Abstract
Purpose
Lung cancer remains the leading cause of cancer death in the United States. Central to the lung-cancer diagnosis and staging process is the assessment of the central-chest lymph nodes. This assessment requires two steps: (1) examination of the lymph-node stations and identification of diagnostically important nodes in a three-dimensional (3D) multidetector computed tomography (MDCT) chest scan; (2) tissue sampling of the identified nodes. We describe a computer-based system for automatically defining the central-chest lymph-node stations in a 3D MDCT chest scan.
Methods
Automated methods first construct a 3D chest model, consisting of the airway tree, aorta, pulmonary artery, and other anatomical structures. Subsequent automated analysis then defines the 3D regional nodal stations, as specified by the internationally standardized TNM lung-cancer staging system. This analysis involves extracting over 140 pertinent anatomical landmarks from structures contained in the 3D chest model. Next, the physician uses data mining tools within the system to interactively select diagnostically important lymph nodes contained in the regional nodal stations.
Results
Results from a ground-truth database of unlabeled lymph nodes identified in 32 MDCT scans verify the system’s performance. The system automatically defined 3D regional nodal stations that correctly labeled 96% of the database’s lymph nodes, with 93% of the stations correctly labeling 100% of their constituent nodes.
Conclusions
The system accurately defines the regional nodal stations in a given high-resolution 3D MDCT chest scan and eases a physician’s burden for analyzing a given MDCT scan for lymph-node station assessment. It also shows potential as an aid for preplanning lung-cancer staging procedures.
Keywords: Lymph node, Lung cancer, MDCT, 3D imaging, Thoracic imaging, Regional nodal station
Introduction
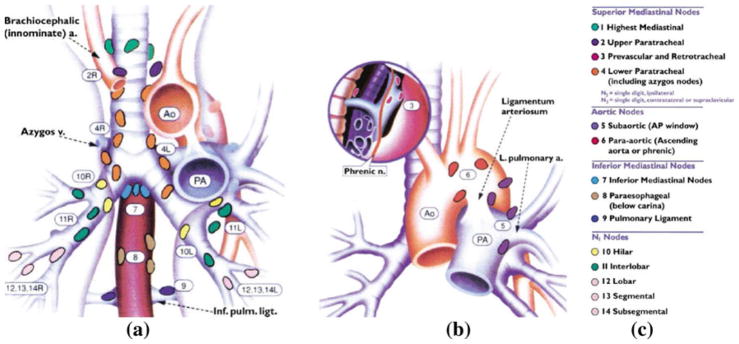
Lung cancer remains the leading cause of cancer death in the United States [1]. For disease diagnosis and staging, physicians rely on the internationally standardized TNM system for selecting suspicious anatomical sites, where T denotes tumor, N denotes central-chest lymph nodes, and M represents distant metastases [2–8]. In particular, the TNM system describes 14 distinct lymph-node stations within the chest, where anatomical and geometric landmarks define the extent and location of each station (Fig. 1; Table 1) [2–4].
Fig. 1.
Standard TNM regional nodal station map (reproduced by permission of the American College of Chest Physicians) [2,3]. a, b Complementary anatomical maps for the 14 TNM regional nodal stations, where a gives a coronal view of most of the stations, while b focuses on stations 3, 5, and 6. c Legend indicating region names and color codes for the TNM stations
Table 1.
Verbatim excerpt of the TNM system’s anatomical criteria for defining regional nodal stations 2, 5, 7, and 13 (reproduced by permission of the American College of Chest Physicians) [2,3]
| Station | Anatomical criteria |
|---|---|
| 2 | Nodes lying above a horizontal line drawn tangential to the upper margin of the aortic arch and below the inferior boundary of station 1 nodes |
| 5 | Subaortic nodes are lateral to the ligamentum arteriosum or the aorta or left pulmonary artery and proximal to the first branch of the left pulmonary artery and lie within the mediastinal pleural envelope |
| 7 | Nodes lying caudal to the carina of the trachea but not associated with the lower lobe bronchi or arteries within the lung |
| 13 | Nodes adjacent to the segmental bronchi |
The complete set describes 14 stations
Using the TNM station-definition criteria as a mental guide, the physician visually searches each station by manually scrolling through the two-dimensional (2D) sections constituting a patient’s three-dimensional (3D) multidetector computed tomography (MDCT) chest scan [4,9,10]; during this search, the physician identifies diagnostically pertinent lymph nodes, if any, contained in the stations. Later, the physician performs tissue sampling on the identified nodes, using a technique such as bronchoscopy, to make a final determination on disease diagnosis and stage [8,11,12]. Unfortunately, the TNM station criteria are complex and open to interpretation, and the direct application of the criteria to a typical 3D MDCT chest scan can be time-consuming. We describe a system for the automatic definition of the central-chest lymph-node stations from 3D MDCT scans.
Modern MDCT scanners produce high-resolution 3D volumetric images that depict an extraordinary level of 3D anatomical detail [13–16]. When considering the pulmonary lymph nodes, the physician need only focus attention on the small volumetric regions associated with the nodal stations. Cymbalista et al. were the first to propose MDCT-based station-definition guidelines, but only gave manually derived anecdotal results on 2D transverse-plane sections for a single MDCT scan [4]. A preliminary method has been proposed for assigning station labels to predefined lymph nodes in an MDCT scan, but it does not give 3D regional nodal station definitions and only provides labels for subgroupings of the TNM stations [17]. Ko et al. and Chapet et al. made more detailed efforts to identify the regional nodal stations depicted in MDCT images [9,10]. They defined the stations using manual region outlining on 2D transverse-plane sections. This manual approach, however, is not practical for general use, as it is extremely time-consuming and does not give reproducible results. In addition, it is dependent on a physician’s skill in translating the TNM anatomical criteria to series of 2D sections comprising a 3D MDCT scan.
Given the sheer volume of current 3D MDCT scans, it is well acknowledged that image assessment is overwhelming in general [14,15]. This is clearly an issue for TNM station assessment, as the limitations of recent work make it difficult to exploit the inherent detail of a given scan and don’t enable the long-term possibility of performing comprehensive lymph-node mapping and staging. Therefore, it is clear that a computer-based approach could be helpful for addressing these issues.
Our proposed computer-based system, referred as the Lymph-Node Station Mapper, performs fully automatic 3D regional nodal station definition. Given a patient’s 3D MDCT chest scan, the system first constructs a 3D chest model, consisting of the airway tree, aorta, lungs, and other anatomical structures. Next, drawing upon the TNM system’s station-definition criteria, further analysis extracts pertinent landmarks from structures constituting the chest model; these landmarks delineate the final 3D TNM regional nodal stations. Following this automated analysis, the system provides interactive tools that enable a physician to perform efficient station examination and semi-automatic selection of diagnostically relevant lymph nodes in the stations. The system’s outputs can then interface to a system we have been devising for the guidance of follow-on bronchoscopy-based tissue sampling [18–22].
“Methods” describes the system in detail. “Results” demonstrate the system’s efficacy for accurately defining the 3D TNM stations, with validation results derived from a large database of human 3D MDCT scans. Finally, “Discussion” offers concluding comments.
Methods
The Lymph Node Station Mapper (LNSM) is a fully integrated computer-based system. It consists of a set of automatic image-processing tools and a graphical user interface (GUI). The automatic tools perform 3D operations leading to TNM station definition, while the GUI facilitates all system interactions, visualization, and data mining. The primary input is a patient’s 3D MDCT chest scan, while the primary outputs are the 3D TNM regional nodal stations. The system also provides the option of semi-automatically segmenting lymph nodes of interest observed in the defined stations.
Let I denote the input 3D MDCT chest scan and I (x, y, z) refer to the intensity value of voxel (x, y, z). We assume that the x dimension increases laterally from right to left, the y dimension increases from anterior (front) to posterior (back), and the z dimension increases from top (superior) to bottom (inferior). Furthermore, we assume the standard chest-scanning protocol for lung-cancer patients, whereby a patient lies roughly horizontal on the scanner table during a scan. With this convention, the trachea, sternum, and spine lie approximately parallel to the z axis and orthogonal to the lateral (x and y) directions in 2D transverse-plane sections.
For our work, we use high-resolution 3D MDCT scans having characteristics similar to those used by other MDCT chest-imaging researchers [14–16]. Because lymph nodes can have long-axis length <10 mm, high resolution is essential. In addition, the near isotropic voxel resolution we use is more suitable for analyzing the chest anatomy, as anatomical structures don’t strictly conform to orthogonal planes [16]. Furthermore, we wish to exploit state-of-the-art chest MDCT, so as to better realize a long-term research goal of exhaustive central-chest lymph-node staging. Thus, the typical input MDCT scan we use consists of several hundred 2D transverse-plane sections, with section spacing 0.5 mm, section thickness 0.75 mm, and transverse-plane resolution Δx = Δy ≈ 0.6 mm (Sect. 3). Also, as is an industry standard for MDCT scanners, voxel intensity values range from −1,000 Hounsfield Units (HU) for pure air, 0 HU for water (soft tissue: [−200 HU, 200 HU]), and +1,000 HU for pure bone [23]. Finally, the LNSM can draw upon scans produced with or without contrast agent.
The general procedure for applying the LNSM consists of three stages:
3D Chest-Model Computation—Automatic image-processing operations extract a set of chest structures from I that are necessary for delineating the TNM stations and for later interactive visualization.
Station Definition—Using the 3D chest model, automatic operations define a set of anatomical landmarks and then use the landmarks to define a set of 3D regions delineating the regional nodal stations.
User Interaction—The user employs interactive tools to examine the 3D TNM stations defined for I. During this examination, the user has the option of identifying diagnostically important lymph nodes within the stations. The system saves all station definitions and selected lymph nodes for follow-on interventional procedure planning.
The LNSM applies the TNM system’s anatomical criteria (Fig. 1; Table 1) to input 3D MDCT scan I by translating the criteria into automatic image-processing operations. These operations follow two stages. During stage 1, key chest structures, such as the airway tree, aorta, etc., are automatically defined to give a 3D chest model. Next, during stage 2, further operations define pertinent anatomical landmarks from 3D chest-model structures, which in turn enable a straightforward construction of 3D volumetric regions delineating the various regional nodal stations. The third stage involves focused user interaction. We emphasize that the LNSM is not a computer-aided detection system for locating candidate central-chest lymph nodes. Rather, its main functions are to automatically define the TNM regional nodal stations and to enable subsequent focused examination of an MDCT scan within the subregions of the computed stations.
The sections “3D chest-model computation” to “User Interaction” further describe the three stages, while “Implementation comments” highlights miscellaneous implementation issues. Reference [24] gives complete system detail.
3D chest-model computation
Stage 1 in the analysis of a given 3D MDCT chest scan I involves the automatic computation of a 3D chest model. This computation involves the following steps:
-
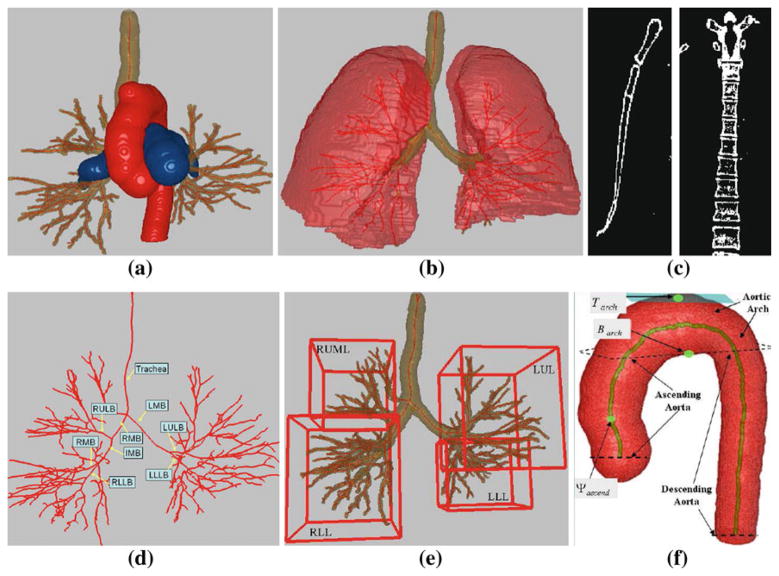
Segment each anatomical chest structure listed in the following set (Fig. 2a–c, f):
(1) where ascend, descend, and arch denote the ascending aorta, descending aorta, and aortic arch, respectively.
Use the airway-tree segmentation to define the airway endoluminal surfaces and centerlines, and use the segmented aorta to obtain the centerline passing through the three parts of the aorta (Fig. 2d, f).
-
Assigns labels to the following set of major airway branches (Fig. 2d):
(2) where RMB = right main bronchus, LMB = left main bronchus, IMB = intermediate bronchus, RULB = right upper lobar bronchus, RMLB = right middle lobar bronchus, etc.
Use the airway-tree segmentation, centerlines, surfaces, and labels to isolate the segmented substructure corresponding to each labeled airway in (2). To do this for a particular airway, we search orthogonally about the air-way’s centerline to locate that portion of the airway-tree surface corresponding to the airway’s region.
-
Delineate each lung lobe structure in the following set (Fig. 2e):
(3) where RUML = union of right upper and right middle lobes, RLL = right lower lobe, LUL = left upper lobe, and LLL = left lower lobe. As an example, the system delineates the RLL by grouping together the RLLB (the RLL’s parent lobar airway) and all of its child airways to isolate the airway subtree constituting the RLL. This is easily done, since labeled set (2) identifies the RLLB, while the child airways arise from the centerlines and corresponding segmented airway regions connected to the RLLB. The other lobar structures are found similarly. Note that, in light of the TNM specifications of stations 12 through 14, we only need a union of two of the right lung’s lobes, RUML.
Fig. 2.
Example structures constituting the 3D chest model. This figure, along with Figs. 3, 4, 5, 6, 7 and 8a are based on MDCT scan 20349_3_3 (578 512×512 transverse-plane sections, resolution Δx = Δy = 0.72 mm and Δz = 0.5 mm). a 3D airway tree (brown), aorta (red), and PA (blue). b Lungs (red) with airway tree (brown) and airway centerlines (red). c Example coronal 2D section of segmented sternum (left) and example sagittal 2D section of segmented spine (right). d Airway-tree centerlines and major-airway labels per (2). e MBCs of four major lung lobar regions per (3). f Aorta, its three constituent parts (ascending aorta, aortic arch, and descending aorta), and centerline (brown); Tarch, Barch, and Ψascend, are landmarks used for station definition (Sect. 2.2)
The final 3D chest model consists of the segmented structures in sets (1) and (3), associated MBCs per (4–6), labeled airways (2), airway/aorta centerlines, and airway endoluminal surfaces.
The analysis above draws upon an extensive set of our group’s previously validated methods for 3D MDCT pulmonary image analysis [20–22,24–32]. In particular, for the set (1), we compute the segmented airway tree using the methods of [22,25], we compute the three sections of the aorta, aorta centerline, and PA using the methods of [27,28], and methods for segmenting the lungs, sternum, and spine appear in [22,24]. References [20,21] describe the methods for airway endoluminal surface definition, while [29] describes the centerline method. Finally, we determine the airway branch labels of set (2) via the methods of [30–32]. We point out that these methods have been extensively validated on a wide range of 3D MDCT chest scans produced by variety of scanners from Siemens, Philips, and Imatron. These methods have been used successfully in our recent human studies on the planning and guidance of bronchoscopy [18–20].
Note that many researchers have presented recent applications that also successfully draw upon a coordinated series of automatic methods to define multiple chest structures depicted in a 3D CT image [33–37]. Hence, specific automatic methods devised by others could conceivably be substituted for the ones we employ.
Station definition
The second stage of analysis for input 3D MDCT scan I involves automatic definition of the 3D regions delineating each TNM regional nodal station.
To define the station regions, the LNSM first extracts a set of anatomical landmarks from structures contained in the 3D chest model. The landmarks in turn then help specify a set of geometric limits defining each station. Each defined station consists of one or more 3D cuboidal regions, with six limits similar to (4–6) delineating each cuboid. The LNSM does not produce curved or irregularly shaped regions. But, as described below, four stations consist of two or more cuboids. Cuboids well represent the desired anatomical zones. They are also especially suitable for user-friendly interaction, as their limits conform precisely to 2D transverse (x–y), coronal (x–z), and sagittal (y–z) multiplanar reformatted (MPR) views [15,16], and, in fact, many of the TNM station criteria also conform to these standard orthogonal viewing orientations.
The station definitions and landmark set we use draw extensively on the criteria specified by the TNM standard (Fig. 1 and Table 1) and by the attempts of past researchers to interpret these criteria in MDCT scans [2–4,9,10]. It is well known that the anatomical criteria given by the TNM system are somewhat vague, open to interpretation, and do not specify numerically precise 3D regions. Instead, the criteria define general region vicinities and fortunately admit considerable latitude and flexibility in station definition.
In addition, researchers have noted that the TNM system criteria do not always readily translate into structures observed in MDCT images [4,9,10]. Thus, in line with the suggestions of these researchers, we made compromises in defining certain stations and landmarks. Finally, we again point out that no one before has attempted to translate the TNM regional nodal station criteria into 3D station regions as depicted in MDCT chest scans, let alone produce a complete automated system for defining the stations.
During our conception of the LNSM, a pulmonologist, chest radiologist, and thoracic surgeon (all senior faculty of our University’s medical school) worked in concert with two imaging scientists to define and confirm the station definitions and landmark set. Following the lead of Chapet et al., we grouped stations 1 and 2 together, because the inferior limit of the highest mediastinal region (station 1), which also equals the superior limit of the upper paratracheal region (station 2), tends to be unclear and varies among patients (see Fig. 1) [10]. Following the studies of Ko et al. and Chapet et al. we also grouped the hilar and interlobar stations (10 and 11) together, because their anatomical definitions result in substantial overlap; also, no explicit boundary appears between them in 3D MDCT images [9,10]. Finally, we grouped the lobar, segmental, and subsegmental stations (12, 13, 14) together, because Chapet et al. stated that these stations cannot be isolated separately as they overlap each other—in fact, they disregarded them in their work [10]. In addition, stations 12 through 14, which are situated in the lung periphery, contain very few lymph nodes relative to the other stations [38]. Hence, the LNSM automatically defines 10 distinct stations.
Before giving the definitions of the stations, we first define the notation denoting the landmarks and then discuss how the LNSM derives the landmarks. For a specified 3D chest-model structure in (1–3), a quantity of the form
| (7) |
refers to either a z, x, or y MBC limit of the structure per (4–6), where T and B specify a z (top/bottom) limit, R and L specify an x (right/left) limit, and A and P specify a y (anterior/posterior) limit, relative to the 3D space of I. As examples, R RMB refers to the right x -coordinate limit xmin of the MBC enclosing the right main bronchus, while Tsternum specifies the top (zmin) of the sternum’s MBC, also referred to as the sternal notch. Per the TNM system criteria, many landmarks specifying right-to-left or anterior-to-posterior limits—i.e., those involving R, L, A, or P in (7)—only consider a structure’s limits over a range of 2D transverse-plane sections of I, I (·, ·, z), i ≤ z ≤ j. Furthermore, when restricted in this way, a quantity can have a minimum or maximum value over the range of sections considered. As an example (Table 3), the posterior limit of the cuboid for station 1–2 is defined as
Table 3.
Limits delineating stations 1–2 through 4
| Limit | 1–2 | 3S | 3I | 4S | 4IR | 4IL | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Top | u | u | v | v | b | c | ||||
| Bottom | v | v | γ | δ | c | b | ||||
| Right | w | w | w | w | θ | w | ||||
| Left | α | α | α | α | α | θ | ||||
| Anterior | β |
|
|
|
o | n | ||||
| Posterior |
|
|
|
|
s | r |
Station 3 consists of two cuboids, 3S and 3I, located superior and inferior to the aortic arch, respectively. Station 4 consists of two cuboids: a superior cuboid 4S and a smaller cuboid, 4IR or 4IL, that shares its superior face with a part of the inferior face delineating 4S. If BRMB ≤ BLMB (i.e., b ≤ c per Table 2), then 4IR completes station 4; otherwise, 4IL completes station 4
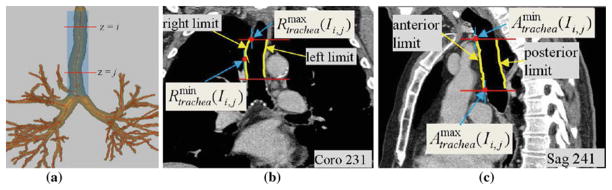
which gives the maximum y value of any point constituting the posterior border of the trachea over 2D transverse sections I (·, ·, z), u ≤ z ≤ v. Figure 3 further illustrates how such landmarks are defined and derived. T and B always denote minimum and maximum z limits, respectively, and are never qualified over a section range.
Fig. 3.
Example landmark definitions for the right, left, anterior, and posterior limits of a structure computed over a range of 2D transverse-plane sections I (·, ·, z), i ≤ z ≤ j. This example focuses on the trachea. All 2D section views in this figure and the figures to follow use the mediastinal window [window width = 400, window level = −160] for display, unless otherwise stated. a Depiction of transverse-plane sections passing through the trachea between z = i and z = j. b 2D coronal-plane section I (x, ·, ·) at x = 231 depicting the right and left surfaces of the trachea between i ≤ z ≤ j (yellow); gives the minimum x value of the right tracheal surface points (middle red dot), while gives the maximum x value of the right tratrachea surface points (upper red dot). c 2D sagittal-plane section I (·, y, ·) at y = 241 depicting the anterior and posterior surfaces of the trachea between i ≤ z ≤ j (yellow); gives the minimum y value of the anterior tracheal surface points (upper red dot), while gives the maximum y value of the anterior tracheal surface points (lower red dot)
Several landmarks draw exclusively on structure centerlines. We represent such landmarks with the function Ψ, where, more specifically, Ψstructure(Ia) denotes the 3D (x,y,z) coordinates of the centerline point of structure intersecting 2D transverse-plane section I (·, ·, a). Again, Ψ is sometimes considered over a range of 2D transverse-plane sections of I. Furthermore, only one coordinate is actually retained to define a landmark. For example (Table 3), the anterior (y) limit of cuboid 4S defining a portion of station 4, denoted by
gives the minimum y coordinate of the ascending aorta’s centerline points between 2D transverse-plane sections I (·, ·, z), v ≤ z ≤ δ. Several landmarks relate to special chest locations:
Used for stations 3, 7, and 8, carina[z] refers the z-coordinate location of the main carina. We define this location as the first 2D transverse-plane section I (·, ·, z) when the airway tree bifurcates from one airway cross-section (trachea) into two separate airway cross-sections (RMB and LMB). This is easily found by scanning the airway-tree segmentation in the vicinity of the trachea, a known labeled airway per (2).
Used by several stations, RootR [x] and RootL [x] denote the lateral (x) locations of the right and left lung roots. RootR [x] is given by the x-coordinate of the 3D location where the centerline of the right main bronchus ΨRMB intersects the surface of the right lung. An analogous definition applies to RootL [x], using ΨLMB and the left lung.
Station 6 must extend beyond the aorta. Thus, following Chapet et al.’s suggestion, we extend the cuboid given in Table 4 by 1 cm in x in the right and left directions and by 1 cm in y in the anterior direction to give the final form of station 6.
-
For stations 8 and 9, Tabdomen defines the approximate beginning of the abdomen. We use the following method to calculate Tabdomen. For each 2D transverse-plane section I (·, ·, z) below z = Tlungs (the top of the lungs), do the following: (a) define the minimum bounding box enclosing both segmented lungs; (b) locate the 2D centroid (xc, yc) of this box; (c) subdivide the box into anterior and posterior sections, using the line y = yc to form the subdivision — this line defines approximate subdivisions of the right and left lungs into anterior and posterior regions on section I (·, ·, z); (d) if
(8) for either the right or left lung, then Tabdomen = z. Relation (8) gives a reasonable approximation to the effective beginning of the abdomen, because the lungs lie superior to the abdomen and largely anterior to abdomen’s base; hence, the inferior regions of the lungs taper more rapidly in the posterior chest region as z increases.
Table 4.
Limits delineating stations 5 through 7
| Limit | 5 | 6 | 7 | ||
|---|---|---|---|---|---|
| Top | λ | v | γ | ||
| Bottom | μ | σ | min (c, d) | ||
| Right |
|
|
f | ||
| Left |
|
|
i | ||
| Anterior |
|
|
min (l, m) | ||
| Posterior |
|
|
max (p, q) |
Following Chapet et al. the final station-6 region spans a 3D region extended 1 cm laterally both left and right and also extended 1 cm anteriorly from the given table limits [10]
Finally, many landmarks are given by the minimum or maximum value of two or more intermediate landmarks. For example (Table 2), u = max(Tlungs, Tsternum) returns the maximum z value of its two arguments.
Table 2.
Landmarks used by multiple station limits
| (a) Landmarks based only on the segmented airways | |||||||||
| a = TLULB | b = BRMB | c = BLMB | d = BIMB | e = BLLLB | |||||
|
|
|
|
|
|
|||||
|
|
|
|
|
|
|||||
|
|
|
|
|
|
|||||
| (b) Landmarks based on other structures and compound landmarks | |||||||||
| u = max(Tlungs, Tsternum) | v = Tarch | w = RootR [x ] | α = RootL [x] |
|
|||||
| γ = carina[z] | δ = min(b, c) | θ = Ψtrachea(Iγ) [x] | λ = Barch | μ = min (LPA [z], a) | |||||
|
|
φ = Tabdomen | ψ = Blungs | |||||||
The large majority of landmarks are straightforward to define, as many tend to be simple limits of 3D chest-model structures. They also tend to be largely data independent, as their definitions arise strictly from the anatomical relationships between structures contained in the 3D chest model. We do acknowledge that there is some dependence on the efficacy of the automatic methods used to compute the chest model, but, as stated earlier, our methods have been shown to be robust over a wide range of human MDCT scans. In addition, it is important to realize that the actual “true” definitions of the regional nodal stations is open to considerable latitude, given the leeway allowed by the TNM criteria. Instead, what is important is the definition of suitable 3D focal regions for later MDCT scan examination. Thus, no clear notion of optimality exists for defining the stations.
Tables 2, 3, 4, 5, 6 give the required landmarks and station definitions. The 10 unique TNM regional nodal stations produced by the LNSM consist of 17 different cuboids, where stations 3, 4, 10–11, and 12–14 consist of two or more disjoint cuboids. The definitions of the cuboids require 102 limits to delineate. Since two or more stations share 21 limits, the system requires only 81 unique cuboid limits. To specify these limits, the LNSM computes 143 different landmarks. Of these, 88 depend on only one structure in (1–3), while 55 depend on multiple chest-model structures. Furthermore, 118 landmarks depend on the airways (2), 17 on the aorta, 7 on the PA, 9 on the lungs, 4 on the spine, and 4 on the sternum. Per Table 2, 33 landmarks occur multiple times in the specifications of other landmarks or limits.
Table 5.
Limits defining stations 8 through 10–11
| Limit | 8 | 9 | 10–11R | 10–11L | |||
|---|---|---|---|---|---|---|---|
| Top | γ | φ | min (TRMB, TRULB) | min (TLMB, a) | |||
| Bottom | φ | ψ | d | e | |||
| Right | f | h |
|
|
|||
| Left | i | k |
|
max (j, k) | |||
| Anterior |
|
Ψtrachea (IBtrachea) [y] | min (l, n) | min (m, o) | |||
| Posterior |
|
|
|
max (q, t) |
Station 10–11 consists of two cuboids, 10–11R and 10–11L, focused on the right and left hilar/interlobar regions, respectively. Refer to Sect. 2 for the definition of Tabdomen
Table 6.
Limits for station 12–14
| Limit | 12–14 RU | 12–14 RL | 12–14 LU | 12–14 LL | ||||
|---|---|---|---|---|---|---|---|---|
| Top | TRUML | min (TRLL, TRLLB) | min (TLUL, a) | min (TLLL, TLLLB) | ||||
| Bottom | BRUML | min (BRLL, BRLLB) | min (BLUL, BLULB) | min (BLLL, e) | ||||
| Right |
|
|
|
|
||||
| Left |
|
|
|
|
||||
| Anterior |
|
|
|
|
||||
| Posterior |
|
|
|
|
This station consists of four cuboids, one for each major lobar region per (3). Thus, 12–14 RU focuses on the right middle/upper lobar region, 12–14 RL focuses on the right lower lobar region, etc.
Figures 4, 5 illustrate example stations and their associated limits, while Fig. 6 gives an additional station example. Referring to Fig. 1, the 3D regions constituting the stations roughly progress in numerical order from superior to inferior, with station 1–2 situated most superior in the chest. As is inherent to the TNM criteria and as can be seen from Tables 3, 4, 5, and 6, many stations share top and bottom (z-direction) transverse-plane boundaries:
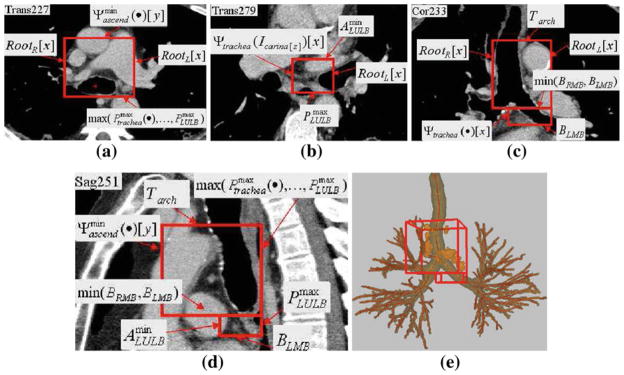
Fig. 4.
Example of station 4, as computed automatically by the LNSM. The red rectangular boxes indicate the location of the station in each view. Labels indicate the landmarks defining the limits of the two cuboids defining station 4, per Table 3. Dimensions of 4S: 65 mm × 70 mm × 63 mm; 4I: 37 mm × 31 mm × 13 mm. a Portion of 2D x–y transverse-plane section I (·, ·, 227) passing through cuboid 4S. b Portion of 2D x–y transverse-plane section I (·, ·, 279) passing through cuboid 4IR. c Portion of 2D x–z coronal-plane section I (·, 233, ·) passing through both adjoining cuboids of station 4. d Portion of 2D y–z sagittal-plane section I (251, ·, ·) passing through both adjoining cuboids of station 4. e 3D surface rendering of station 4 with respect to the airway tree, with complete set of lymph nodes depicted in orange per TNM system convention (Fig. 1)
Fig. 5.
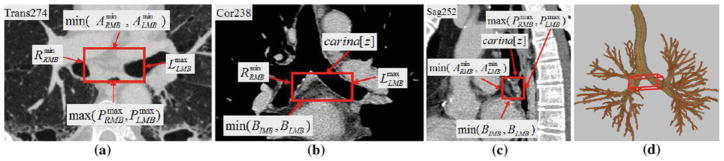
Example of station 7, as defined automatically by the LNSM. All 2D section views use the mediastinal window except a, which uses the lung window [window width = 1,600, window level = −1150]. The red rectangular boxes indicate the location of the station in each view. Labels indicate the landmarks defining the limits of the cuboid defining station 7, per Table 4. Dimensions of cuboid: 57 mm×24 mm × 21 mm. a Portion of transverse-plane section I (·, ·, 274). b Portion of coronal-plane section I (·, 238, ·). c Portion of sagittal-plane section I (252, ·, ·). d 3D surface rendering of station 7 with respect to the airway tree
Fig. 6.
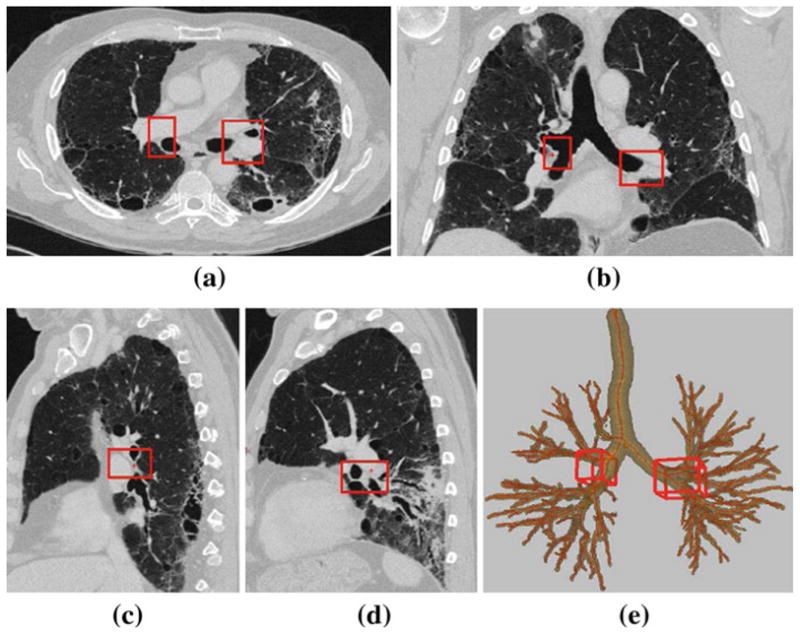
Example of station 10–11, as computed automatically by the LNSM per Table 5; this station consists of two cuboid regions, 10–11R and 10–11L, as highlighted by the red boxes. 2D section views displayed using the lung window. Dimensions of 10–11R: 22 mm × 39 mm × 24 mm. 10–11L: 36 mm × 44 mm × 27 mm. a Transverse-plane section I (·, ·, 266). b Coronal-plane section I (·, 238, ·). c Sagittal-plane section I (204, ·, ·) passing through cuboid 10–11R in the right lung. d Sagittal-plane section I (315, ·, ·) passing through cuboid 10–11L in the left lung. e 3D surface rendering of station 10–11 with respect to the airway tree
Top and bottom of station 1–2 = top and bottom of cuboid 3S for station 3.
For station 3, bottom of cuboid 3S = top of cuboid 3I.
Bottom of cuboid 3S = tops of stations 4 and 6.
Bottom of cuboid 3I = tops of stations 7 and 8.
Bottom of station 8 = top of station 9.
Bottom for stations 4 and 7 ≈ top of station 10–11.
Finally, because of the ingrained impreciseness of the TNM system and the complexity of the 3D chest anatomy, the stations in general do not occupy mutually exclusive zones, and overlaps occur between the following pairs of stations: 1–2 and 3 (with cuboid 3S); 4 and 7; 5 and 6; and 10–11 and 12–14. References [24,39] present a complete pictorial set of station examples, similar to Figs. 4, 5, 6, and give more complete insight into the anatomical considerations of the stations.
User Interaction
After automatic 3D chest-model computation and lymph-node station definition, the user next interacts with I and the computed results through the extensive set of tools integrated into the GUI of the LNSM. User interaction entails two tasks:
Visualization of the defined stations through various graphical manifestations of the 3D MDCT image data and chest-model structures.
Optional semi-automatic identification of diagnostically important lymph nodes.
Upon invocation of interactive visualization, the LNSM GUI presents the following (Fig. 7): (a) the TNM station map and legend (Fig. 1); (b) a flexible display window for visualization; and (c) various controls for display manipulation, semi-automatic lymph-node segmentation, and other functions.
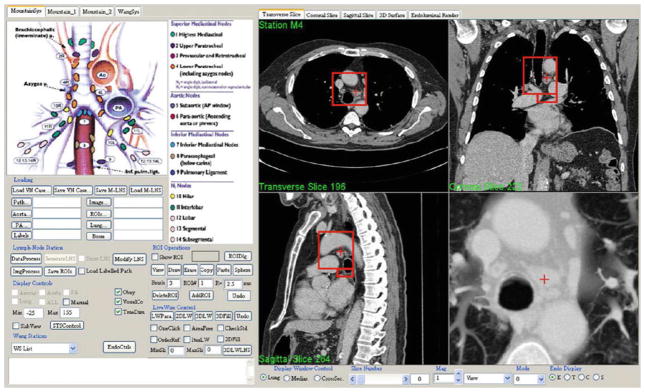
Fig. 7.
Top-level form of LNSM GUI. The user selects a desired computed TNM station by pointing to the station on the TNM system legend (top left center). Many controls exist for interacting with the stations and selecting/defining lymph nodes of interest (bottom left). The right side of the GUI is dedicated to visualization, which generally depicts four linked interactive views. In this example, transverse section z = 196, coronal section x = 222, sagittal section y = 264, and a magnified thin slab view of station 4 appears. The red boxes indicate the 3D location of the station on the presented sections, while the red crosses indicate the current 3D viewing position
To begin station visualization, the user points to a station in the GUI’s TNM legend—this activates the selected station, and views corresponding to the station appear in the display. As a default, the display depicts the 2D transverse, sagittal, and coronal sections of input MDCT scan I situated at the station’s centroid (right side of Fig. 7). Other viewers are available as options: (a) magnified sliding thin slab, which is especially revealing for internal soft-tissue structures (Fig. 7) [15,40]; (b) magnified 2D section; (c) virtual bronchoscopy (VB) endoluminal rendering [15,16,41]; (d) 3D surface rendering of the segmented airway tree and optional views of the aorta and PA (e.g., Fig. 2a, e).
In all views, the active station’s region appears as red boxes and the global 3D viewing position appears as a cross. To examine data within the station, the user selects a master viewer for focusing exploration and then navigates through the viewer’s data in 3D, either by scrolling through 2D sections in the master viewer’s orientation (transverse, sagittal, or coronal) or by traveling through the airways if the VB viewer serves as the master. During station exploration, all other viewers follow the master viewer in synchrony, abiding by the system’s global 3D viewing position established by the master. Because the stations consist of one or more cuboidal regions oriented parallel to the 3D coordinate axes of I, data exploration is convenient and user friendly, as it conforms well to the natural 2D orthogonal section-viewing mode to which physicians have grown accustomed [15,16].
During station exploration, the user can select lymph nodes deemed to have diagnostic significance for follow-on tissue sampling. Each selected node receives a station label, as denoted by the currently active station. The label takes the form of both a number and a color assignment, following the TNM system conventions (Fig. 1c); the colors are useful for visualization. For example, station 4 nodes are all colored orange (e.g., Fig. 4e) and receive unique numerical labels 4a, 4b, etc.
Lymph-node selection can involve simple manual identification of a 3D target node position or full semi-automatic 3D segmentation. Because central-chest lymph nodes vary considerably in size, shape, and intensity, and because they may cluster, which results in complex compound structures, fully automatic segmentation of the 3D central-chest lymph nodes remains an open problem, as no recent effort has yet to be successful for typical human 3D MDCT data [37,42]. Therefore, in light of this gap, the LNSM provides three semi-automatic segmentation methods based on the interactive live wire [43–45]. These methods, listed in order of ease of use, are as follows: (a) the single-click live wire, which requires the user to click on a voxel located inside a node; (b) the single-section live wire, which requires the user to apply the 2D interactive live wire to a single section of a node; and (c) general 2D/3D live-wire interactive segmentation, which requires the user to apply the 2D live wire to two or more sections of a candidate node. In our studies, the single-click live wire succeeded in segmenting 80% of candidate nodes, while the single-section method successfully segmented 86% of candidate nodes [45]. The general live-wire method serves as a fail-safe approach, as it can always lead to a satisfactory result [44].
To improve the efficacy of semi-automatic segmentation and to also facilitate visualization, the LNSM allows the user to interactively remove 3D chest-model structures from the raw image data. By first removing vascular structures such as the aorta and PA, for example, the live-wire methods can become more effective. Furthermore, the user can be less deceived and distracted by the myriad anatomical structures generally visible in an MDCT scan and focus attention on areas likely to be lymph nodes. After the user completes analysis of a selected station, he/she then selects another station for exploration and analysis. When the physician completes all desired interaction, the results can be saved and applied to follow-on procedure planning. Reference [24] gives a complete discussion of the LNSM’s interactive capabilities.
Implementation comments
The LNSM runs on a standard Windows-based PC. The software is implemented in C++, with some graphics functions drawing upon OpenGL [46].
The 3D chest volumes produced by modern MDCT scanners give vivid detail for the large majority of anatomical structures necessary for automatic station definition. To this end, the methods drawn upon by the LNSM for automatic 3D chest-model computation have undergone substantial prior development and validation using 3D human data and in most instances were first developed for other pulmonary imaging applications [20–22,24–32]. As stated earlier, the system assumes a standard MDCT chest-scanning protocol. Additionally, we assume that a patient has a “typical” airway tree, in that he/she has both lungs and at least a few well-defined airway generations. If it happens that a part of the airway tree or a lung is missing, then the LNSM may not able to obtain sufficient landmark information for defining nodal stations in the missing regions; this has not occurred in our experience to date. As Sect. 3 demonstrates, the LNSM’s automatic processing is consistently reliable and robust for a wide variety of human data.
The computation time required to manage a typical 3D MDCT human chest scan (500 512×512 2D sections; data set size ≈ 200 MB) for the two stages of automatic analysis is as follows:
3D Chest-Model Computation: <15 min
-
Station Definition
landmark computation: <10 s
station limit calculation: virtually instantaneous
The times are based on a Dell Precision 390 PC (dual-core x6800 2.93 GHz, 4 GB RAM). These stages are managed by a technician, prior to physician involvement.
Results
To assess the accuracy of the LNSM’s automatically computed TNM stations, we constructed an unlabeled database of central-chest lymph nodes contained in a series of human 3D MDCT chest scans. We then ran the LNSM’s automated analysis procedures on each raw MDCT scan to generate the TNM stations. Our research team then ascertained the correctness of each station by determining which predefined lymph nodes were situated correctly in their proper stations. We give complete detail and results of this study below.
To begin, we first constructed a ground-truth database of unlabeled lymph nodes depicted in a set of human MDCT chest scans. We selected 32 consecutive MDCT scans from a series of patients undergoing diagnostic evaluation of possible lung cancer. All patients were enrolled under informed consent per Penn State IRB protocols #20349 or 21405. The scans followed a standard lung-cancer scanning regimen and were generated by either a Siemens Sensation 16, Emotion 16, or Sensation 40 MDCT scanner. The scans had the following characteristics: 16 produced with contrast agent (7 male, 9 female), 16 were not (5 male, 10 female, 1 unknown); age range of patients, [28, 85]; scan current range, [134 mA, 539 mA]; scan voltage range, [120 V, 130 V]; 0.5 mm section spacing (Δz); 0.75 mm section thickness; transverse-plane (Δx, Δy) resolution range, [0.52 mm, 0.92 mm]. All scans were reconstructed with kernels that gave smooth images, as smooth images are preferable for soft tissue analysis (B31, 27 scans; B25, 1 scan; B41, 2 scans; unspecified, 2 scans).
Given this set of patient scans, our research team, consisting of a pulmonologist, chest radiologist, and two imaging scientists worked in collaboration to exhaustively locate and define all observable pulmonary lymph nodes; we included all nodes having a long-axis length ≥ 3 mm (≈ 4 to 6 voxels across). To perform this task, we scrolled through 2D transverse, sagittal, or coronal sections of a scan (lung and mediastinal windows used for display). The 2D section views were linked, as in Fig. 7, to enable multiview verification of a node. We then used the fail-safe 2D/3D live wire to segment each lymph node and verified each segmentation for correctness [44]. The team identified 852 lymph nodes over the 32 patient scans, with the average patient scan containing 27 nodes (range 3–59).
Given this database of patient scans and associated unlabeled nodes, we then validated the efficacy of the LNSM in defining regional nodal stations. To do this, we first ran 3D Chest-Model Computation and Station Definition on all 32 raw scans to give a total of 320 automatically defined 3D regional nodal stations. Next, the research team examined each patient scan in conjunction with the database of previously identified lymph nodes to determine if the automatically defined stations properly covered constituent lymph nodes. Armed with their knowledge of the TNM system’s anatomical criteria (e.g., Fig. 1; Table 1), the research team again worked in close collaboration and drew upon the LNSM’s visualization tools (Fig. 7) to make judgments on nodal coverage.
Note that the LNSM’s automatic analysis has no connection to the previously identified lymph nodes. Hence, there is no “system training” per se to run the automatic analysis. Furthermore, as discussed in Sect. 2.1, the methods employed by 3D Chest-Model Computation were built previously and independent of this study. Finally, the automatically defined landmarks depend strictly on anatomy and hence are independent of the data.
Tables 7, 8, 9 and Fig. 8 give the results. Table 7 summarizes overall lymph-node coverage, Table 8 gives a detailed breakdown by station, and Table 9 examines missed nodes. Figure 8 depicts complete sets of labeled lymph nodes for two patients, with [24,38] giving additional complete-scan results. To the best of our knowledge, this is the first time anyone has ever depicted complete sets of lymph nodes in a human 3D MDCT chest scan.
Table 7.
Summary performance of the LNSM over 32-scan database
| Overall performance | By station over 32 scans |
||
|---|---|---|---|
| Worst | Best | ||
| Database nodes covered | 96% (816/852) | 83% (station 8) | 100% (stations 4, 5, 7) |
| Missing nodes per scan | 1.13 | – | – |
| Stations covering all nodes | 93% (299/320) | 81% (station 3) | 100% (stations 4, 5, 7) |
| Missing nodes per station | 0.11 | 0.41 (station 3) | 0 (stations 4, 5, 7) |
The average patient scan contained 27 nodes (range 3–59)
Table 8.
Performance of the LNSM broken down by station number and scan type (contrast-enhanced, no contrast)
| TNM Regional nodal station number |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1–2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10–11 | 12–14 | Total | |
| Contrast-enhanced (16 scans) | |||||||||||
| Nc | 49 | 53 | 100 | 24 | 38 | 26 | 8 | 6 | 39 | 29 | 372 |
| Nm | 2 | 3 | 0 | 0 | 5 | 0 | 4 | 1 | 3 | 0 | 18 |
| Ng | 51 | 56 | 100 | 24 | 43 | 26 | 12 | 7 | 42 | 29 | 390 |
| % | 96% | 95% | 100% | 100% | 88% | 100% | 67% | 86% | 93% | 100% | 95% |
| No contrast (16 scans) | |||||||||||
| Nc | 44 | 65 | 147 | 33 | 48 | 34 | 21 | 10 | 35 | 7 | 444 |
| Nm | 0 | 10 | 0 | 0 | 0 | 0 | 2 | 2 | 3 | 1 | 18 |
| Ng | 44 | 75 | 147 | 33 | 48 | 34 | 23 | 12 | 38 | 8 | 462 |
| % | 100% | 87% | 100% | 100% | 100% | 100% | 91% | 83% | 92% | 88% | 96% |
| All 32 scans | |||||||||||
| Nc | 93 | 118 | 247 | 57 | 86 | 60 | 29 | 16 | 75 | 36 | 816 |
| Nm | 2 | 13 | 0 | 0 | 5 | 0 | 6 | 3 | 6 | 1 | 36 |
| Ng | 95 | 131 | 247 | 57 | 91 | 60 | 35 | 19 | 80 | 37 | 852 |
| % | 98% | 90% | 100% | 100% | 95% | 100% | 83% | 84% | 93% | 97% | 96% |
| Station success | |||||||||||
| Ns | 31 | 26 | 32 | 32 | 30 | 32 | 28 | 30 | 27 | 31 | 299 |
| % | 97% | 81% | 100% | 100% | 94% | 100% | 88% | 94% | 84% | 97% | 93% |
The following quantities were computed as an aggregate over 32 scans: Nc = total number of nodes covered by LNSM’s automatically defined stations. Nm = total number of nodes missed by LNSM’s stations. Ng = total number of ground-truth database nodes in indicated station. Ns = number of scans where LNSM gave 100% nodal coverage. Finally, % gives percentage measures, whereby % = 100 × Nc/Ng or 100 × Ns/32
Table 9.
Characteristics of the lymph nodes missed by the system’s automatically defined stations
| Nodes missed by LNSM (36 nodes) |
Ground-truth database (852 nodes) |
|||||
|---|---|---|---|---|---|---|
| Mean | Min | Max | Mean | Min | Max | |
| Long-axis length (mm) | 9.0 | 4.2 | 21.3 | 13.1 | 3.3 | 83.8 |
| Short-axis length (mm) | 4.7 | 2.5 | 12.1 | 5.9 | 1.3 | 35.4 |
| Volume (mm3) | 191 | 21 | 1,473 | 689 | 12 | 60,095 |
| Distance from station (mm) | 4.2 | 0.5 | 27.0 | – | – | – |
The portion of the table labeled “Nodes Missed by LNSM” profiles the 36 nodes mislabeled by the LNSM’s automatically defined stations, while the portion of the table labeled “Ground-Truth Database” profiles the characteristics of the 852 nodes contained in the ground-truth database. The parameters “long-axis length,” “short-axis length,” and “volume” refer to the long-axis length, short-axis length, and volume of a node, while “distance from station” refers to the shortest distance of a missed node to its proper nodal station
Fig. 8.
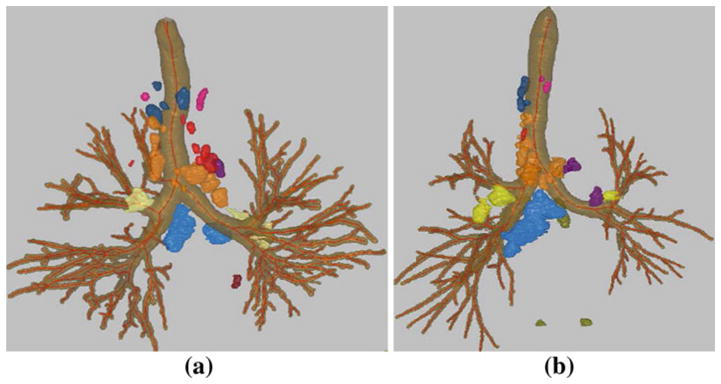
Surface-rendered view of the segmented central-chest lymph nodes for human MDCT chest scans: a 20349_3_3 (31 nodes), which contains three nodes in station 1–2, two in station 3, eleven in station 4, one in station 5, seven in station 6, two in station 7, zero in station 8, one in station 9, four in station 10–11, and zero in station 12–14. b 20349_3_27 (35 nodes), which contains three nodes in station 1–2, two in station 3, seventeen in station 4, three in station 5, one in station 6, one in station 7, three in station 8, zero in station 9, four in station 10–11, and one in station 12–14; scan consists of 752, 512 × 512 transverse-plane sections and has resolution Δx = Δy = 0.67 mm and Δz = 0.5 mm. The segmented lymph nodes (blobs outside the airway tree) are displayed in colors derived from the TNM system’s color codes, per Fig. 1
By convention, an automatically defined station is said to cover a node if any portion of the node lies within the station’s limits. Note that it is typical for a diseased lymph node to be enlarged and have much of its volume extend outside a station’s nominal anatomical zone. For example, note the very large station-7 (blue) nodes in Fig. 8b. Also, Table 9 shows that a lymph node can have a long axis that is several cm in length, a length that can easily make a node lie partially outside a given station.
Overall, 93% of the automatically defined stations (299/320) covered 100% of their respective nodes, with 96% of the stations (307/320) missing 1 or fewer nodes (Tables 7, 8). Stated another way, 96% of the nodes (816/852) were covered by the automatically defined stations. Furthermore, the LNSM produced stations that covered 100% of constituent database nodes across all station 4, 5, and 7 locations; these stations accounted for 43% of all nodes.
Note that a particular station need not necessarily contain any verifiable lymph nodes; in fact, 85 of 320 stations (32 scans, 10 TNM stations per scan) contained 0 nodes. This is because most healthy nodes are typically very small and, hence, in line with our size criterion, are too small to ascertain their existence in an MDCT scan. Also, some stations, such as 9 and 12–14, usually exhibit few lymph nodes (only 6.5% of our database’s nodes); Chapet et al. attested to this general phenomenon, as they ignored stations 9 and 12–14 in their work [10].
The 36 mislabeled nodes appeared in 21 stations over 15 scans (Table 9). Thus, the LNSM successfully labeled all lymph nodes appearing in the scans for 17/32 patients (53%). But, again, per Table 7, note that on average 96% of the nodes were properly labeled per patient scan; this translates to 1.13 missed nodes per patient scan and 0.11 missed nodes per station (2.67 nodes per station on average; range 0–20). For the 21 stations missing nodes, the number of missed nodes ranged from 1 to 4. Regarding worst cases,
Scan 21405_66 had three stations missing a total of 6 nodes (33 nodes overall in the scan; 82% coverage by the LNSM).
4/20 nodes were missed in station 4 for scan 21405_67 (59 nodes overall; 93% coverage).
Table 9 shows that the average missed node had a substantially smaller volume than the average database node (volume = 191 mm3 vs. 689 mm3). In addition, the mean distance of a missed lymph node from its correct station was only 4.2 mm, with 20 missed nodes ≤ 2.5 mm from their respective stations—this amounts to only a few image voxels. Such close-by nodes, while not located inside their proper automatically defined stations, were still readily observable in the LNSM’s focused station view (Fig. 7). Given the latitude provided by the TNM station criteria, it would seem to be a prudent and reasonable design decision to add an extra “guard band” region around all stations to make them slightly larger and more encompassing. (In fact, the LNSM has an option for interactively changing the dimensions of a station.) The relatively low station-3 performance (81% of scans had all station-3 nodes properly labeled, Table 8) may imply that the LNSM’s definition of this station could be improved. Station 3 is a challenge, as pointed out by Chapet et al., because it involves the great vessels emanating from the top of the aortic arch, which are not readily identifiable in MDCT [10]. Nevertheless, 90% of station-3 nodes were properly covered by the LNSM and all remaining nodes appeared close by.
Per Table 8, the use of contrast agent had no significant influence on the likelihood of correctly labeling a node (p = 0.61, two-tailed Fisher exact test). This is expected, because the LNSM’s automated procedures leading to the defined stations are not affected by contrast agent. (Our method for aorta/PA segmentation automatically detects the use of contrast agent [28].) Again, the LNSM does not detect lymph nodes, but instead labels manually identified nodes. Interestingly, it also appears that contrast agent did not help our research team better locate lymph nodes in a high-resolution 3D MDCT scan, as described in a separate study [38].
Discussion
For the first time, we propose a system—the Lymph Node Station Mapper—that enables automatic 3D TNM regional nodal station definition from a patient’s 3D MDCT chest scan. Results show that the system is very promising for this purpose. In addition to providing station labels automatically, the LNSM’s stations also greatly reduce the amount of image data that the physician need examine to identify lymph nodes of interest. For example, for case 21405_3_3 (Figs. 4, 5, 6), stations 4, 7, and 10–11 covered a total volume of 304 cc and the remaining seven stations covered under 3,000 cc, while the complete scan covered 39,200 cc—this represented over a 90% reduction in data to examine.
User interaction with the LNSM may entail anywhere from a few minutes to 30 min, depending on the physician’s assessment of a patient’s needs. Yet, the appropriate amount of interaction time is very unclear for two major reasons. First, the number of verifiable nodes in a given MDCT scan can vary considerably (range 3–59 nodes per scan in our database). Second, it is still an open long-term research issue as to just how many nodes should be considered to effectively stage a patient. Note that current lung-cancer staging practice focuses on a small number of enlarged lymph nodes (1 to 4 nodes per patient) having long-axis length ≥ 10 mm [5,6]. But given the poor survival rate of lung-cancer patients, it may be more effective to stage the nodes more exhaustively.
It is important to realize that current practice requires the physician to mentally apply the TNM nodal classification system during chest CT interpretation and bronchoscopy planning. The LNSM system offers the possibility of automatic, reproducible, and patient-specific identification of the standard TNM stations. With the computer-based assistance afforded by the LNSM, the physician can better focus attention during the assessment of a high-resolution MDCT scan. While the system performs considerable automated analysis, it is imperative that the physician verify results. For this purpose, the LNSM provides many interactive visualization and data mining tools. As an intriguing possibility, the LNSM could conceivably be used as an educational tool for learning the 3D chest anatomy and its relationship to nodal station locations.
A major impetus for our development of the LNSM has been to help fill the scientific void in the medical community’s lack of understanding of the characteristics and distribution of lymph nodes in 3D MDCT chest scans. To this end, we have used the LNSM to perform a detailed quantitative study of the lymph nodes depicted in our database [38,45, 47]. In addition, we have performed studies to determine the ability of both radiologists and pulmonologists to identify lymph nodes in MDCT, and we have made early efforts to apply the LNSM to planning follow-on staging bronchoscopy [24,48–50].
We caution that our results apply only to a 32-scan database (containing 852 verifiable nodes have long-axis length ≥ 3 mm, currently the largest such database in existence to our knowledge). Furthermore, despite our care in constructing the database, the team could have missed lymph nodes. Also, we mention again that the TNM station criteria are vague and open to interpretation. This could conceivably make it easier to achieve high station-definition accuracy. While we endeavored to select scans in an unbiased manner, it would be beneficial to consider a larger set of scans, possibly derived from multiple institutions, to further ascertain system efficacy.
As an additional point, all of our test scans were produced by scanners from a single vendor. We do not believe this to be a limitation, as the chest radiology community in general only works with MDCT scanners calibrated to a standard HU scale [23]. Furthermore, as nearly all of our methods employed for 3D Chest-Model computation have been tested with scans from multiple vendors (landmark extraction is virtually data independent), we believe that the LNSM’s approach for station definition is essentially independent of scanner manufacturer. Finally, we only considered sub-millimeter-resolution scans in our work. While many current practitioners use thicker-slice data, most chest imaging-science researchers only consider thin-slice data, as such data better represents the current state-of-the-art [14–16]. In addition, we believe that high-resolution—i.e., a resolution fine enough to enable confirmation of lymph nodes < 10 mm in long-axis length—is essential for achieving a long-term goal of exhaustive nodal staging.
While the identification and analysis of suspect lung-cancer tumors (the “T” in the TNM staging system) in MDCT chest scans have received considerable attention from the research community, little research has been dedicated to the lymph-node problem (the “N” in TNM) and no computer-aided tools exist currently for this purpose [3,5,7,11]. This is probably true for two reasons: (1) the identification of a tumor, any tumor, could result in a diagnosis of cancer—hence, the management of tumors is vital; (2) unlike tumors, lymph nodes are plentiful and part of the normal anatomy—and a large majority of identifiable nodes will not result in a cancer diagnosis. Furthermore, as stated earlier, it is not standard practice or yet practical, to consider all lymph nodes during cancer diagnosis and staging—this is a long-term research question in the problem of trying to improve the still abysmal survival rate of lung cancer [1]. It is likely that the use of combined PET/CT will be important to this problem [5,6,8]. Nevertheless, the future study of lymph nodes for lung-cancer assessment is vital. Some preliminary success has been reported in the identification of possible lymph-node candidates in 3D MDCT chest scans, yet no method exists that can successfully and reliably identify/segment central-chest lymph nodes [37]. It is possible that a combination of candidate identification and the LNSM’s station calculation could greatly facilitate reliable lymph-node detection.
Acknowledgments
This work was partially supported by grants R01-CA074325, R44-CA091534, and R01-CA151433 from the National Cancer Institute of the National Institutes of Health. Portions of this work appeared earlier in a preliminary form in the conference presentation [51].
Contributor Information
Kongkuo Lu, Departments of Electrical Engineering and Computer Science and Engineering, Penn State University, University Park, Hershey, PA, USA.
Pinyo Taeprasartsit, Departments of Electrical Engineering and Computer Science and Engineering, Penn State University, University Park, Hershey, PA, USA.
Rebecca Bascom, Division of Pulmonary, Allergy, and Critical Care Medicine, Department of Medicine, Penn State University, University Park, Hershey, PA, USA.
Rickhesvar P. M. Mahraj, Department of Radiology, Penn State University, University Park, Hershey, PA, USA
William E. Higgins, Email: weh2@psu.edu, Departments of Electrical Engineering and Computer Science and Engineering, Penn State University, University Park, Hershey, PA, USA
References
- 1.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Murray T, Thun MJ. Cancer statistics 2008. CA Cancer J Clin. 2008;58:71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 2.Mountain C, Dresler C. Regional lymph node classification for lung cancer staging. Chest. 1997;111(6):1718–1723. doi: 10.1378/chest.111.6.1718. [DOI] [PubMed] [Google Scholar]
- 3.Mountain C. Staging classification of lung cancer. A critical evaluation. Clin Chest Med. 2002;23(1):103–121. doi: 10.1016/s0272-5231(03)00063-7. [DOI] [PubMed] [Google Scholar]
- 4.Cymbalista M, Waysberg A, Grenier P, et al. CT demonstration of the 1996 AJCC-UICC regional lymph node classification for lung cancer staging. Radiographics. 1999;19:899. doi: 10.1148/radiographics.19.4.g99jl09899. [DOI] [PubMed] [Google Scholar]
- 5.Silvestri GA, Tanoue LT, Margolis ML, Barker J, Detterbeck F. The noninvasive staging of non-small cell lung cancer: the guidelines. Chest. 2003;123(1):147S–156S. doi: 10.1378/chest.123.1_suppl.147s. [DOI] [PubMed] [Google Scholar]
- 6.Torabi M, Aquino SL, Harisinghani MG. Current concepts in lymph node imaging. J Nucl Med. 2004;45(9):1509–1518. [PubMed] [Google Scholar]
- 7.Goldstraw P, Crowley J, Chansky K, Giroux DJ, Groome PA, et al. The IASLC lung cancer staging project: proposals for the revision of the TNM stage groupings in the forthcoming (seventh) edition of the TNM classification of malignant tumours. J Thorac Oncol. 2007;2(8):706–714. doi: 10.1097/JTO.0b013e31812f3c1a. [DOI] [PubMed] [Google Scholar]
- 8.Quint LE. Staging non-small cell lung cancer. Cancer Imaging. 2007;7(1):148–159. doi: 10.1102/1470-7330.2007.0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ko JP, Drucker EA, Shepard JO, Mountain CF, Dresler C, Sabloff B, McLoud TC. CT depiction of regional nodal stations for lung cancer staging. Am J Roentgenol. 2000;174(3):775–782. doi: 10.2214/ajr.174.3.1740775. [DOI] [PubMed] [Google Scholar]
- 10.Chapet O, Kong F-M, Quint LE, Chang AC, Ten-Haken RK, Eisbruch A, Hayman JA. CT-based definition of thoracic lymph node stations: an atlas from the University of Michigan. Int J Radiat Oncol Biol Phys. 2005;63(1):170–178. doi: 10.1016/j.ijrobp.2004.12.060. [DOI] [PubMed] [Google Scholar]
- 11.Spira A, Ettinger DS. Multidisciplinary management of lung cancer. New Engl J Med. 2004;350(4):379–392. doi: 10.1056/NEJMra035536. [DOI] [PubMed] [Google Scholar]
- 12.Wang KP, Mehta AC, Turner JF, editors. Flexible bronchoscopy. 2. Blackwell; Cambridge: 2003. [Google Scholar]
- 13.Kazerooni EA. High resolution CT of the lungs. Am J Roentgenol. 2001;177(3):501–519. doi: 10.2214/ajr.177.3.1770501. [DOI] [PubMed] [Google Scholar]
- 14.Rubin GD. 3-D imaging with MDCT. Eur J Radiol. 2003;45(Suppl):S37–S41. doi: 10.1016/s0720-048x(03)00035-4. [DOI] [PubMed] [Google Scholar]
- 15.Dalrymple NC, Prasad SR, Freckleton MW, Chintapalli KN. Introduction to the language of three-dimensional imaging with multidetector CT. Radiographics. 2005;25(5):1409–1428. doi: 10.1148/rg.255055044. [DOI] [PubMed] [Google Scholar]
- 16.Ueno J, Murase T, Yoneda K, Tsujikawa T, Sakiyama S, Kondoh K. Three-dimensional imaging of thoracic diseases with multidetector row CT. J Med Investigat. 2004;51:163–170. doi: 10.2152/jmi.51.163. [DOI] [PubMed] [Google Scholar]
- 17.Kiraly AP, Naidich DP, Guendel L, Zhang L, Novak CL. Novel method and applications for labeling and identifying lymph nodes. In: Manduca A, Hu CP, editors. SPIE medical imaging 2007: physiology, function, and structure from medical images. Vol. 6511. 2007. pp. 651111–651119. [Google Scholar]
- 18.Helferty JP, Sherbondy AJ, Kiraly AP, Higgins WE. Computer-based system for the virtual-endoscopic guidance of bronchoscopy. Comput Vis Image Underst. 2007;108(1–2):171–187. doi: 10.1016/j.cviu.2006.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Merritt SA, Gibbs JD, Yu KC, Patel V, Rai L, Cornish DC, Bascom R, Higgins WE. Real-time image-guided bronchoscopy for peripheral lung lesions: a phantom study. Chest. 2008;134(5):1017–1026. doi: 10.1378/chest.08-0603. [DOI] [PubMed] [Google Scholar]
- 20.Higgins WE, Helferty JP, Lu K, Merritt SA, Rai L, Yu KC. 3D CT-video fusion for image-guided bronchoscopy. Comput Med Imaging Graph. 2008;32(3):159–173. doi: 10.1016/j.compmedimag.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gibbs JD, Graham MW, Higgins WE. 3D MDCT-based system for planning peripheral bronchoscopic procedures. Comput Biol Med. 2009;39(3):266–279. doi: 10.1016/j.compbiomed.2008.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Graham MW, Gibbs JD, Cornish DC, Higgins WE. Robust 3D airway-tree segmentation for image-guided peripheral bronchoscopy. IEEE Trans Med Imaging. 2010;29(4):982–997. doi: 10.1109/TMI.2009.2035813. [DOI] [PubMed] [Google Scholar]
- 23.Kalender W. Computed tomography: fundamentals, system technology, image quality, applications. Publicis MCD Verlag; Munich: 2000. [Google Scholar]
- 24.Lu K. PhD dissertation. The Pennsylvania State University, Department of Electrical Engineering; 2010. Multidimensional image segmentation and pulmonary lymph-node analysis. [Google Scholar]
- 25.Kiraly AP, Hoffman EA, McLennan G, Higgins WE, Reinhardt JM. 3D human airway segmentation methods for clinical virtual bronchoscopy. Acad Radiol. 2002;9(10):1153–1168. doi: 10.1016/s1076-6332(03)80517-2. [DOI] [PubMed] [Google Scholar]
- 26.Kiraly AP, Helferty JP, Hoffman EA, McLennan G, Higgins WE. 3D path planning for virtual bronchoscopy. IEEE Trans Med Imaging. 2004;23(9):1365–1379. doi: 10.1109/TMI.2004.829332. [DOI] [PubMed] [Google Scholar]
- 27.Taeprasartsit P, Higgins WE. System for definition of the central-chest vasculature. In: Pluim JP, Dawant BM, editors. SPIE medical imaging 2009: image processing. Vol. 7259 2009. [Google Scholar]
- 28.Taeprasartsit P, Higgins WE. Robust extraction of the aorta and pulmonary artery from 3D MDCT images. In: Dawant B, Haynor D, editors. SPIE medical imaging 2010: image processing. Vol. 7623. 2010. pp. 76230H1–76230H17. [Google Scholar]
- 29.Yu KC, Ritman EL, Higgins WE. 3D model-based vasculature analysis using differential geometry. IEEE international symposium on biomedical imaging; Arlington, VA. 15–18 April; 2004. pp. 177–180. [Google Scholar]
- 30.Graham M, Higgins WE. Globally optimal model-based matching of anatomical trees. In: Reinhardt JM, Pluim JPW, editors. SPIE medical imaging 2006: image processing. Vol. 6144. 2006. pp. 373–388. [Google Scholar]
- 31.Graham MW, Higgins WE. Optimal graph-theoretic approach to 3D anatomical tree matching. IEEE international symposium on biomedical imagings; 2006. pp. 109–112. [Google Scholar]
- 32.Graham MW, Dolina MY, Bascom R, Higgins WE. Graphics-based computer system for the anatomical labeling of the major airways: initial development. Am J Respir Crit Care Med. 2007;175:A333. abstracts issue. [Google Scholar]
- 33.Brown MS, McNitt-Gray MF, Mankovich NJ, Goldin JG, Hiller J, Wilson LS, Aberle DR. Method for segmenting chest CT image data using an anatomical model: preliminary results. IEEE Trans Med Imaging. 1997;16:828–839. doi: 10.1109/42.650879. [DOI] [PubMed] [Google Scholar]
- 34.Tozaki T, Kawata Y, Niki N, Ohmatsu H, Kakinuma R, Eguchi K, Kaneko M, Moriyama N. Pulmonary organs analysis for differential diagnosis based on thoracic thin-section CT images. IEEE Trans Nucl Sci. 2001;45(6):3075–3082. [Google Scholar]
- 35.Tozaki T, Kawata Y, Niki N, Ohmatsu H. Extraction and classification of pulmonary organs based on thoracic 3D CT images. Syst Comput Jpn. 2001;32(9):42–53. [Google Scholar]
- 36.Suter M, Tschirren J, Reinhardt JM, Sonka M, Hoffman EA, Higgins WE, McLennan G. Evaluation of the human airway with multi-detector X-ray-computed tomography and optical imaging. Physiol Measure. 2004;25:837–847. doi: 10.1088/0967-3334/25/4/005. [DOI] [PubMed] [Google Scholar]
- 37.Feuerstein M, Deguchi D, Kitasaka T, Iwano S, Imaizumi K, Hasegawa Y, Suenaga Y, Mori K. Automatic mediastinal lymph node detection in chest CT. In: Karssemeijer N, Giger ML, editors. SPIE medical imaging 2009: computer-aided diagnosis. Vol. 7260. 2009. pp. 72600V1–72600V11. [Google Scholar]
- 38.Lu K, Higgins WE, Bascom R, Mahraj RP. Quantitative analysis of the central-chest lymph nodes based on 3D MDCT image data. In: Karssemeijer N, Giger ML, editors. SPIE medical imaging 2009: computer-aided diagnosis. Vol. 7260. 2009. pp. 72600U1–72600U9. [Google Scholar]
- 39.Lu K, Bascom R, Mahraj RPM, Higgins WE. 3D MDCT-based presentation of the TNM regional nodal stations. Proceedings of American thoracic society. 2011 (under submission) [Google Scholar]
- 40.Turlington JZ, Higgins WE. New techniques for efficient sliding thin-slab volume visualization. IEEE Trans Med Imaging. 2001;20(8):823–835. doi: 10.1109/42.938250. [DOI] [PubMed] [Google Scholar]
- 41.Haponik EF, Aquino SL, Vining DJ. Virtual bronchoscopy. Clin Chest Med. 1999;20(1):201–217. doi: 10.1016/s0272-5231(05)70135-0. [DOI] [PubMed] [Google Scholar]
- 42.Vining DJ, Ferretti G, Stelts DR, Ahn D, Ge Y, Haponik EF. Mediastinal lymph node mapping using spiral CT and three-dimensional reconstructions in patients with lung cancer: preliminary observations. J Bronchol Intervent Pulmonol. 1997;4(1):18–25. [Google Scholar]
- 43.Mortensen EN, Barrett WA. Interactive segmentation with intelligent scissors. Graph Models Image Process. 1998;60(5):349–384. [Google Scholar]
- 44.Lu K, Higgins WE. Interactive segmentation based on the live wire for 3D CT chest image analysis. Int J Comput Assist Radiol Surg. 2007;2(3–4):151–167. [Google Scholar]
- 45.Lu K, Higgins WE. Central-chest lymph nodes definition from 3D MDCT image data. In: Karssemeijer N, Summers R, editors. SPIE medical imaging 2010: computer-aided diagnosis. Vol. 7624. 2010. pp. 76240T1–76240T10. [Google Scholar]
- 46.Wright RS, Jr, Lipchak B. OpenGL super bible. 3. SAMS Publishing; Indianapolis: 2005. [Google Scholar]
- 47.Lu K, Bascom R, Mahraj RP, Higgins WE. Assessment of the 3D characteristics of central-chest lymph nodes in 3D MDCT images. Am J Respirat Crit Care Med. 2009;179:A4369. [Google Scholar]
- 48.Higgins WE, Gibbs JD, Bascom R, Khare R, Taeprasartsit P. Planning and visualization of accessible, safe bronchoscopy routes: application to central-chest lymph node biopsy. Am J Respir Crit Care Med. 2009;179:A6148. [Google Scholar]
- 49.Higgins WE, Lu K, Bascom R, Mahraj RPM, Toth J, Campbell D. Selection of central-chest lymph nodes in 3D MDCT images: an observer study. Am J Respir Crit Care Med. 2010;181:A5518. [Google Scholar]
- 50.Taeprasartsit P. PhD dissertation. The Pennsylvania State University, Department of Computer Science and Engineering; 2010. 3D image analysis for definition of the chest vasculature. [Google Scholar]
- 51.Lu K, Merritt SA, Higgins WE. Extraction and visualization of the central chest lymph-node stations. In: Giger ML, Karssemeijer N, editors. SPIE medical imaging 2008: computer-aided diagnosis. Vol. 6915. 2008. pp. 69151B1–69151B15. [Google Scholar]