditure
- FAAH
- fatty acid amide hydrolase
- FAE
- fatty acid ethanolamide
- GOX
- carbohydrate oxidation
- LOX
- lipid oxidation
- NAAA
- N-acylethanolamine-hydrolyzing acid amidase
- OEA
- oleoylethanolamide
- OEA
- oleoylethanolamide
- PLS
- partial least-square regression
- PLS-DA
- partial least squares discriminant analysis
- PPAR
- peroxisome proliferator-activated receptor
- VIP
- variable importance index
This study was sponsored by Nestlé Research Centre (Lausanne, Switzerland) through the Ph.D. fellowship of C.T. G.G.M. is a recipient of a Fonds spéciaux de recherches subsidy (UCL, Belgium) and a Belgian National Fund for Scientific Research grant (FRFC 2.4555.08). D.T.F., F.D., and J.C.M. designed the research. M.M. and J.C.M. performed statistical analyses. D.L. and G.G.M. performed FAAH assays and participated in manuscript writing, with C.T. and J.C.M. T.H. tested the lipase activity. Oleoylethanolamide, used to supplement the mice diet, was synthesized at the Nestlé Research Centre (Lausanne, Switzerland) with participation of Jean-Baptiste Bezelgues and Laurent Crosset. Geoffray Labar produced the recombinant FAAH.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of nine figures and supplementary data and references.
REFERENCES
- 1.Bisogno T., Delton-Vandenbroucke I., Milone A., Lagarde M., Di Marzo V. 1999. Biosynthesis and inactivation of N-arachidonoylethanolamine (anandamide) and N-docosahexaenoylethanolamine in bovine retina. Arch. Biochem. Biophys. 370: 300–307. [DOI] [PubMed] [Google Scholar]
- 2.Yang Y., Chen M., Georgeson K. E., Harmon C. M. 2007. Mechanism of oleoylethanolamide on fatty acid uptake in small intestine after food intake and body weight reduction. Am. J. Physiol. Regul. Integr. Comp. Physiol. 292: R235–R241. [DOI] [PubMed] [Google Scholar]
- 3.Sun Y. X., Tsuboi K., Zhao L. Y., Okamoto Y., Lambert D. M., Ueda N. 2005. Involvement of N-acylethanolamine-hydrolyzing acid amidase in the degradation of anandamide and other N-acylethanolamines in macrophages. Biochim. Biophys. Acta. 1736: 211–220. [DOI] [PubMed] [Google Scholar]
- 4.Fegley D., Gaetani S., Duranti A., Tontini A., Mor M., Tarzia G., Piomelli D. 2005. Characterization of the fatty acid amide hydrolase inhibitor cyclohexyl carbamic acid 3′-carbamoyl-biphenyl-3-yl ester (URB597): effects on anandamide and oleoylethanolamide deactivation. J. Pharmacol. Exp. Ther. 313: 352–358. [DOI] [PubMed] [Google Scholar]
- 5.Thabuis C., Tissot-Favre D., Bezelgues J. B., Martin J. C., Cruz-Hernandez C., Dionisi F., Destaillats F. 2008. Biological functions and metabolism of oleoylethanolamide. Lipids. 43: 887–894. [DOI] [PubMed] [Google Scholar]
- 6.Lo Verme J., Gaetani S., Fu J., Oveisi F., Burton K., Piomelli D. 2005. Regulation of food intake by oleoylethanolamide. Cell. Mol. Life Sci. 62: 708–716. [DOI] [PubMed] [Google Scholar]
- 7.Rodriguez de Fonseca F., Navarro M., Gomez R., Escuredo L., Nava F., Fu J., Murillo-Rodriguez E., Giuffrida A., LoVerme J., Gaetani S., et al. 2001. An anorexic lipid mediator regulated by feeding. Nature. 414: 209–212. [DOI] [PubMed] [Google Scholar]
- 8.Fu J., Gaetani S., Oveisi F., Lo Verme J., Serrano A., Rodriguez de Fonseca F., Rosengarth A., Luecke H., Di Giacomo B., Tarzia G., et al. 2003. Oleoylethanolamide regulates feeding and body weight through activation of the nuclear receptor PPAR-. Nature. 425: 90–93. [DOI] [PubMed] [Google Scholar]
- 9.Fu J., Oveisi F., Gaetani S., Lin E., Piomelli D. 2005. Oleoylethanolamide, an endogenous PPAR-[alpha] agonist, lowers body weight and hyperlipidemia in obese rats. Neuropharmacology. 48: 1147–1153. [DOI] [PubMed] [Google Scholar]
- 10.Desvergne B., Wahli W. 1999. Peroxisome proliferator-activated receptors: nuclear control of metabolism. Endocr. Rev. 20: 649–688. [DOI] [PubMed] [Google Scholar]
- 11.Guzman M., Lo Verme J., Fu J., Oveisi F., Blazquez C., Piomelli D. 2004. Oleoylethanolamide stimulates lipolysis by activating the nuclear receptor peroxisome proliferator-activated receptor alpha (PPAR-alpha). J. Biol. Chem. 279: 27849–27854. [DOI] [PubMed] [Google Scholar]
- 12.Diep T. A., Madsen A. N., Holst B., Kristiansen M. M., Wellner N., Hansen S. H., Hansen H. S. 2011. Dietary fat decreases intestinal levels of the anorectic lipids through a fat sensor. FASEB J. 25: 765–774. [DOI] [PubMed] [Google Scholar]
- 13.Thabuis C., Destaillats F., Landrier J. F., Tissot-Favre D., Martin J. C. 2010. Analysis of gene expression pattern reveals potential targets of dietary oleoylethanolamide in reducing body fat gain in C3H mice. J. Nutr. Biochem. 21: 922–928. [DOI] [PubMed] [Google Scholar]
- 14.Navarro V., Portillo M. P., Margotat A., Landrier J. F., Macarulla M. T., Lairon D., Martin J. C. 2009. A multi-gene analysis strategy identifies metabolic pathways targeted by trans-10, cis-12-conjugated linoleic acid in the liver of hamsters. Br. J. Nutr. 102: 537–545. [DOI] [PubMed] [Google Scholar]
- 15.Oltvai Z. N., Barabasi A. L. 2002. Systems biology. Life's complexity pyramid. Science. 298: 763–764. [DOI] [PubMed] [Google Scholar]
- 16.Kleemann R., van Erk M., Verschuren L., van den Hoek A. M., Koek M., Wielinga P. Y., Jie A., Pellis L., Bobeldijk-Pastorova I., Kelder T., et al. 2010. Time-resolved and tissue-specific systems analysis of the pathogenesis of insulin resistance. PLoS ONE. 5: e8817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wold S., Kettaneh N., Tjessem K. 1996. Hierarchical multiblock PLS and PC models for easier model interpretation and as an alternative to variable selection. J. Chemom. 10: 463–482. [Google Scholar]
- 18.Shannon P., Markiel A., Ozier O., Baliga N. S., Wang J. T., Ramage D., Amin N., Schwikowski B., Ideker T. 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13: 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Astarita G., Di Giacomo B., Gaetani S., Oveisi F., Compton T. R., Rivara S., Tarzia G., Mor M., Piomelli D. 2006. Pharmacological characterization of hydrolysis-resistant analogs of oleoylethanolamide with potent anorexiant properties. J. Pharmacol. Exp. Ther. 318: 563–570. [DOI] [PubMed] [Google Scholar]
- 20.Terpstra A. H. M. 2001. Differences between humans and mice in efficacy of the body fat lowering effect of conjugated linoleic acid: role of metabolic rate. J. Nutr. 131: 2067–2068. [DOI] [PubMed] [Google Scholar]
- 21.McGarry J. D., Foster D. W. 1980. Regulation of hepatic fatty acid oxidation and ketone body production. Annu. Rev. Biochem. 49: 395–420. [DOI] [PubMed] [Google Scholar]
- 22.Kotz C. M., Teske J. A., Billington C. J. 2008. Neuroregulation of nonexercise activity thermogenesis and obesity resistance. Am. J. Physiol. Regul. Integr. Comp. Physiol. 294: R699–R710. [DOI] [PubMed] [Google Scholar]
- 23.Cani P. D., Montoya M. L., Neyrinck A. M., Delzenne N. M., Lambert D. M. 2004. Potential modulation of plasma ghrelin and glucagon-like peptide-1 by anorexigenic cannabinoid compounds, SR141716A (rimonabant) and oleoylethanolamide. Br. J. Nutr. 92: 757–761. [DOI] [PubMed] [Google Scholar]
- 24.Nielsen M. J., Petersen G., Astrup A., Hansen H. S. 2004. Food intake is inhibited by oral oleoylethanolamide. J. Lipid Res. 45: 1027–1029. [DOI] [PubMed] [Google Scholar]
- 25.Popeijus H. E., Saris W. H., Mensink R. P. 2008. Role of stearoyl-CoA desaturases in obesity and the metabolic syndrome. Int. J. Obes. (Lond). 32: 1076–1082. [DOI] [PubMed] [Google Scholar]
- 26.Ntambi J. M., Miyazaki M., Stoehr J. P., Lan H., Kendziorski C. M., Yandell B. S., Song Y., Cohen P., Friedman J. M., Attie A. D. 2002. Loss of stearoyl-CoA desaturase-1 function protects mice against adiposity. Proc. Natl. Acad. Sci. U S A. 99: 11482–11486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Terrazzino S., Berto F., Dalle Carbonare M., Fabris M., Guiotto A., Bernardini D., Leon A. 2004. Stearoylethanolamide exerts anorexic effects in mice via down-regulation of liver stearoyl-coenzyme A desaturase-1 mRNA expression. FASEB J. 18: 1580–1582. [DOI] [PubMed] [Google Scholar]
- 28.Serrano A., Del Arco I., Javier Pavon F., Macias M., Perez-Valero V., Rodriguez de Fonseca F. 2008. The cannabinoid CB1 receptor antagonist SR141716A (Rimonabant) enhances the metabolic benefits of long-term treatment with oleoylethanolamide in Zucker rats. Neuropharmacology. 54: 226–234. [DOI] [PubMed] [Google Scholar]
- 29.Miyazaki M., Kim Y. C., Ntambi J. M. 2001. A lipogenic diet in mice with a disruption of the stearoyl-CoA desaturase 1 gene reveals a stringent requirement of endogenous monounsaturated fatty acids for triglyceride synthesis. J. Lipid Res. 42: 1018–1024. [PubMed] [Google Scholar]
- 30.Van Wymelbeke V., Louis-Sylvestre J., Fantino M. 2001. Substrate oxidation and control of food intake in men after a fat-substitute meal compared with meals supplemented with an isoenergetic load of carbohydrate, long-chain triacylglycerols, or medium-chain triacylglycerols. Am. J. Clin. Nutr. 74: 620–630. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.