Abstract
Successful transcription of specific genes required for long-term memory processes involves the orchestrated effort of not only transcription factors, but also very specific enzymatic protein complexes that modify chromatin structure. Chromatin modification has been identified as a pivotal molecular mechanism underlying certain forms of synaptic plasticity and memory. The best-studied form of chromatin modification in the learning and memory field is histone acetylation, which is regulated by histone acetyltransferases and histone deacetylases (HDACs). HDAC inhibitors have been shown to strongly enhance long-term memory processes, and recent work has aimed to identify contributions of individual HDACs. In this review, we focus on HDAC3 and discuss its recently defined role as a negative regulator of long-term memory formation. HDAC3 is part of a corepressor complex and has direct interactions with class II HDACs that may be important for its molecular and behavioral consequences. And last, we propose the “molecular brake pad” hypothesis of HDAC function. The HDACs and associated corepressor complexes may function in neurons, in part, as “molecular brake pads.” HDACs are localized to promoters of active genes and act as a persistent clamp that requires strong activity-dependent signaling to temporarily release these complexes (or brake pads) to activate gene expression required for long-term memory formation. Thus, HDAC inhibition removes the “molecular brake pads” constraining the processes necessary for long-term memory and results in strong, persistent memory formation.
Introduction
Gene transcription is essential for long-term synaptic plasticity and long-term memory formation (Alberini, 2009). Chromatin modifications can modulate the transcription required for long-term memory processes by providing transient and potentially stable epigenetic marks that activate and/or maintain transcriptional processes. Histone acetylation is a chromatin modification that modulates histone-DNA interactions via two different classes of enzymes: histone acetyltransferases (HATs), which acetylate histone tails and promote gene transcription; and histone deacetylases (HDACs), which remove acetyl groups and lead to gene silencing (Kouzarides, 2007). A learning event that produces long-term memory enhances histone acetylation, by increasing HAT and decreasing HDAC activity, to induce specific patterns of gene expression (Levenson et al., 2004; Federman et al., 2009).
Numerous studies have shown that a potent HAT, cyclicAMP response element binding protein (CREB)-binding protein (CBP), is necessary for long-lasting forms of synaptic plasticity and long-term memory (reviewed in Barrett and Wood, 2008). Mouse models with a loss of CBP’s HAT function all show attenuated histone acetylation and impaired long-term memory formation (Oike et al., 1999; Bourtchouladze et al., 2003; Alarcon et al., 2004; Korzus et al., 2004; Wood et al., 2005; Wood et al., 2006; Vecsey et al., 2007; Stefanko et al., 2009; Chen et al., 2010; Barrett et al., 2011).
In contrast, HDACs have been shown to be powerful negative regulators of long-term memory processes. Nonspecific HDAC inhibitors enhance histone acetylation, synaptic plasticity, as well as long-term memory (Levenson et al., 2004; Bredy and Barad, 2008; Lattal et al., 2007; Vescey et al., 2007; Guan et al., 2009; Malvaez et al., 2010; Roozendaal et al., 2010). Using subthreshold training conditions, HDAC inhibition can transform a learning event that does not lead to memory formation into a learning event that does result in significant long-term memory (Stefanko et al., 2009). Further, HDAC inhibition can also generate a form of long-term memory that persists beyond the point at which normal memory fails (Stefanko et al., 2009). This body of work implicates histone acetylation as a potential mechanism by which a learning event can result in encoding of a long-term memory, whereas deacetylation likely inhibits this process.
Classification and Contribution of Individual HDACs
HDACs are grouped into four classes based on sequence homology with yeast factors and domain organization. All classes are dependent on zinc for their catalytic activity except for the sirtuins (Class III) which are structurally unrelated NAD-dependent enzymes and will not be discussed in this review. Class I, comprised of HDACs 1, 2, 3, and 8, share homology with yeast RPD3 protein. This group contains nuclear localization signal (NLS) and lack a nuclear export signal (NES), with the exception of HDAC3 which can be found in the nucleus and cytoplasm (Gregoretti et al., 2004). Class II HDACs resemble yeast protein HDA1 and are separated by domain organization into IIa (HDACs 4, 5, 7, and 9) and IIb (HDACs 6 and 10). This class contains NLS and NES for phosphorylation-regulated shuttling between the cytoplasm and nucleus as well as additional regulatory domains. HDAC3 has been shown to interact with most of the class II proteins (HDAC4, 5, 7, and 10; Fischle et al., 2002; Tong et al., 2002). HDAC11 is the sole member of Class IV, and has been found primarily in the nucleus in complexes with HDAC6 (Gao et al., 2002). HDAC11 has similarities with both Class I and II HDACs but likely has a unique physiological role.
Currently, the role of individual HDACs in long-term memory formation remains largely unexplored except for few recent studies. HDAC5 was the first discrete HDAC to be implicated as a negative regulator of long-term synaptic plasticity. Recruitment of HDAC5 to the C/EBP promoter repressed transcription and blocked long-term facilitation in aplysia (Guan et al., 2002). Further, mice lacking HDAC5 show enhanced reward learning in cocaine conditioned place preference (Renthal et al., 2007). Conversely, overexpression of HDAC4 or HDAC5 attenuated the expression of cocaine conditioned place preference, further supporting their role as negative regulators of reward-associated memory (Kumar et al., 2005; Renthal et al., 2007). However, it was recently shown that purified HDAC4 and HDAC5 have little to no catalytic activity on canonical HDAC substrates containing acetyl-lysines (Lahm et al., 2007). There is mounting evidence that class IIa HDACs function in vivo by interacting with class I HDACs, which have very potent HDAC activity, and other corepressors to form multi-protein complexes (Fischle et al., 2002; Lahm et al., 2007). These findings strongly suggest that future studies should include examination of not only an individual HDAC, but consider the corepressor complex as a whole to determine how gene expression required for long-term memory formation is being modulated.
Most of the evidence for the contribution of HDACs in learning and memory comes from pharmacological studies. HDAC inhibitors sodium butyrate (NaBut), valproate and suberoylanilide hydroxamic acid (SAHA) were thought to non-specifically block class I, IIa and IIb, but not class III, HDACs. However, a recent biochemical analysis of the in vitro activities of recombinantly expressed, purified HDACs 1–9 demonstrated that those drugs are potent inhibitors of class I, but not class IIa and IIb, HDACs (Kilgore et al., 2010). These findings suggest that it is the class I HDACs that are critical for regulating long-term memory processes. Indeed, recent work on individual class I HDACs supports this hypothesis. Forebrain overexpression of HDAC2, but not HDAC1, caused impaired memory formation and synapse formation (Guan et al., 2009). Conversely, loss of HDAC2 resulted in enhanced memory formation and synaptic plasticity. Further, HDAC2, but not HDAC1, was shown to be associated with the promoters of several genes implicated in plasticity and learning, and so it is likely that removal of this negative regulator would allow for greater learning-induced gene expression. We have shown recently that another class I HDAC, HDAC3, negatively regulates memory formation (McQuown et al., 2011), which will be the focus of the rest of this review.
HDAC3 Expression and Function
HDAC3 is found in many tissues throughout the body, including the brain (Mahlknecht et al., 1999). It is the most highly expressed Class I HDAC in the brain with greatest expression in the hippocampus, cortex, and cerebellum (Broide et al., 2007). HDAC3 is predominantly expressed in neurons, but it is also one of the few HDACs localized in oligodendrocytes (Shen et al., 2005; Broide et al., 2007). While its primary localization is in the nucleus, HDAC3 can also be found in the cytoplasm and at the plasma membrane (Takami and Nakayama, 2000; Longworth and Laimins, 2006).
HDAC3 catalytic activity can be regulated by phosphorylation at the serine 424 residue of the C-terminal domain (Zhang et al., 2005). Casein kinase 2 phosphorylation of HDAC3 at this site has been shown to increase the basal enzymatic activity, whereas protein phosphatase 4 has the inverse effect (Zhang et al., 2005). Although phosphorylation can alter activity of HDAC3, it has not been found to alter subcellular localization or protein interactions (Zhang et al., 2005; Jeyakumar et al., 2007). Also, an oligomerization domain has been identified in the N-terminal by which the protein can self-associate to form dimers and trimers (Yang et al., 2002). However, recombinant HDAC3 alone has no HDAC function (Guenther et al., 2001). HDAC3 must be properly folded by TCP-1 ring complex and then bound to corepressors NCoR (nuclear receptor corepressor) or SMRT (silencing mediator of retinoic acid and thyroid hormone receptor) to form an active enzyme complex (Guenther et al., 2001, 2002).
HDAC3 forms a stable multiprotein complex with corepressors NCoR and SMRT in order to regulate transcription of genes as well as other nontranscriptional functions. Three different binding sites on NCoR/SMRT are associated with HDAC3 (Wen et al., 2000). One site important for HDAC3 activity is the deacetylase domain (DAD) of NCoR/SMRT, which binds both the amino and carboxy termini of HDAC3 and transforms HDAC3 into a four-helical structure (Codina et al., 2005; Guenther et al., 2001). HDAC3 is the primary HDAC enzyme in NCoR/SMRT complexes, however other HDACs or HDAC complexes can be recruited in a transcription factor-specific or context-specific manner by less stable interactions with NCoR/SMRT (Huang et al., 2000; Fischle et al., 2001; 2002). This has been best described of the class II HDACs.
Class IIa HDACs (HDAC4 and 5) are found to directly interact with the RD3 domain of NCoR/SMRT, a distinct domain from HDAC3, and become part of the repressor complex (Huang et al., 2000; Wen et al., 2000; Fischle et al., 2001). In addition, class II HDACs (4, 5, 7, and 10) have been shown to interact with HDAC3, but not HDAC1 or 2 (Huang et al., 2000; Fischle et al., 2001; 2002). Specifically, HDAC4 coimmunoprecipitates with HDAC3 via its C-terminal domain and disruption of this interaction results in loss of observed HDAC activity. Further, it has been suggested that the enzymatic activities of Class IIa HDACs rely on interactions with HDAC3 and NCoR/SMRT (Huang et al., 2000; Fischle et al., 2001). Purified HDAC4 and 5 have little to no catalytic activity on canonical HDAC substrates containing acetyl-lysines (Lahm et al., 2007). However, HDAC4 or 5 associated with HDAC3 and/or NCoR results in observable deacetylase activity which is disrupted by mutations in these interaction domains. Thus, Class IIa HDACs likely function in vivo by interacting with HDAC3, which has potent HDAC activity, as part of a corepressor multi-protein complex (Fischle et al., 2002; Lahm et al., 2007).
Histone substrates of HDAC3
Histone deacetylation by HDAC3 and the NCoR/SMRT complex have been shown to cause transcriptional repression in two different ways. First, the removal of acetyl groups increases the affinity of the histone tail for the negatively charged DNA backbone, which yields a closed chromatin conformation intractable to transcription. Second, histone deacetylation reduces the recruitment of bromo-domain coactivators, such as chromatin-associated proteins and histone acetyltranferases that typically bind the acetyl-lysine motifs and promote transcription (Zeng and Zhou, 2002). In this fashion, HDAC3/NCoR/SMRT mediates the repression of unliganded nuclear receptors (e.g., PPAR, RAR, RXR, GR, NR4A2 and Rev-Erb; Fajas et al., 2002; Jepsen and Rosenfeld, 2002; Yin and Lazar, 2005; Lammi et al., 2008). HDAC3 repression is lost when a ligand binds and induces a conformational change to release the corepressor complex and allowing a coactivator to bind. In other cases, HDAC3 complexes interact with transcription factors to keep their target genes in an inactive state (Heinzel et al., 1997; Lutterbach et al., 1998).
Some research has aimed to identify HDAC3-preferred lysine residues on histone tails and has yielded conflicting results. The first studies of HDAC3 identified it as a potent deacetylase of both histones H3 and H4, however HDAC3 deacetylated histone H4 more completely than HDAC1 (Yang et al., 1997; Dangond et al., 1998; Emiliani et al., 1998). In contrast, Vermeulen et al. (2004) found histone deacetylation of SMRT/NCoR complexes preferred histone H3 when targeted to preacetylated nucleosomal templates. More detailed studies began to identify specific lysine residues on the histone tails targeted by HDAC3. One in vitro study suggested that HDAC3 completely deacetylated H2AK5, H4K5, and H4K12, but only partially deacetylated H3, H2B, H4K8, and H4K16 (Johnson et al., 2002). Using the retinoic acid-regulated gene as a prototype, recruitment of HDAC3-NCoR/SMRT complex showed deacetylation of H4 lysines in order of K5, K8, K12, and K16 (Hartman et al., 2005). Other models of genetic deletion and pharmacological inhibitor administration supported these histone H4 acetylation sites as targets and also found increased histone H3 K9/K14 acetylation (Bhaskara et al., 2010; Knutson et al., 2008; Xu et al., 2009; McQuown et al., 2011).
HDAC3 deacetylation occurs in concert with complementary enzymes (e.g., methyltransferases, phosphotases) to allow for coordinated epigenetic modifications. Thus, knockdown of HDAC3 has revealed interdependent changes in histone modifications. Aurora B kinase has been shown to prefer a hypoacetylated H3 tail in order to phosphorylate H3S10 residues. When HDAC3 is removed, histone H3 is hyperacetylated and a corresponding decrease in phosphorylation at H3S10 is observed (Li et al., 2006). Other HDAC3 interdependent modifications identified are phosphoryation of H3S28, methylation of H4K20, and dimethylation of H3K4 (Eot-Houllier et al., 2008; Yoo et al., 2010). The histone demethylase JMJD2A HDAC3 is an essential component for the coordination of these histone modification profiles to regulate specific gene expression profiles and to bring about distinct downstream events. Therefore, future studies should consider that the specific composition of modifications on a histone can further influence chromatin modification and ultimately establish specific transcription profiles that dictate cellular function.
Nonhistone substrates of HDAC3
In addition to histone targets, HDAC3 can deacetylate other proteins and regulate their localization and activity. One such target that is relevant to learning and memory is nuclear factor-kappaB (NF-κB) protein RelA. NF-κB is a transcription factor required for induction of genes necessary for long-term memory formation (Freudenthal and Romano, 2000; Merlo et al., 2005). HDAC3 has been shown to terminate NF-κB signaling by deacetylating RelA which triggers its nuclear export via interaction with inhibitory-κB (Chen et al., 2001). Removal of HDAC3 may extend NF-κB-dependent transcription following a learning event to enhance long-term memory formation.
Another nonhistone substrate is myocyte enhancer factor 2 (MEF2), a transcription factor important for the regulation of structural plasticity genes. HDAC4 and other class IIa HDACs have been shown to repress MEF2-dependent transcription, however, only HDAC3 has demonstrated the ability to deacetylate MEF2 (Gregoire et al., 2007). HDAC3 has a physical association with MEF2 at a DNA-binding domain that is distinct from where class IIa HDACs bind (Gregoire et al., 2007). Deacetylation of MEF2 by HDAC3 terminates transcription of plasticity genes. Strikingly, HDAC3 also has been shown to deacetylate the acetyltransferases PCAF and p300/CBP for MEF2 and other proteins (Chuang et al., 2006; Gregoire et al., 2007). The NCoR-HDAC3 complex has been shown to directly interact with CBP, such that increasing concentrations of NCoR lead to decreasing amounts of CBP HAT activity (Cowger and Torchia, 2006). Deacetylation of these coactivators, which are also HATs, has been shown to inhibit their function and shut down the activation of these pathways. Through deacetylation of histone and nonhistone substrates critical for learning, it can be predicted that HDAC3 would negatively regulate long-term memory formation.
HDAC3 and memory formation
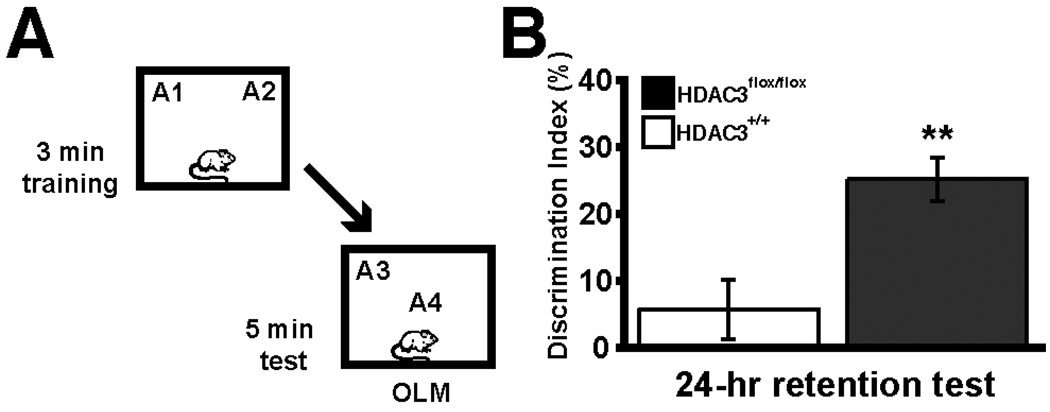
We examined the role of HDAC3 in learning and memory using three different approaches (McQuown et al., 2011). First, HDAC3floχ/floχ mice were infused with AAV-Cre recombinase into the dorsal hippocampus to create a homozygous focal deletion of HDAC3. Another genetic approach used was the DADm mouse that has a single amino acid substitution in the DAD domain that disrupts HDAC3 binding to NCoR (Alenghat et al., 2008). And last, we used a pharmacological inhibitor with greatest inhibition of HDAC3, RGFP136 (Rai et al., 2010). All three approaches lead to facilitated long-term memory formation after a subthreshold training period in the novel object recognition task (McQuown et al., 2011). This subthreshold training period failed to yield 24-hr long-term memory in control animals. The schematic in Fig. 1A shows the experimental design, such that the mice received subthreshold training (3 min) in an environment with 2 identical objects and received a retention test 24 hrs later in which one object is moved to a new location (object location memory). HDAC3floχ/floχ mice spent significantly more time with the object in the new location, whereas wildtype controls had no preference (Fig. 1B). Further, memory in animals with a loss of HDAC3 persisted at least seven days, a point beyond where normal object memory fails (see Stefanko et al., 2009). These behavioral findings suggest that HDAC3 is a critical negative regulator of long-term memory formation.
Figure 1.
Deletion of Hdac3 in the dorsal hippocampus leads to enhanced memory for object location (OLM). (A) Mice received subthreshold training (3 min) in an environment with 2 identical objects and received a retention test 24 hrs later in which one object is moved to a new location. (B) HDAC3flox/flox mice exhibited significant long-term memory for object location 24 hours after subthreshold training (n=8/group, ** indicates p<0.005). Adapted from McQuown et al. (2011) with permission.
Despite the consistent enhancements of long-term memory by loss of HDAC3 function, short-term memory for the object recognition task was unaffected. A major difference between these forms of memory is that, in general, transcription is essential for the formation of long-term memory, but not short-term memory. We and others have found that HDAC inhibition by sodium butyrate, TSA, or SAHA have no effect on short-term memory (Yeh et al., 2004; Levenson et al., 2004; Korzus et al., 2004; Vecsey et al., 2007; Stefanko et al., 2009). Therefore, HDAC3 inhibition is likely enhancing memory through increased gene expression.
As described above, HDAC3 can repress CBP function by deacetylation (Chuang et al., 2006; Gregoire et al., 2007). It is likely that HDAC3 inhibition allows greater CREB-CBP interactions to enhance gene transcription necessary for memory formation. We tested this hypothesis using genetically modified CBP mutant mice carrying a triple point mutation in the phospho-CREB (KIX) binding domain of CBP (CBPKIX/KIX mice; Kasper et al., 2002). Previously we have demonstrated that HDAC inhibition enhances hippocampal synaptic plasticity in wildtype but not CBPKIX/KIX mice, suggesting enhancement via HDAC inhibition requires hippocampal CREB:CBP interaction (Vecsey et al., 2007). Also, CBPKIX/KIX mice have deficits in long-term memory formation of a hippocampus-dependent task (Haettig et al., 2011). Intrahippocampal delivery of RGFP136 resulted in long-term memory after subthreshold training in CBP+/+ mice, but not CBPKIX/KIX littermates (McQuown et al., 2011). These results indicate that RGFP136, like sodium butyrate and trichostatin A, enhances long-term memory through a CBP-dependent mechanism. This appears to be a fundamental mechanism by which HDAC inhibitors modulate hippocampal synaptic plasticity and hippocampus-dependent long-term memory, and strongly suggest that HDAC inhibitors (even only class I specific inhibitors) modulate memory via a specific mechanism.
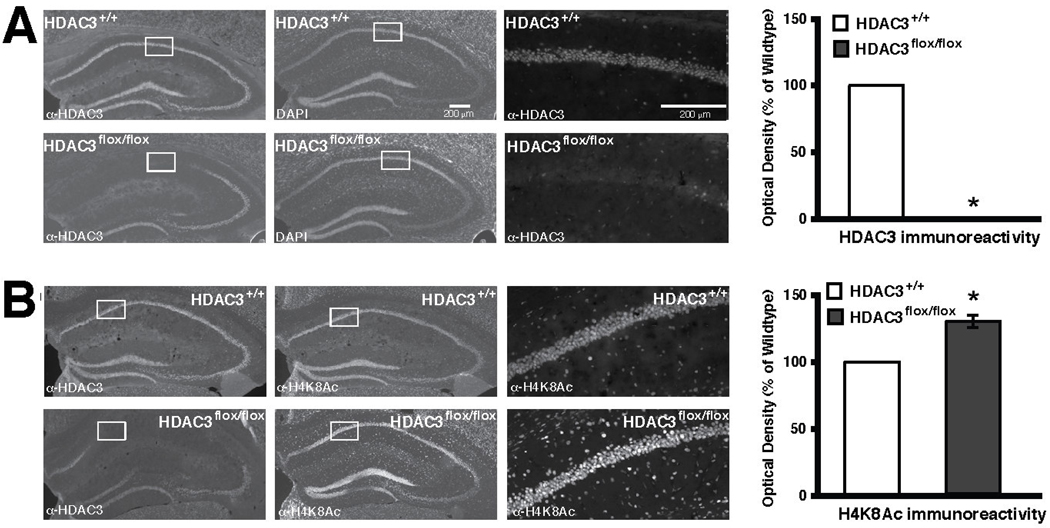
Immunohistochemical studies were conducted to examine the effects of HDAC3 disruption on other histone modifying enzymes and modifications. HDAC3 is highly expressed in the dorsal hippocampus, and HDAC3floχ/floχ mice infused with AAV-Cre showed a complete focal deletion of HDAC3 primarily in the CA1 of the dorsal hippocampus (Fig. 2A). In the area of the HDAC3 deletion, we observed a decrease in HDAC4, but not HDAC2, immunoreactivity (McQuown et al., 2011). This suggests that the behavioral effects were not a consequence of HDAC2, but rather the disruption of HDAC3 corepressor complex is responsible and that may lead to mislocalization or degradation of HDAC4. Again, these findings imply that examination of individual HDACs cannot exclude their effects on the corepressor complex which may incorporate other HDACs.
Figure 2.
Intrahippocampal AAV2/1-Cre infusion in HDAC3flox/flox mice results in a complete, focal deletion of HDAC3 that correlates with increased histone acetylation. Images are 4X except the right panels which are 20X magnifications of the regions boxed in white. Histograms depict quantification of optical density as a percent of wildtype. (A) Representative images showing DAPI labeling and HDAC3 immunoreactivity in hippocampi of AAV2/1-Cre infused HDAC3+/+ and HDAC3flox/flox mice. HDAC3 labeling is found throughout CA1, CA3 and the dentate gyrus, and no immunoreactivity is found in the AAV2/1-Cre infusion site of HDAC3flox/flox mice. * indicates p<0.05. (B) Representative images show that acetylation at H4K8 is increased specifically in the AAV2/1-Cre infusion site of HDAC3flox/flox mice. * indicates p<0.05. Adapted from McQuown et al. (2011) with permission.
We also observed increased acetylation of histone H4 at K8 (H4K8Ac) in the area of HDAC3 deletion (Fig. 2B; McQuown et al., 2011). Acetylation at H4K8 has been shown to increase after the dissociation of the NCoR/HDAC3 complex from promoter regions and consequently leads to an increase in transcriptional activity (Guenther et al., 2000; Li et al., 2000; Wang et al., 2010). Intrahippocampal infusion of RGFP136 showed the same alterations in immunoreactivity for HDAC4 and H4K8Ac as HDAC3floχ/floχ mice. Increased histone acetylation in the absence of HDAC3 likely suggests corresponding increases in gene expression.
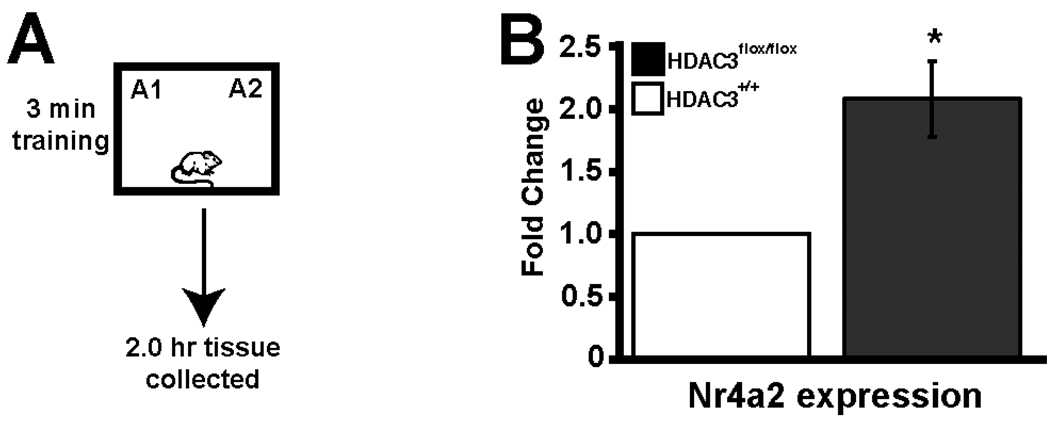
We investigated gene expression of Nr4a2 and c-Fos, immediate early genes, in the HDAC3flox/flox mice. Nr4a2, also known as nurr1, is a CREB-dependent gene implicated in long-term memory (Pena de Ortiz et al., 2000; von Hertzen and Giese, 2005; Colon-Cesario et al., 2006; Vecsey et al., 2007). We have previously demonstrated that Nr4a2 expression is enhanced by the HDAC inhibitor TSA during memory consolidation (Vecsey et al., 2007). Indeed, we observed increased Nr4a2 and c-Fos expression in the area of the focal HDAC3 deletion in the dorsal hippocampus 2 hours after subthreshold training compared to wildtype mice (Fig. 3; McQuown et al., 2011). However, there was no effect of HDAC3 deletion on Nr4a2 expression in naïve handled mice, which suggests that this enhanced expression is triggered by learning event.
Figure 3.
Gene expression is increased in the area of focal homozygous deletion of Hdac3 in HDAC3flox/flox mice. (A) Mice received subthreshold training (3 min) in an environment with 2 identical objects. (B) 2 hrs following training, tissue was collected by taking 1mm punches from dorsal hippocampal slices in the area of the focal deletion in HDAC3flox/flox mice as confirmed by immunohistochemistry for HDAC3 and equivalent regions in HDAC3+/+ mice. Quantitative RT-PCR shows that Nr4a2 expression is significantly increased in the dorsal hippocampus of HDAC3flox/flox mice as compared to wildtype littermates (n=3/group, * indicates p<0.02). Adapted from McQuown et al. (2011) with permission.
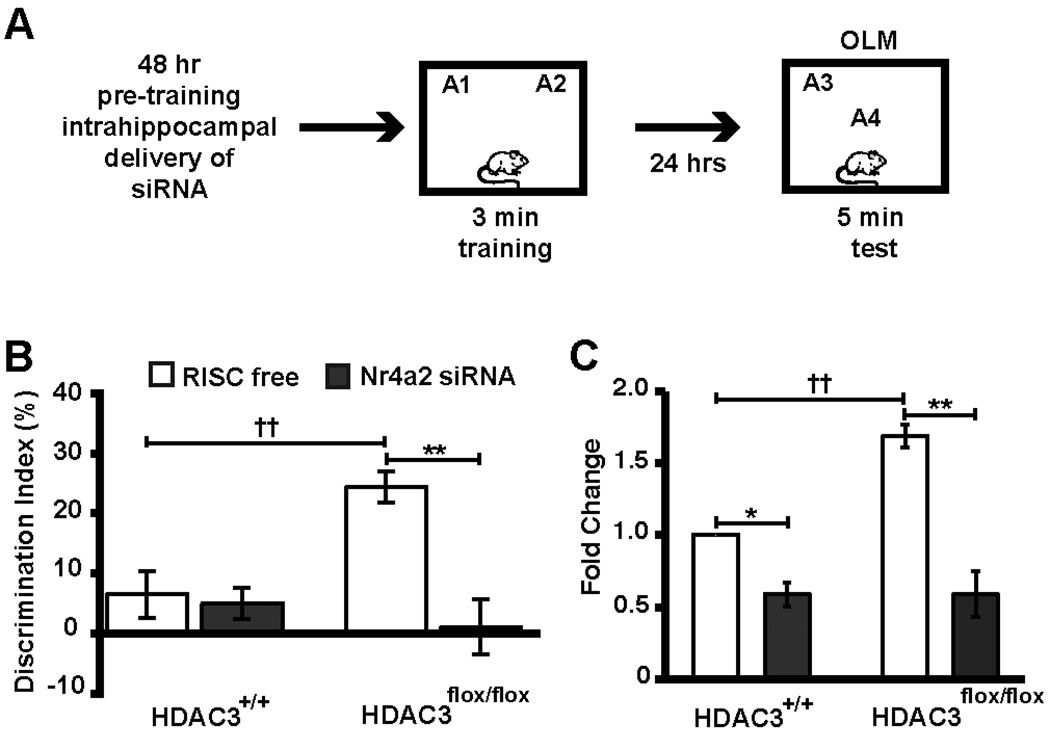
To determine if increased Nr4a2 expression is necessary for enhanced long-term memory in HDAC3flox/flox mice, we infused siRNA targeting Nr4a2 48 hrs prior to the subthreshold training in novel object recognition task (Fig. 4A). HDAC3flox/flox mice with a homozygous deletion of HDAC3 in the dorsal hippocampus failed to exhibit enhanced long-term memory when Nr4a2 siRNA, but not RISC free siRNA, was infused into the area of HDAC3 deletion prior to training (Fig. 4B; McQuown et al., 2011). Further, quantitative RT-PCR showed that Nr4a2 siRNA treatment significantly reduced Nr4a2 expression in both HDAC3flox/flox and HDAC3+/+ mice, and RISC free-treated HDAC3flox/flox mice also exhibited an increased induction of Nr4a2 mRNA after the long-term memory test (Fig. 4C). These data suggest a mechanism by which the loss of HDAC3 enhances long-term memory by allowing increased and/or prolonged CREB:CBP dependent transcription of Nr4a2. These data also suggest that the transcription factor and immediate early gene Nr4a2 may be a key molecular component of the machinery underlying synaptic plasticity and memory formation.
Figure 4.
Nr4a2 siRNA completely blocks the long-term memory enhancement observed in HDAC3flox/flox mice. (A) 48 hours after infusions of Nr4a2 or RISC free siRNA, HDAC3flox/flox and HDAC3+/+ mice received subthreshold training (3 min) in an environment with 2 identical objects and received a retention test 24 h later in which one object is moved to a new location. (B) HDAC3flox/flox mice infused with RISC free (n=10) exhibited significant memory for object location compared to HDAC3+/+ mice (†† indicates p=0.001), which was blocked by Nr4a2 siRNA treatment (n=9–10/group, ** indicates p<0.001). (C) 2 hrs following testing, quantitative RT-PCR shows that Nr4a2 siRNA treatment significantly reduced Nr4a2 expression in both HDAC3flox/flox and HDAC3+/+ mice (n=3/group, ** p<0.001 and *p<0.05 vs. respective RISC-free siRNA controls). HDAC3flox/flox mice also exhibited an increased induction of Nr4a2 mRNA after the long-term memory test (†† indicates p = 0.002 vs. HDAC3+/+ RISC free). Adapted from McQuown et al. (2011) with permission.
Molecular Brake Pad Hypothesis
In the learning and memory field, a prevailing question that drives numerous research programs, including our own, is what are the molecular mechanism underlying long-term memory formation? Although mechanisms underlying memory are of great importance, an equally important question is by what molecular mechanisms are long-term memories prevented from forming? This is an old question, and has taken several manifestations, including a recent version in which genes that prevent memory formation were described as memory suppressor genes, relating to their counterparts (tumor suppressor genes) in cell cycle regulation (Abel et al., 1998). Below we present yet another version, but modified to reflect the current understanding of the contribution of HDACs and associated corepressors in regulating transcription required for long-term memory processes.
In this last section of the review, we would like to put forth a hypothesis aimed at describing how long-term memory processes are prevented at the level of gene regulation. The hypothesis, which we term the “molecular brake pad” hypothesis, states that HDACs and associated corepressors form complexes (or molecular brake pads) that normally maintain specific genes in a silent state and sufficiently strong activity-dependent signaling is required to temporarily remove these complexes (or brake pads) to activate gene expression required for long-term memory formation. Thus, these repressor complexes (or brake pads) are always on, except during important signaling events triggering specific gene expression profiles for cellular function. Below we explore a few predictions made by this hypothesis.
Genomic DNA in its relaxed form would extend approximately two meters, which needs to fit into a six micron diameter nucleus. To achieve this incredible level of compaction, genomic DNA goes through multiple levels of organization resulting in approximately a 10,000 fold compaction. “10,000 fold” is an extremely difficult idea to grasp, but it becomes readily clear that accessing and indexing genes required for long-term memory processes is a remarkable achievement. The point is that the molecular machinery involved in this organization and compaction of genomic DNA is part and parcel to accessing and indexing genes. Again, not a novel idea, but it helps to consider this before exploring how genes are turned on for long-term memory formation---it’s not just as simple as loading RNA pol II.
One simple prediction is that HDACs and associated corepressors forming “molecular brake pads” are normally engaged in silencing gene expression because they are normally involved in the compaction of chromatin structure. However, there are many mechanisms of genomic DNA compaction (polycomb, etc.), so what makes HDACs and associated corepressors unique? Part of the answer may be that HDACs and associated corepressors are preferentially found at actively transcribed genes in a constant interplay with HATs and RNA pol II to regulate gene expression. A recent genome-wide mapping of HATs and HDACs found that both are found at active genes with acetylated histones and both are targeted to transcribed regions of active genes by phosphorylated RNA pol II (Wang et al., 2009). The authors extend the interpretation of their findings to conclude that the majority of HDACs function to reset chromatin by removing acetylation at active genes. These results support the idea of HDACs and associated corepressors functioning as “molecular brake pads” at actively transcribed genes as they are primarily found at actively transcribed genes and reset their state of expression.
Another simple prediction is that inhibition of these molecular brake pad complexes should have specific consequences on activity-dependent transcription and long-term memory (see Fig. 5). We’ll consider the former first. If these molecular brake pad complexes serve to reset chromatin and silence gene expression following activity-dependent signaling, then prohibiting the molecular brake pads from re-engaging may be predicted to prolong gene expression beyond the point it would normally following a learning event. This has been observed in a study by Vecsey et al. (2007) in which mice were fit with intrahippocampal cannulae, subject to contextual fear conditioning, and then immediately after training injected with an HDAC inhibitor. Two hours after training, hippocampi were collected and gene expression was examined. At a point when immediate early genes are normally turned off, the immediate early gene and transcription factor Nr4a2 had maintained expression, which was associated with increased histone acetylation at its promoter (Vecsey et al., 2007). HDAC inhibition alone had no effect on the genes examined and contextual fear conditioning alone did not result in maintained gene expression at two hours post-training. Furthermore, out of about a dozen genes examined, only Nr4a2 and Nr4a1 had maintained gene expression. These results demonstrate that HDAC inhibition may prevent the resetting of chromatin structure by molecular brake pad complexes, resulting in maintained gene expression beyond the point normally observed after learning.
Figure 5.
Illustration of the molecular brake pad hypothesis. This is a simplified schematic diagram of the transcriptional regulation by interactions with HATs and HDACs. Cartoon depicts the nucleosome as blue cylinders with DNA tightly wound around them in black. Learning- induced activation releases HDAC/Corepressor complexes and allows HAT/Coactivator complexes to recruit transcriptional machinery to induce gene expression. Although depicted as separate protein complexes, it is quite likely that HATs and HDACs may be found in the same complex. Dotted lines represent theoretical immediate early gene expression levels after learning with an HDAC inhibitor.
Does maintained gene expression result in enhanced long-term memory? Does maintained gene expression transform a learning event that does not normally result in short- or long-term memory into an event that does? We don’t have the answers to those specific questions, but studies have demonstrated remarkable effects on the modulation of memory by HDAC inhibition. In particular, one simple prediction of the molecular brake pad hypothesis is that if the brake pads are removed, then a subthreshold stimulus should result in long-term potentiation (LTP) and long-term memory. With regard to synaptic plasticity, Vecsey et al. (2007) showed that a stimulus that normally induces a transient, transcription-independent form of LTP, can be transformed into a stable, transcription-dependent form of LTP in the presence of HDAC inhibition. With regard to long-term memory, Stefanko et al. (2009) showed that a subthreshold learning period of three minutes, which does not result in observable short- or long-term memory, does result in robust long-term memory in the presence of HDAC inhibition. Similarly, Hdac3-FLOX mice with a focal homozygous deletion of HDAC3 in the dorsal hippocampus also exhibit robust long-term memory for object location following a subthreshold training period (McQuown et al., 2011).
Is this via any specific mechanism? Indeed, Haettig et al. (2011) and McQuown et al (2011) recently showed that HDAC inhibition modulates hippocampus-dependent long-term memory in a CBP-dependent manner. This supports the interplay between HDACs and HATs as suggested by Wang et al. (2009) in regulating actively transcribed genes. More importantly, there is strong evidence demonstrating that removal of molecular brake pad complexes results in remarkable effects on long-term memory predicted by this hypothesis.
There remains an immense amount of work to fully define and understand how these HDACs and associated corepressor complexes function as molecular brake pads to regulate gene expression required for long-term memory processes, but we believe that this hypothesis may provide a useful conceptual framework to understand how these molecular mechanisms contribute to learning and memory. Future work in understanding how these molecular brake pads interact with HAT complexes, target non-histone substrates, regulate specific gene expression profiles, and receive and integrate incoming upstream signaling for specific neural function is pivotal. It may even help, in part, elucidate aspects of post-traumatic stress disorder, humans with excellent autobiographical memory, and contribute to novel therapeutic agents for cognitive disorders.
Acknowledgements
This work was supported by the Whitehall Foundation, NIMH (R01MH081004), and NIDA (R01DA025922) to MAW.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abel T, Martin KC, Bartsch D, Kandel ER. Memory suppressor genes: inhibitory constraints on the storage of long-term memory. Science. 1998;279:338–341. doi: 10.1126/science.279.5349.338. [DOI] [PubMed] [Google Scholar]
- Alarcon JM, Malleret G, Touzani K, Vronskaya S, Ishii S, Kandel ER, Barco A. Chromatin acetylation, memory, and LTP are impaired in CBP+/− mice: a model for the cognitive deficit in Rubinstein-Taybi syndrome and its amelioration. Neuron. 2004;42:947–959. doi: 10.1016/j.neuron.2004.05.021. [DOI] [PubMed] [Google Scholar]
- Alberini CM. Transcription factors in long-term memory and synaptic plasticity. Physiological Review. 2009;89:121–145. doi: 10.1152/physrev.00017.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alenghat T, Meyers K, Mullican SE, Leitner K, Adeniji-Adele A, Avila J, Bucan M, Ahima RS, Kaestner KH, Lazar MA. Nuclear receptor corepressor and histone deacetylase 3 govern circadian metabolic physiology. Nature. 2008;456:997–1000. doi: 10.1038/nature07541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett RM, Wood MA. Beyond transcription factors: the role of chromatin modifying enzymes in regulating transcription required for memory. Learning and Memory. 2008;15:460–467. doi: 10.1101/lm.917508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett RM, Malvaez M, Kramar E, Matheos DP, Arrizon A, Cabrera SM, Lynch G, Greene RW, Wood MA. Hippocampal focal knockout of CBP affects specific histone modifications, long-term potentiation, and long-term memory. Neuropsychopharmacology. 2011 doi: 10.1038/npp.2011.61. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaskara S, Knutson SK, Jiang G, Chandrasekharan MB, Wilson AJ, et al. Hdac3 is essential for the maintenance of chromatin structure and genome stability. Cancer Cell. 2010;18:436–447. doi: 10.1016/j.ccr.2010.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourtchouladze R, Lidge R, Catapano R, Stanley J, Gossweiler S, Romashko D, Scott R, Tully T. A mouse model of Rubinstein-Taybi syndrome: defective long-term memory is ameliorated by inhibitors of phosphodiesterase 4. Proceedings of the National Academy of Sciences U S A. 2003;100:10518–10522. doi: 10.1073/pnas.1834280100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredy TW, Barad M. The histone deacetylase inhibitor valproic acid enhances acquisition, extinction, and reconsolidation of conditioned fear. Learning and Memory. 2008;15:39–45. doi: 10.1101/lm.801108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broide RS, Redwine JM, Aftahi N, Young W, Bloom FE, Winrow CJ. Distribution of histone deacetylases 1–11 in the rat brain. Journal of Molecular Neuroscience. 2007;31:47–58. doi: 10.1007/BF02686117. [DOI] [PubMed] [Google Scholar]
- Chen G, Zou X, Watanabe H, van Deursen JM, Shen J. CREB binding protein is required for both short-term and long-term memory formation. Journal of Neuroscience. 2010;30:13066–13077. doi: 10.1523/JNEUROSCI.2378-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L-f, Fischle W, Verdin E, Greene WC. Duration of nuclear NF-kappaB action regulated by reversible acetylation. Science. 2001;293:1653–1657. doi: 10.1126/science.1062374. [DOI] [PubMed] [Google Scholar]
- Chuang HC, Chang CW, Chang GD, Yao TP, Chen H. Histone deacetylase 3 binds to and regulates the GCMa transcription factor. Nucleic Acids Research. 2006;34:1459–1469. doi: 10.1093/nar/gkl048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Codina A, Love JD, Li Y, Lazar MA, Neuhaus D, Schwabe JW. Structural insights into the interaction and activation of histone deacetylase 3 by nuclear receptor corepressors. Proceedings of the National Academy of Sciences U S A. 2005;102:6009–6014. doi: 10.1073/pnas.0500299102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowger JJ, Torchia J. Direct association between the CREB-binding protein (CBP) and nuclear receptor corepressor (N-CoR) Biochemistry. 2006;45:13150–13162. doi: 10.1021/bi060562g. [DOI] [PubMed] [Google Scholar]
- Dangond F, Hafler DA, Tong JK, Randall J, Kojima R, Utku N, Gullans SR. Differential display cloning of a novel human histone deacetylase (HDAC3) cDNA from PHA-activated immune cells. Biochemical and Biophysical Research Communications. 1998;242:648–652. doi: 10.1006/bbrc.1997.8033. [DOI] [PubMed] [Google Scholar]
- Emiliani S, Fischle W, Van Lint C, Al-Abed Y, Verdin E. Characterization of a human RPD3 ortholog, HDAC3. Proceedings of the National Academy of Sciences U S A. 1998;95:2795–2800. doi: 10.1073/pnas.95.6.2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eot-Houllier G, Fulcrand G, Watanabe Y, Magnaghi-Jaulin L, Jaulin C. Histone deacetylase 3 is required for centromeric H3K4 deacetylation and sister chromatid cohesion. Genes & Development. 2008;22:2639–2644. doi: 10.1101/gad.484108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fajas L, Egler V, Reiter R, Hansen J, Kristiansen K, Debril MB, Miard S, Auwerx J. The retinoblastoma-histone deacetylase 3 complex inhibits PPARgamma and adipocyte differentiation. Developmental Cell. 2002;3:903–910. doi: 10.1016/s1534-5807(02)00360-x. [DOI] [PubMed] [Google Scholar]
- Federman N, Fustiñana MS, Romano A. Histone acetylation is recruited in consolidation as a molecular feature of stronger memories. Learning and Memory. 2009;16:600–606. doi: 10.1101/lm.1537009. [DOI] [PubMed] [Google Scholar]
- Fischle W, Dequiedt F, Hendzel MJ, Guenther MG, Lazar MA, Voelter W, Verdin E. Enzymatic activity associated with class II HDACs is dependent on a multiprotein complex containing HDAC3 and SMRT/N-CoR. Molecular Cell. 2002;9:45–57. doi: 10.1016/s1097-2765(01)00429-4. [DOI] [PubMed] [Google Scholar]
- Fischle W, Dequiedt F, Fillion M, Hendzel MJ, Voelter W, Verdin E. Human HDAC7 histone deacetylase activity is associated with HDAC3 in vivo. Journal of Biological Chemistry. 2001;276:35826–35835. doi: 10.1074/jbc.M104935200. [DOI] [PubMed] [Google Scholar]
- Freudenthal R, Romano A. Participation of Rel/NF-kappaB transcription factors in long-term memory in the crab Chasmagnathus. Brain Research. 2000;855:274–281. doi: 10.1016/s0006-8993(99)02358-6. [DOI] [PubMed] [Google Scholar]
- Gao L, Cueto MA, Asselbergs F, Atadja P. Cloning and functional characterization of HDAC11, a novel member of the human histone deacetylase family. Journal of Biological Chemistry. 2002;277:25748–25755. doi: 10.1074/jbc.M111871200. [DOI] [PubMed] [Google Scholar]
- Grégoire S, Xiao L, Nie J, Zhang X, Xu M, Li J, Wong J, Seto E, Yang XJ. Histone deacetylase 3 interacts with and deacetylates myocyte enhancer factor 2. Molecular and Cellular Biology. 2007;27:1280–1295. doi: 10.1128/MCB.00882-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregoretti IV, Lee YM, Goodson HV. Molecular evolution of the histone deacetylase family: functional implications of phylogenetic analysis. Journal of Molecular Biology. 2004;338:17–31. doi: 10.1016/j.jmb.2004.02.006. [DOI] [PubMed] [Google Scholar]
- Guan JS, Haggarty SJ, Giacometti E, Dannenberg JH, Joseph N, Gao J, Nieland TJ, Zhou Y, Wang X, Mazitschek R, Bradner JE, DePinho RA, Jaenisch R, Tsai LH. HDAC2 negatively regulates memory formation and synaptic plasticity. Nature. 2009;459:55–60. doi: 10.1038/nature07925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Z, Giustetto M, Lomvardas S, Kim JH, Miniaci MC, Schwartz JH, Thanos D, Kandel ER. Integration of long-term-memory-related synaptic plasticity involves bidirectional regulation of gene expression and chromatin structure. Cell. 2002;111:483–493. doi: 10.1016/s0092-8674(02)01074-7. [DOI] [PubMed] [Google Scholar]
- Guenther MG, Yu J, Kao GD, Yen TJ, Lazar MA. Assembly of the SMRT- histone deacetylase 3 repression complex requires the TCP-1 ring complex. Genes & Development. 2002;16:3130–3135. doi: 10.1101/gad.1037502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther MG, Barak O, Lazar MA. The SMRT and N-CoR corepressors are activating cofactors for histone deacetylase 3. Molecular and Cellular Biology. 2001;21:6091–6101. doi: 10.1128/MCB.21.18.6091-6101.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haettig J, Stefanko DP, Multani ML, Figueroa DX, McQuown SC, Wood MA. HDAC inhibition modulates hippocampus-dependent long-term memory for object location in a CBP-dependent manner. Learning and Memory. 2011 doi: 10.1101/lm.1986911. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman HB, Yu J, Alenghat T, Ishizuka T, Lazar MA. The histone-binding code of nuclear receptor co-repressors matches the substrate specificity of histone deacetylase 3. EMBO Reports. 2005;6:445–451. doi: 10.1038/sj.embor.7400391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinzel T, Lavinsky RM, Mullen TM, Söderstrom M, Laherty CD, Torchia J, Yang WM, Brard G, Ngo SD, Davie JR, Seto E, Eisenman RN, Rose DW, Glass CK, Rosenfeld MG. A complex containing N-CoR, mSin3 and histone deacetylase mediates transcriptional repression. Nature. 1997;387:43–48. doi: 10.1038/387043a0. [DOI] [PubMed] [Google Scholar]
- Huang EY, Zhang J, Miska EA, Guenther MG, Kouzarides T, Lazar MA. Nuclear receptor corepressors partner with class II histone deacetylases in a Sin3- independent repression pathway. Genes & Development. 2000;14:45–54. [PMC free article] [PubMed] [Google Scholar]
- Jepsen K, Rosenfeld MG. Biological roles and mechanistic actions of co-repressor complexes. Journal of Cell Science. 2002;115:689–698. doi: 10.1242/jcs.115.4.689. [DOI] [PubMed] [Google Scholar]
- Jeyakumar M, Liu XF, Erdjument-Bromage H, Tempst P, Bagchi MK. Phosphorylation of thyroid hormone receptor-associated nuclear receptor corepressor holocomplex by the DNA-dependent protein kinase enhances its histone deacetylase activity. Journal of Biological Chemistry. 2007;282:9312–9322. doi: 10.1074/jbc.M609009200. [DOI] [PubMed] [Google Scholar]
- Johnson CA, White DA, Lavender JS, O'Neill LP, Turner BM. Human class I histone deacetylase complexes show enhanced catalytic activity in the presence of ATP and co-immunoprecipitate with the ATP-dependent chaperone protein Hsp70. Journal of Biological Chemistry. 2002;277:9590–9597. doi: 10.1074/jbc.M107942200. [DOI] [PubMed] [Google Scholar]
- Kilgore M, Miller CA, Fass DM, Hennig KM, Haggarty SJ, Sweatt JD, Rumbaugh G. Inhibitors of class 1 histone deacetylases reverse contextual memory deficits in a mouse model of Alzheimer's disease. Neuropsychopharmacology. 2010;35:870–880. doi: 10.1038/npp.2009.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korzus E, Rosenfeld MG, Mayford M. CBP histone acetyltransferase activity is a critical component of memory consolidation. Neuron. 2004;42:961–972. doi: 10.1016/j.neuron.2004.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Knutson SK, Chyla BJ, Amann JM, Bhaskara S, Huppert SS, Hiebert SW. Liver-specific deletion of histone deacetylase 3 disrupts metabolic transcriptional networks. EMBO Journal. 2008;27:1017–1028. doi: 10.1038/emboj.2008.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Choi KH, Renthal W, Tsankova NM, Theobald DE, Truong HT, Russo SJ, Laplant Q, Sasaki TS, Whistler KN, Neve RL, Self DW, Nestler EJ. Chromatin remodeling is a key mechanism underlying cocaine-induced plasticity in striatum. Neuron. 2005;48:303–314. doi: 10.1016/j.neuron.2005.09.023. [DOI] [PubMed] [Google Scholar]
- Lahm A, Paolini C, Pallaoro M, Nardi MC, Jones P, Neddermann P, Sambucini S, Bottomley MJ, Lo Surdo P, Carfi A, Koch U, De Francesco R, Steinkuhler C, Gallinari P. Unraveling the hidden catalytic activity of vertebrate class IIa histone deacetylases. Proceedings of the National Academy of Sciences U S A. 2007;104:17335–17340. doi: 10.1073/pnas.0706487104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammi J, Perlmann T, Aarnisalo P. Corepressor interaction differentiates the permissive and non-permissive retinoid X receptor heterodimers. Archives of Biochemistry and Biophysics. 2008;472:105–114. doi: 10.1016/j.abb.2008.02.003. [DOI] [PubMed] [Google Scholar]
- Lattal KM, Barrett RM, Wood MA. Systemic or intrahippocampal delivery of histone deacetylase inhibitors facilitates fear extinction. Behavioral Neuroscience. 2007;121:1125–1131. doi: 10.1037/0735-7044.121.5.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levenson JM, O'Riordan KJ, Brown KD, Trinh MA, Molfese DL, Sweatt JD. Regulation of histone acetylation during memory formation in the hippocampus. Journal of Biological Chemistry. 2004;279:40545–40559. doi: 10.1074/jbc.M402229200. [DOI] [PubMed] [Google Scholar]
- Li Y, Kao GD, Garcia BA, Shabanowitz J, Hunt DF, Qin J, Phelan C, Lazar MA. A novel histone deacetylase pathway regulates mitosis by modulating Aurora B kinase activity. Genes & Development. 2006;20:2566–2579. doi: 10.1101/gad.1455006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longworth MS, Laimins LA. Histone deacetylase 3 localizes to the plasma membrane and is a substrate of Src. Oncogene. 2006;25:4495–4500. doi: 10.1038/sj.onc.1209473. [DOI] [PubMed] [Google Scholar]
- Lutterbach B, Westendorf JJ, Linggi B, Patten A, Moniwa M, Davie JR, Huynh KD, Bardwell VJ, Lavinsky RM, Rosenfeld MG, Glass C, Seto E, Hiebert SW. ETO, a target of t(8;21) in acute leukemia, interacts with the N-CoR and mSin3 corepressors. Molecular and Cellular Biology. 1998;18:7176–7184. doi: 10.1128/mcb.18.12.7176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahlknecht U, Hoelzer D, Bucala R, Verdin E. Cloning and characterization of the murine histone deacetylase (HDAC3) Biochemical and Biophysical Research Communications. 1999;263:482–490. doi: 10.1006/bbrc.1999.1389. [DOI] [PubMed] [Google Scholar]
- Malvaez M, Sanchis-Segura C, Vo D, Lattal KM, Wood MA. Modulation of chromatin modification facilitates extinction of cocaine-induced conditioned place preference. Biological Psychiatry. 2010;67:36–43. doi: 10.1016/j.biopsych.2009.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQuown SC, Barrett RM, Matheos DP, Post RJ, Rogge GR, Alenghat T, Mullican SE, Jones S, Rusche JR, Lazar MA, Wood MA. HDAC3 is a critical negative regulator of long-term memory formation. Journal of Neuroscience. 2011 doi: 10.1523/JNEUROSCI.5052-10.2011. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlo E, Freudenthal R, Maldonado H, Romano A. Activation of the transcription factor NF-kappaB by retrieval is required for long-term memory reconsolidation. Learning and Memory. 2005;12:23–29. doi: 10.1101/lm.82705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oike Y, Hata A, Mamiya T, Kaname T, Noda Y, Suzuki M, Yasue H, Nabeshima T, Araki K, Yamamura K. Truncated CBP protein leads to classical Rubinstein- Taybi syndrome phenotypes in mice: implications for a dominant-negative mechanism. Human Molecular Genetics. 1999;8:387–396. doi: 10.1093/hmg/8.3.387. [DOI] [PubMed] [Google Scholar]
- Perissi V, Jepsen K, Glass CK, Rosenfeld MG. Deconstructing repression: evolving models of co-repressor action. Nature Reviews Genetics. 2010;11:109–123. doi: 10.1038/nrg2736. [DOI] [PubMed] [Google Scholar]
- Rai M, Soragni E, Chou CJ, Barnes G, Jones S, Rusche JR, Gottesfeld JM, Pandolfo M. Two new pimelic diphenylamide HDAC inhibitors induce sustained frataxin upregulation in cells from Friedreich's ataxia patients and in a mouse model. PLoS One. 2010;5:e8825. doi: 10.1371/journal.pone.0008825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renthal W, Maze I, Krishnan V, Covington HE, 3rd, Xiao G, Kumar A, Russo SJ, Graham A, Tsankova N, Kippin TE, Kerstetter KA, Neve RL, Haggarty SJ, McKinsey TA, Bassel-Duby R, Olson EN, Nestler EJ. Histone deacetylase 5 epigenetically controls behavioral adaptations to chronic emotional stimuli. Neuron. 2007;56:517–529. doi: 10.1016/j.neuron.2007.09.032. [DOI] [PubMed] [Google Scholar]
- Roozendaal B, Hernandez A, Cabrera SM, Hagewoud R, Malvaez M, Stefanko DP, Haettig J, Wood MA. Membrane-associated glucocorticoid activity is necessary for modulation of long-term memory via chromatin modification. Journal of Neuroscience. 2010;30:5037–5046. doi: 10.1523/JNEUROSCI.5717-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen S, Li J, Casaccia-Bonnefil P. Histone modifications affect timing of oligodendrocyte progenitor differentiation in the developing rat brain. Journal of Cell Biology. 2005;169:577–589. doi: 10.1083/jcb.200412101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanko DP, Barrett RM, Ly AR, Reolon GK, Wood MA. Modulation of long-term memory for object recognition via HDAC inhibition. Proceedings of the National Academy of Sciences U S A. 2009;106:9447–9452. doi: 10.1073/pnas.0903964106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takami Y, Nakayama T. N-terminal region, C-terminal region, nuclear export signal, and deacetylation activity of histone deacetylase-3 are essential for the viability of the DT40 chicken B cell line. Journal of Biological Chemistry. 2000;275:16191–16201. doi: 10.1074/jbc.M908066199. [DOI] [PubMed] [Google Scholar]
- Tong JJ, Liu J, Bertos NR, Yang XJ. Identification of HDAC10, a novel class II human histone deacetylase containing a leucine-rich domain. Nucleic Acids Research. 2002;30:1114–1123. doi: 10.1093/nar/30.5.1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vecsey CG, Hawk JD, Lattal KM, Stein JM, Fabian SA, Attner MA, Cabrera SM, McDonough CB, Brindle PK, Abel T, Wood MA. Histone deacetylase inhibitors enhance memory and synaptic plasticity via CREB:CBP-dependent transcriptional activation. Journal of Neuroscience. 2007;27:6128–6140. doi: 10.1523/JNEUROSCI.0296-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeulen M, Carrozza MJ, Lasonder E, Workman JL, Logie C, Stunnenberg HG. In vitro targeting reveals intrinsic histone tail specificity of the Sin3/histone deacetylase and N-CoR/SMRT corepressor complexes. Molecular and Cellular Biology. 2004;24:2364–2372. doi: 10.1128/MCB.24.6.2364-2372.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Zang C, Cui K, Schones DE, Barski A, Peng W, Zhao K. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell. 2009;138:1019–1031. doi: 10.1016/j.cell.2009.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen YD, Perissi V, Staszewski LM, Yang WM, Krones A, Glass CK, Rosenfeld MG, Seto E. The histone deacetylase-3 complex contains nuclear receptor corepressors. Proceedings of the National Academy of Sciences U S A. 2000;97:7202–7207. doi: 10.1073/pnas.97.13.7202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood MA, Attner MA, Oliveira AM, Brindle PK, Abel T. A transcription factor-binding domain of the coactivator CBP is essential for long-term memory and the expression of specific target genes. Learning and Memory. 2006;13:609–617. doi: 10.1101/lm.213906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood MA, Kaplan MP, Park A, Blanchard EJ, Oliveira AM, Lombardi TL, Abel T. Transgenic mice expressing a truncated form of CREB-binding protein (CBP) exhibit deficits in hippocampal synaptic plasticity and memory storage. Learning and Memory. 2005;12:111–119. doi: 10.1101/lm.86605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Soragni E, Chou CJ, Herman D, Plasterer HL, Rusche JR, Gottesfeld JM. Chemical probes identify a role for histone deacetylase 3 in Friedreich's ataxia gene silencing. Chemistry & Biology. 2009;16:980–989. doi: 10.1016/j.chembiol.2009.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang WM, Tsai SC, Wen YD, Fejer G, Seto E. Functional domains of histone deacetylase-3. Journal of Biological Chemistry. 2002;277:9447–9454. doi: 10.1074/jbc.M105993200. [DOI] [PubMed] [Google Scholar]
- Yang WM, Yao YL, Sun JM, Davie JR, Seto E. Isolation and characterization of cDNAs corresponding to an additional member of the human histone deacetylase gene family. Journal of Biological Chemistry. 1997;272:28001–28007. doi: 10.1074/jbc.272.44.28001. [DOI] [PubMed] [Google Scholar]
- Yin L, Lazar MA. The orphan nuclear receptor Rev-erbalpha recruits the N- CoR/histone deacetylase 3 corepressor to regulate the circadian Bmal1 gene. Molecular Endocrinology. 2005;19:1452–1459. doi: 10.1210/me.2005-0057. [DOI] [PubMed] [Google Scholar]
- Yoo JY, Choi KC, Kang H, Kim YJ, Lee J, Jun WJ, Kim MJ, Lee YH, Lee OH, Yoon HG. Histone deacetylase 3 is selectively involved in L3MBTL2- mediated transcriptional repression. FEBS Letters. 2010;584:2225–2230. doi: 10.1016/j.febslet.2010.03.048. [DOI] [PubMed] [Google Scholar]
- Zeng L, Zhou MM. Bromodomain: an acetyl-lysine binding domain. FEBS Letters. 2002;513:124–128. doi: 10.1016/s0014-5793(01)03309-9. [DOI] [PubMed] [Google Scholar]
- Zhang X, Ozawa Y, Lee H, Wen YD, Tan TH, Wadzinski BE, Seto E. Histone deacetylase 3 (HDAC3) activity is regulated by interaction with protein serine/threonine phosphatase 4. Genes & Development. 2005;19:827–839. doi: 10.1101/gad.1286005. [DOI] [PMC free article] [PubMed] [Google Scholar]