Abstract
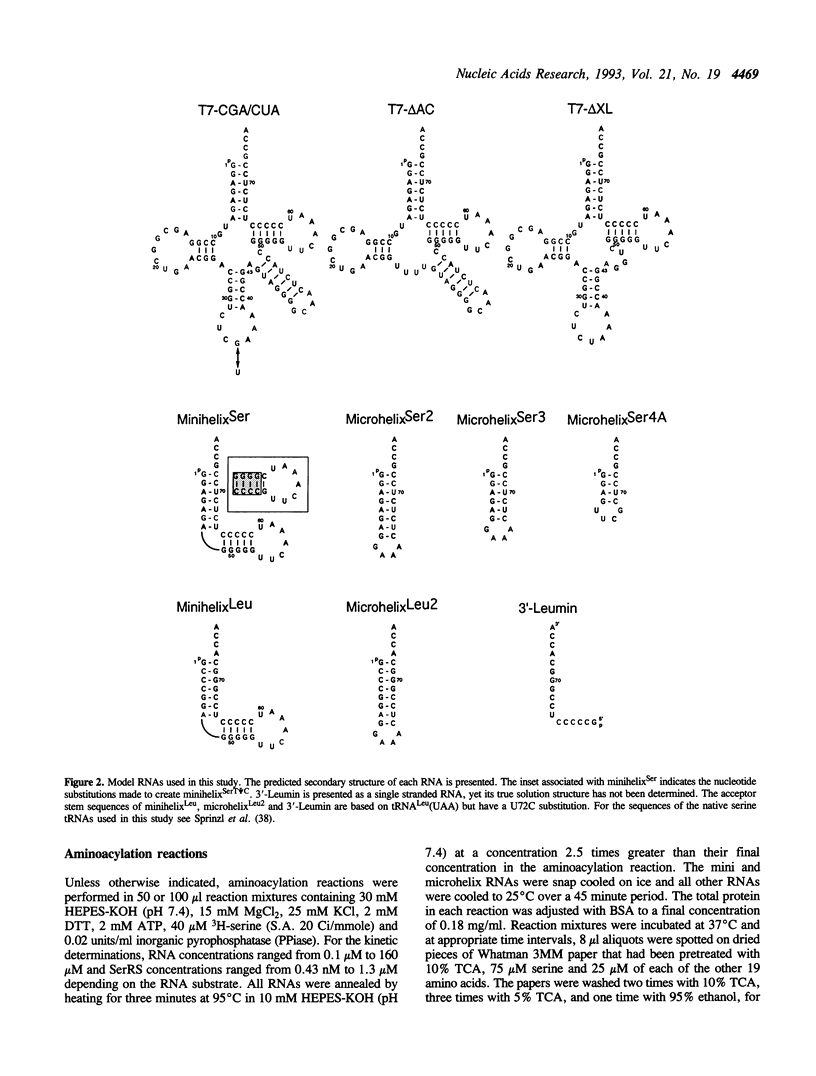
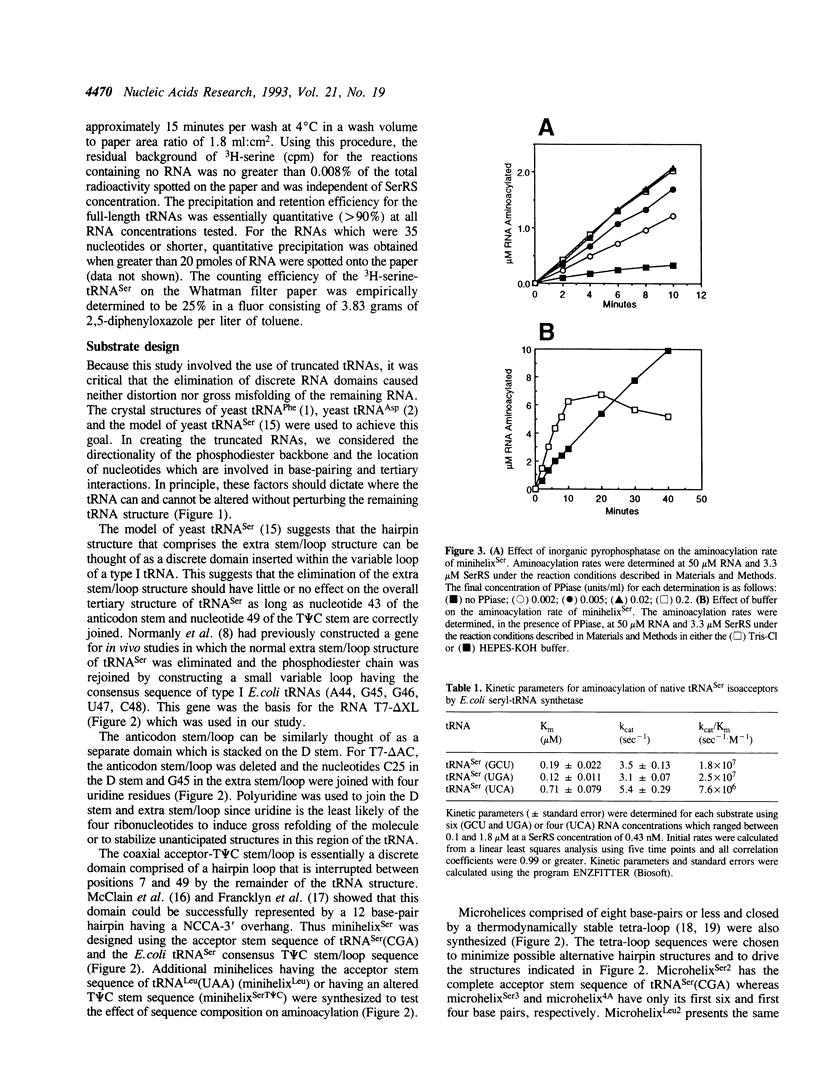
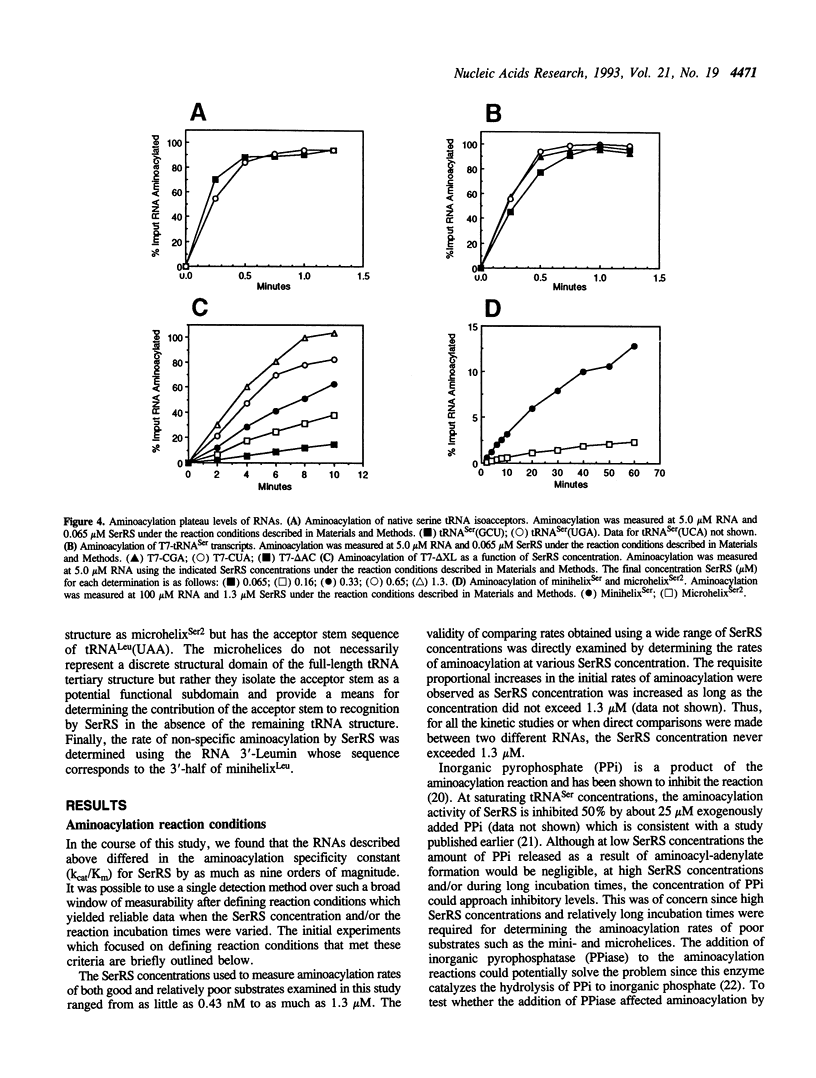
The aminoacylation kinetics of T7 transcripts representing defined regions of Escherichia coli serine tRNAs were determined using purified E.coli seryl-tRNA synthetase (SerRS) and the kinetic values were used to estimate the relative contribution of various tRNA(Ser) domains to recognition by SerRS. The analysis revealed that the extra stem/loop structure, characteristic of type II tRNAs such as tRNA(Ser), is the domain which makes the largest contribution to kcat/Km of aminoacylation. Moreover, Km of aminoacylation was increased by a factor of about 1000 when the extra stem/loop was changed to the consensus sequence of type I tRNA extra loops indicating that the stem structure contributes significantly to the binding of tRNA(Ser) to SerRS. A model RNA, which represents only the tRNA(Ser) coaxial acceptor-T psi C stem/loop domain, was also specifically aminoacylated by SerRS having a kcat/Km about 1000-fold greater than background levels. A significant portion of the contribution of this domain to aminoacylation is attributable to the acceptor stem sequence making the acceptor stem the second most important domain for recognition by SerRS. Finally, kcat/Km was essentially unchanged when the entire anticodon stem/loop of tRNA(Ser) was deleted indicating that neither the anticodon nucleotides nor the surrounding stem/loop structure are important for recognition by SerRS.
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Behlen L. S., Sampson J. R., DiRenzo A. B., Uhlenbeck O. C. Lead-catalyzed cleavage of yeast tRNAPhe mutants. Biochemistry. 1990 Mar 13;29(10):2515–2523. doi: 10.1021/bi00462a013. [DOI] [PubMed] [Google Scholar]
- Behlen L. S., Sampson J. R., Uhlenbeck O. C. An ultraviolet light-induced crosslink in yeast tRNA(Phe). Nucleic Acids Res. 1992 Aug 11;20(15):4055–4059. doi: 10.1093/nar/20.15.4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooperman B. S. The mechanism of action of yeast inorganic pyrophosphatase. Methods Enzymol. 1982;87:526–548. doi: 10.1016/s0076-6879(82)87030-4. [DOI] [PubMed] [Google Scholar]
- Crothers D. M., Seno T., Söll G. Is there a discriminator site in transfer RNA? Proc Natl Acad Sci U S A. 1972 Oct;69(10):3063–3067. doi: 10.1073/pnas.69.10.3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cusack S., Berthet-Colominas C., Härtlein M., Nassar N., Leberman R. A second class of synthetase structure revealed by X-ray analysis of Escherichia coli seryl-tRNA synthetase at 2.5 A. Nature. 1990 Sep 20;347(6290):249–255. doi: 10.1038/347249a0. [DOI] [PubMed] [Google Scholar]
- Davanloo P., Rosenberg A. H., Dunn J. J., Studier F. W. Cloning and expression of the gene for bacteriophage T7 RNA polymerase. Proc Natl Acad Sci U S A. 1984 Apr;81(7):2035–2039. doi: 10.1073/pnas.81.7.2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignam J. D., Deutscher M. P. Aminoacyl-tRNA synthetase stimulatory factors and inorganic pyrophosphatase. Biochemistry. 1979 Jul 10;18(14):3165–3170. doi: 10.1021/bi00581a039. [DOI] [PubMed] [Google Scholar]
- Dock-Bregeon A. C., Westhof E., Giegé R., Moras D. Solution structure of a tRNA with a large variable region: yeast tRNASer. J Mol Biol. 1989 Apr 20;206(4):707–722. doi: 10.1016/0022-2836(89)90578-0. [DOI] [PubMed] [Google Scholar]
- Eigner E. A., Loftfield R. B. Kinetic techniques for the investigation of amino acid: tRNA ligases (aminoacyl-tRNA synthetases, amino acid activating enzymes). Methods Enzymol. 1974;29:601–619. doi: 10.1016/0076-6879(74)29053-0. [DOI] [PubMed] [Google Scholar]
- Francklyn C., Schimmel P. Aminoacylation of RNA minihelices with alanine. Nature. 1989 Feb 2;337(6206):478–481. doi: 10.1038/337478a0. [DOI] [PubMed] [Google Scholar]
- Francklyn C., Shi J. P., Schimmel P. Overlapping nucleotide determinants for specific aminoacylation of RNA microhelices. Science. 1992 Feb 28;255(5048):1121–1125. doi: 10.1126/science.1546312. [DOI] [PubMed] [Google Scholar]
- Freist W., Sternbach H., Cramer F. Isoleucyl-tRNA synthetase from Escherichia coli MRE 600. Different pathways of the aminoacylation reaction depending on presence of pyrophosphatase, order of substrate addition in the pyrophosphate exchange, and substrate specificity with regard to ATP analogs. Eur J Biochem. 1982 Nov 15;128(2-3):315–329. [PubMed] [Google Scholar]
- Grodberg J., Dunn J. J. ompT encodes the Escherichia coli outer membrane protease that cleaves T7 RNA polymerase during purification. J Bacteriol. 1988 Mar;170(3):1245–1253. doi: 10.1128/jb.170.3.1245-1253.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himeno H., Hasegawa T., Ueda T., Watanabe K., Shimizu M. Conversion of aminoacylation specificity from tRNA(Tyr) to tRNA(Ser) in vitro. Nucleic Acids Res. 1990 Dec 11;18(23):6815–6819. doi: 10.1093/nar/18.23.6815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y. M., Schimmel P. Modeling with in vitro kinetic parameters for the elaboration of transfer RNA identity in vivo. Biochemistry. 1989 Jun 13;28(12):4942–4947. doi: 10.1021/bi00438a005. [DOI] [PubMed] [Google Scholar]
- Härtlein M., Madern D., Leberman R. Cloning and characterization of the gene for Escherichia coli seryl-tRNA synthetase. Nucleic Acids Res. 1987 Feb 11;15(3):1005–1017. doi: 10.1093/nar/15.3.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katze J. R., Konigsberg W. Purification and properties of seryl transfer ribonucleic acid synthetase from Escherichia coli. J Biol Chem. 1970 Mar 10;245(5):923–930. [PubMed] [Google Scholar]
- Khvorova A. M., Motorin YuA, Wolfson A. D. Crucial role of pyrophosphate in the aminoacylation of E. coli tRNA(Phe) by yeast phenylalanyl-tRNA synthetase. FEBS Lett. 1992 Oct 19;311(2):139–142. doi: 10.1016/0014-5793(92)81385-y. [DOI] [PubMed] [Google Scholar]
- Kim S. H., Suddath F. L., Quigley G. J., McPherson A., Sussman J. L., Wang A. H., Seeman N. C., Rich A. Three-dimensional tertiary structure of yeast phenylalanine transfer RNA. Science. 1974 Aug 2;185(4149):435–440. doi: 10.1126/science.185.4149.435. [DOI] [PubMed] [Google Scholar]
- Martinis S. A., Schimmel P. Enzymatic aminoacylation of sequence-specific RNA minihelices and hybrid duplexes with methionine. Proc Natl Acad Sci U S A. 1992 Jan 1;89(1):65–69. doi: 10.1073/pnas.89.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClain W. H., Guerrier-Takada C., Altman S. Model substrates for an RNA enzyme. Science. 1987 Oct 23;238(4826):527–530. doi: 10.1126/science.2443980. [DOI] [PubMed] [Google Scholar]
- Milligan J. F., Groebe D. R., Witherell G. W., Uhlenbeck O. C. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987 Nov 11;15(21):8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normanly J., Ogden R. C., Horvath S. J., Abelson J. Changing the identity of a transfer RNA. Nature. 1986 May 15;321(6067):213–219. doi: 10.1038/321213a0. [DOI] [PubMed] [Google Scholar]
- Normanly J., Ollick T., Abelson J. Eight base changes are sufficient to convert a leucine-inserting tRNA into a serine-inserting tRNA. Proc Natl Acad Sci U S A. 1992 Jun 15;89(12):5680–5684. doi: 10.1073/pnas.89.12.5680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson E. T., Uhlenbeck O. C. Determination of recognition nucleotides for Escherichia coli phenylalanyl-tRNA synthetase. Biochemistry. 1992 Oct 27;31(42):10380–10389. doi: 10.1021/bi00157a028. [DOI] [PubMed] [Google Scholar]
- Sampson J. R., Uhlenbeck O. C. Biochemical and physical characterization of an unmodified yeast phenylalanine transfer RNA transcribed in vitro. Proc Natl Acad Sci U S A. 1988 Feb;85(4):1033–1037. doi: 10.1073/pnas.85.4.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SantaLucia J., Jr, Kierzek R., Turner D. H. Context dependence of hydrogen bond free energy revealed by substitutions in an RNA hairpin. Science. 1992 Apr 10;256(5054):217–219. doi: 10.1126/science.1373521. [DOI] [PubMed] [Google Scholar]
- Schatz D., Leberman R., Eckstein F. Interaction of Escherichia coli tRNA(Ser) with its cognate aminoacyl-tRNA synthetase as determined by footprinting with phosphorothioate-containing tRNA transcripts. Proc Natl Acad Sci U S A. 1991 Jul 15;88(14):6132–6136. doi: 10.1073/pnas.88.14.6132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimmel P. R., Söll D. Aminoacyl-tRNA synthetases: general features and recognition of transfer RNAs. Annu Rev Biochem. 1979;48:601–648. doi: 10.1146/annurev.bi.48.070179.003125. [DOI] [PubMed] [Google Scholar]
- Schulman L. H. Recognition of tRNAs by aminoacyl-tRNA synthetases. Prog Nucleic Acid Res Mol Biol. 1991;41:23–87. [PubMed] [Google Scholar]
- Sprinzl M., Dank N., Nock S., Schön A. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 1991 Apr 25;19 (Suppl):2127–2171. doi: 10.1093/nar/19.suppl.2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundharadas G., Katze J. R., Söll D., Konigsberg W., Lengyel P. On the recognition of serine transfer RNA's specific for unrelated codons by the same seryl-transfer RNA synthetase. Proc Natl Acad Sci U S A. 1968 Oct;61(2):693–700. doi: 10.1073/pnas.61.2.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuerk C., Gauss P., Thermes C., Groebe D. R., Gayle M., Guild N., Stormo G., d'Aubenton-Carafa Y., Uhlenbeck O. C., Tinoco I., Jr CUUCGG hairpins: extraordinarily stable RNA secondary structures associated with various biochemical processes. Proc Natl Acad Sci U S A. 1988 Mar;85(5):1364–1368. doi: 10.1073/pnas.85.5.1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westhof E., Dumas P., Moras D. Crystallographic refinement of yeast aspartic acid transfer RNA. J Mol Biol. 1985 Jul 5;184(1):119–145. doi: 10.1016/0022-2836(85)90048-8. [DOI] [PubMed] [Google Scholar]


