Abstract
A β-hairpin conformation has been characterized in crystals of the decapeptide t-butoxycarbonyl-Leu-Val-βPhe-Val-DPro-Gly-Leu-βPhe-Val-Val-methyl ester [βPhe; (S)-β3 homophenylalanine] by x-ray diffraction. The polypeptide chain reversal is nucleated by the centrally positioned DPro-Gly segment, which adopts a type-I′ β-turn conformation. Four intramolecular cross-strand hydrogen bonds stabilize the peptide fold. The βPhe(3) and βPhe(8) residues occupy facing positions on the hairpin, with the side chains projecting on opposite faces of the β-sheet. At the site of insertion of β-residues, the polarity of the peptide units along each strand reverses, as compared with the α-peptide segments. In this analog, a small segment of a polar sheet is observed, where adjacent CO and NH groups line up in opposite directions in each strand. In the crystal, an extended β-sheet is formed by hydrogen bonding between strands of antiparallel pairs of β-hairpins. The crystallographic parameters for C65H102N10O13⋅ 3H2O are: space group P212121; a = 19.059(8) Å, b = 19.470(2) Å, c = 21.077(2) Å; Z = 4; agreement factor R1 = 9.12% for 3,984 data observed >4σ(F) and a resolution of 0.90 Å.
The formation of novel folded polypeptide structures by oligomers of β-amino acids has stimulated considerable recent interest in the conformation of designed peptides containing the higher homologs of naturally occurring α-amino acids (1–6). The ready availability of the β-analogs of the naturally occurring protein amino acids by Arndt–Eistert homologation has facilitated the incorporation of these residues into designer peptides (7–9). The insertion of an additional carbon atom into the polypeptide backbone enhances the conformational versatility of these residues. In principle, the backbone conformation of β-amino acid residues in peptides is determined by three torsional variables, dihedral angles φ (N-Cβ), θ (Cβ-Cα), and ψ (Cα-CO) (3). Although novel folded structures, foldamers, have been well characterized in oligomeric sequences of cyclic β-amino acids (10–12), only limited information is available on the accommodation of the β-residues into the classical secondary structural motifs formed by α-amino acid sequences. The incorporation of β-amino acids into helical and β-sheet structures is of considerable importance in the design of analogs of biologically active peptides, in view of the reported stability of the β-peptides to proteolytic attack (13). Both β- and γ-amino acids have been incorporated into the backbone of a polar helical peptide without disruption of overall folded structure (14). In this report, we describe crystallographic characterization of a designed β-hairpin peptide containing two facing (S) β3-homophenylalanine [(S)-β3-HPhe or β-Phe used for simplicity in this paper] (1) residues on antiparallel strands.
Experimental Methods.
The 10-residue peptide BBH-10 was synthesized by conventional solution-phase procedures by using a fragment condensation strategy. t-butoxycarbonyl (Boc) and methyl groups are used for N- and C-terminal protection. The Boc-(S)-β-Phe was synthesized by Arndt–Eistert homologation of Boc-Phe (7). Peptide couplings were mediated by N,N′-dicyclohexylcarbodiimide and 1-hydroxybenzotriazole. The peptide was purified by medium-pressure liquid chromatography on a reverse-phase C18 (10–60 μm) column followed by HPLC on a C18 (5–10 μm) column with methanol–water gradients. Circular dichroism spectra were recorded on a Jasco (Easton, MD) J-715 spectropolarimeter by using a 1-mm path length cell. The identity of the final peptide was confirmed by matrix-assisted laser desorption ionization mass spectrometry [M + Na (observed) = 1,254.2, M (calculated) = 1,231.5 and 1H NMR by using 500 MHz Bruker (Madison, WI) DRX 500 spectrometer.
Crystals suitable for x-ray diffraction were obtained by slow evaporation from methanol. A crystal in the form of a prism was coated with microscope oil, and x-ray data were obtained at −60°C on a four-circle diffractometer (Bruker P4) by using CuKα radiation (λ = 1.54178 Å). The θ-2θ scan mode was used with a 1.9° + 2θ(α1-α2) scan width, 13°/min scan speed with 2θmax = 115° (0.90 Å resolution). The crystal size was 1.05 × 0.25 × 0.22 mm. The crystal data are C65H102N10O13 ⋅ 3H2O, space group P212121 with a = 19.059(8) Å, b = 19.470(2) Å, c = 21.077(2) Å; V = 7,821 Å3, Z = 4 and calculated density 1.087 gm/cm3. The structure was solved by direct-phase determination with the use of the tangent formula (15). Three water molecules were located in a difference map. Hydrogen atoms were placed in idealized positions and were allowed to ride with the C or N atom, to which they were bonded in a least-squares refinement on F2 values of 6,234 reflections. The R1 factor is 9.12% for 3,984 data with |Fo| > 4σ and 821 variables. The least-squares program used was Siemens SHELXTL, version 5.03 (Iselin, NJ).
Results and Discussion
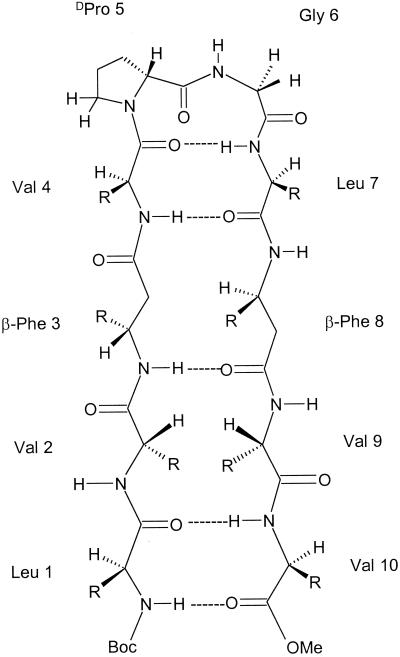
The decapeptide (Boc-Leu-Val-βPhe-Val-DPro-Gly-Leu-βPhe-Val-Val-methyl ester) BBH-10 was designed with a centrally positioned DPro-Gly segment to facilitate type II′ or type I′ β-turn formation, which are stereochemically favorable for registry of interstrand hydrogen bonds (16–19) (Table 1). The β-Phe residues were inserted at positions 3 and 8, where it was anticipated that interstrand hydrogen bonds would be maintained. The sole difference as compared with conventional all α-amino acid peptide β-hairpins is that the inward- and outward-facing CO and NH groups would not strictly alternate along the strands (Fig. 1). Indeed, two possible β-sheet structures have been considered for all β-peptide chains. In the “polar” sheet, the NH groups face outwards on one side, whereas the CO groups are similarly positioned on the opposite face. In an “apolar” sheet, alternation of NH and CO groups similar to that in an α-peptide sheet is expected. The difference between the two forms is that the former structure has θ ≈ ±180°, whereas the latter is obtained by θ ≈ ±60° (20, 21).
Table 1.
Hydrogen bonds
| Type | Donor | Acceptor | D⋯A(Å) | H*⋯A(Å) | D⋯O=C angle (degrees) |
|---|---|---|---|---|---|
| Intermol. | N1 | O6† | 2.810 | 2.01 | 136 |
| Intermol. | N2 | O6† | 3.079 | 2.20 | 152 |
| Intramol. | N3 | O8 | 2.883 | 2.06 | 155 |
| Intramol. | N4 | O7 | 2.922 | 2.04 | 149 |
| N5 (Proline) | |||||
| Pept.-solv. | N6 | W1 | 2.902 | 2.05 | |
| Intramol. | N7 | O4 | 2.895 | 2.03 | 124 |
| Intermol. | N8 | O2‡ | 2.840 | 1.97 | 166 |
| Intermol. | N9 | O3‡ | 2.928 | 2.12 | 171 |
| Intramol. | N10 | O1 | 2.874 | 1.98 | 150 |
| Solv.-solv. | W1 | W2 | 2.717 | ||
| Solv.-pept. | W2 | O9† | 2.814 | ||
| Solv.-solv. | W2 | W3§ | 2.877 | ||
| Solv.-pept. | W3 | O5 | 3.015 |
Hydrogen atoms were placed in idealized positions with N-H = 0.90 Å.
At symmetry equivalent −1/2 + x, 1/2 − y, 2 − z.
At symmetry equivalent 1/2 + x, 1/2 − y, 2 − z.
At symmetry equivalent −x, −1/2 + y, 1.5 − z.
Figure 1.
Schematic representation of the β-hairpin in peptide BBH10.
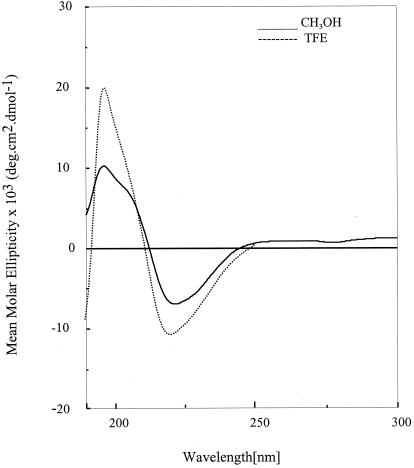
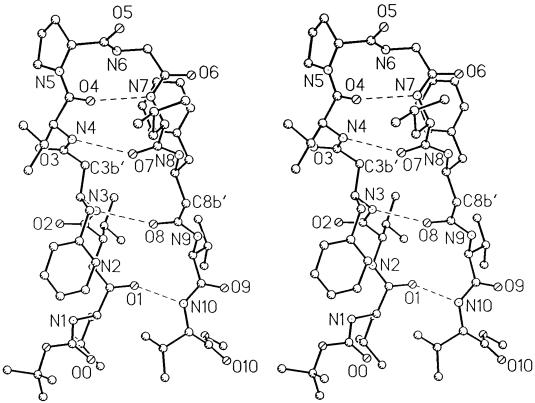
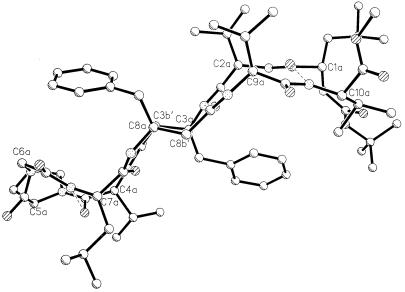
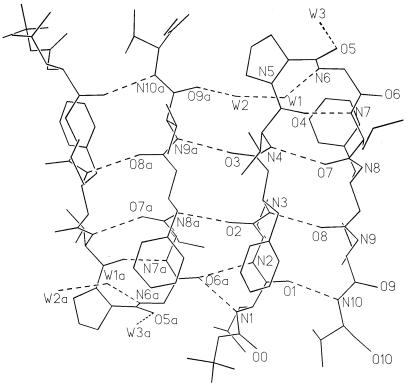
Fig. 2 shows circular dichroism (CD) spectra of peptide BBH-10 in methanol and trifluoroethanol. In both solvents, the negative CD band at approximately 220 nm is observed, characteristic of peptide β-hairpins (16, 22, 23). The 500 MHz 1H NMR spectrum reveals the wide dispersion of the backbone NH and CαH chemical shifts, diagnostic of a well-structured peptide in solution. Crystals of BBH-10 obtained in methanol solution yield the structure shown in Fig. 3. The molecule adopts a β-hairpin conformation (Table 1) exactly as anticipated in Fig. 1. Inspection of the DPro-Gly segment shows that it adopts a type I′ conformation (Table 2). It is noted that in previous examples of the DPro-Gly segment in peptide β-hairpins, type II′ β-turns were observed (24–26). From a cursory inspection, it is not apparent why a different β-turn occurs in the present peptide. The Val(2), Val(4), Leu(7), and Val(9) residues all adopt backbone conformations characteristic of a β-sheet. Both β-Phe residues adopt very similar backbone conformations, β-Phe(3) (φ = −116°, θ = +168°, ψ = +106°) and β-Phe(8) (φ = −109°, θ = +170°, ψ = +128°). The trans-geometry about the Cα-Cβ bond (θ ≈ 180) facilitates accommodation of an additional carbon atom into the backbone without disrupting the cross β-hairpin structure. Interestingly, the φ and ψ values for β-Phe residues have values very similar to those observed for α-amino acids in the strands. The Phe side chains of the two β-Phe residues point toward opposite sides of the β-sheet (Fig. 4), in contrast to the situation in all α-amino acids, where facing residues will have the side chains on the same face of the β-sheet. The observed hairpin conformation is stabilized by four cross-strand hydrogen bonds Leu(1) CO⋅⋅⋅NH Val(10), β-Phe(3) NH⋅⋅⋅CO β-Phe(8), Val(4) NH⋅⋅⋅CO Leu(7), and Val(4) CO⋅⋅⋅NH Leu(7). The fifth hydrogen bond (Fig. 1) is not observed in the crystal structure because of fraying of strands at the N and C termini, a feature also evident in dihedral angles, Leu(1) φ = −124°, ψ = −51° and Val(10) φ = −76°, ψ = −36°. A notable feature observed in the structure is the polarity of the peptide bond orientations observed at the site of β-amino acid insertion. From the structure depicted in Fig. 1 and the crystal structure shown in Fig. 3, it is seen that the β-Phe(3) NH and Val(4) NH point inwards, making hydrogen bonds with Leu(7) CO and β-Phe(8) CO, which also point inwards. In the classical α-amino acid β-hairpin, there is strict alternation in the direction of adjacent NH and CO groups in a single strand. The observed local structure at the β-Phe(3) and β-Phe(8) residues corresponds to a “polar sheet” (27). Furthermore, extended β-sheets are formed in the crystal of BBH-10 in an antiparallel motif (Fig. 5) that is very similar to the extended sheets formed by hairpins with all α-amino residues. The interhairpin hydrogen bonds Val(2)CO⋅⋅⋅NH β-Phe(8a) and β-Phe(3) CO⋅⋅⋅NH Val(9a) continue the “polar sheet.”
Figure 2.
Circular dichroism spectra of BBH10 in MeOH and 2,2,2-trifluoroethanol. Peptide concentration, 0.813 × 10−4M. Molar ellipticities ([θ]m) are expressed as degrees cm2 dmol−1.
Figure 3.
Stereo diagram of the x-ray structure of the β-hairpin in peptide BBH10. Note the positioning of C3b′ and C8b′, the additional CH2 moieties in the β-amino residues 3 and 8.
Table 2.
Torsional angles* with two β-Phe residues
| Backbone | Side
chains
|
||||||
|---|---|---|---|---|---|---|---|
| χ1 | χ2 | χ3 | χ4 | χ5 | |||
| Boc0 | ϕO | 168 | |||||
| ωO | −166 | ||||||
| Leu1 | φ | −124 | −59 | −62 | |||
| ϕ | −51 | +176 | |||||
| ω | −175 | ||||||
| Val2 | φ | −137 | −57 | ||||
| ϕ | +127 | +179 | |||||
| ω | −173 | ||||||
| βHPhe3† | φ | −116 | −70 | −92 | |||
| θ | +168 | +87 | |||||
| ϕ | +106 | ||||||
| ω | −177 | ||||||
| Val4 | φ | −137 | −61 | ||||
| ϕ | +113 | +179 | |||||
| ω | +170 | ||||||
| d-Pro5 | φ | +56 | −10 | +20 | −21 | +12 | −2 |
| ϕ | +37 | ||||||
| ω | −179 | ||||||
| Gly6 | φ | +88 | |||||
| ϕ | −1 | ||||||
| ω | +175 | ||||||
| Leu7 | φ | −100 | −51 | 173 | |||
| ϕ | +114 | −60 | |||||
| ω | −175 | ||||||
| βHPhe8† | φ | −109 | −63 | 100 | |||
| θ | +172 | −77 | |||||
| ϕ | +128 | ||||||
| ω | +177 | ||||||
| Val9 | φ | −118 | +177 | ||||
| ϕ | +131 | −61 | |||||
| ω | 180 | ||||||
| Val10 | φ | −76 | +175 | ||||
| ϕ* | −36 | −63 | |||||
| ω* | +178 | ||||||
Figure 4.
Side view of the pleated sheet. The segments C3a-C3b′ and C8a-C8b′ form an additional pleat that is quite narrow. The Phe side chains of the two β-Phe residues point in opposite directions.
Figure 5.
Two molecules in an extended antiparallel β-sheet in the crystal of BBH10.
Minimal β-hairpin structures by using β2,3-disubstituted amino acids in the strands have been crystallographically characterized in two tetrapeptides containing different turn segments (20, 21). Solution NMR data have been used to support a population of β-hairpin conformations in a hexapeptide containing β2,3, β2, and β3 residues (27).
The crystal structure of peptide BBH-10 demonstrates that β-amino acids can be inserted into peptide hairpins without disturbing the overall fold of the molecule. In an earlier report, we demonstrated that a two-residue segment containing contiguous β-alanine and γ-amino butyric acid residues can be inserted into a polypeptide helix without destruction of the secondary structure. These results emphasize that the insertion of additional atoms into polypeptide backbones is indeed possible, without significant disruption of classical secondary structure motifs. The use of chiral β-amino acid residues in peptide chemistry greatly enhances the scope of analog design in the case of biologically active peptides. Moreover, sequences containing α- and β-amino acid residues should also be useful in generating new polypeptide scaffolds for arraying functional side chains.
Acknowledgments
This research was supported at Bangalore by a program support grant in the area of drug and molecular design by the Department of Biotechnology, Government of India. H.N.G. is a recipient of a senior research fellowship of the Council of Scientific and Industrial Research, Government of India. The work at the Naval Research Laboratory was supported by National Institutes of Health Grant GM30902 and the Office of Naval Research.
Abbreviations
- Boc
t-butoxycarbonyl
- β-Phe
(S)-β3-homophenylalanine
Footnotes
Data deposition: The atomic coordinates have been deposited in the Cambridge Data Centre, 12 Union Road, Cambridge CB2 1EZ, U.K. (ref. no. 155411).
References
- 1.Seebach D, Matthews J L. J. Chem. Soc. Chem. Commun. 1997. , 2015–2022. [Google Scholar]
- 2.Gellman S H. Acc Chem Res. 1998;31:173–180. [Google Scholar]
- 3.Banerjee A, Balaram P. Curr Sci. 1997;73:1067–1077. [Google Scholar]
- 4.Iverson B L. Nature (London) 1997;385:113–115. doi: 10.1038/385113a0. [DOI] [PubMed] [Google Scholar]
- 5.Koert U. Angew Chem Int Ed Engl. 1997;36:1836–1837. [Google Scholar]
- 6.Gademann K, Hintermann T, Schreiber J V. Curr Med Chem. 1999;6:905–925. [PubMed] [Google Scholar]
- 7.Seebach D, Overhand M, Kuhnle F N M, Martinoni B, Oberer L, Hommel U, Widmer H. Helv Chim Acta. 1996;79:913–941. [Google Scholar]
- 8.Hintermann T, Seebach D. Synlett. 1997. 437–438. [Google Scholar]
- 9.Guichard G, Abele S, Seebach D. Helv Chim Acta. 1998;81:187–206. [Google Scholar]
- 10.Appella D H, Christianson L A, Klein D A, Powell D R, Huang X, Barchi J J, Jr, Gellman S H. Nature (London) 1997;387:381–382. doi: 10.1038/387381a0. [DOI] [PubMed] [Google Scholar]
- 11.Barchi J J, Jr, Huang X, Appella D H, Christianson L A, Durell S R, Gellman S H. J Am Chem Soc. 2000;122:2711–2718. [Google Scholar]
- 12.Appella D H, Christianson L A, Karle I L, Powell D R, Gellman S H. J Am Chem Soc. 1996;118:13071–13072. [Google Scholar]
- 13.Hinterman T, Seebach D. Chimia. 1997;50:244–247. [Google Scholar]
- 14.Karle I L, Pramanik A, Banerjee A, Bhattachajya S, Balaram P. J Am Chem Soc. 1997;119:9087–9095. [Google Scholar]
- 15.Karle J, Karle I L. Acta Crystallogr. 1966;21:849–859. [Google Scholar]
- 16.Awasthi S K, Ragothama S, Balaram P. Biochem Biophys Res Commun. 1995;216:375–381. doi: 10.1006/bbrc.1995.2634. [DOI] [PubMed] [Google Scholar]
- 17.Haque T S, Little J C, Gellman S H. J Am Chem Soc. 1996;118:6975–6985. [Google Scholar]
- 18.Haque T S, Gellman S H. J Am Chem Soc. 1997;119:2303–2304. [Google Scholar]
- 19.Struthers M D, Cheng R P, Imperiali B. Science. 1996;271:342–345. doi: 10.1126/science.271.5247.342. [DOI] [PubMed] [Google Scholar]
- 20.Krauthauser S, Christianson L A, Powell D R, Gellman S H. J Am Chem Soc. 1997;119:11719–11720. [Google Scholar]
- 21.Chung Y J, Christianson L A, Stanger H E, Powell D R, Gellman S H. J Am Chem Soc. 1998;120:10555–10556. [Google Scholar]
- 22.Alvarado M R, Blanco F J, Serrano L. Nat Struct Biol. 1996;3:604–612. doi: 10.1038/nsb0796-604. [DOI] [PubMed] [Google Scholar]
- 23.Nesloney C L, Kelly J W. J Am Chem Soc. 1996;118:5836–5845. [Google Scholar]
- 24.Ragothama S, Awasthi S K, Balaram P. J Chem Soc Perkin. 1998;2:137–143. [Google Scholar]
- 25.Karle I L, Awasthi S K, Balaram P. Proc Natl Acad Sci USA. 1996;93:8189–8193. doi: 10.1073/pnas.93.16.8189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karle I L, Das C, Balaram P. Proc Natl Acad Sci USA. 2000;97:3034–3037. doi: 10.1073/pnas.070042697. . (First Published March 21, 2000; 10.1073/pnas.070042697) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seebach D, Abele S, Gademann K, Jaun B. Angew Chem Int Ed Engl. 1999;38:1595–1597. doi: 10.1002/(SICI)1521-3773(19990601)38:11<1595::AID-ANIE1595>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 28.IUPAC–IUB Commission on Biochemical Nomenclature. Biochemistry. 1970;9:3471–3479. doi: 10.1021/bi00820a001. [DOI] [PubMed] [Google Scholar]