Abstract
Insertional mutagenesis in a haploid background can lead to complete disruption of gene function1. Here we generate a population of human cells that contain insertions in >98% of their expressed genes. We established Phenotypic Interrogation via Tag Sequencing (PhITSeq) as a method to examine millions of mutant alleles through selection and parallel sequencing. Analysis of pools of selected cells rather than individual clones provides a rapid assessment of the spectrum of genes involved in phenotypes under study. This facilitates comparative screens as illustrated here for the family of cytolethal distending toxins (CDTs). CDTs are virulence factors secreted by a variety of pathogenic gram-negative bacteria that cause tissue damage at distinct anatomical sites2. We identified 743 mutations distributed over 12 human genes important for intoxication by four different CDTs. While related CDTs may share host factors, they also exploit unique host factors yielding a characteristic profile for each CDT.
The ability to remove or inactivate single genes in cells or intact organisms has revolutionized all aspects of modern biology. Gene disruption in human cells is hampered by their diploid nature, the inability to set up genetic crosses at will and the low rates of homologous recombination. As a result, only very few traits in man have been subjected to detailed mutagenesis-based analysis by conventional methods. We have recently developed a mutagenesis-based screening approach in human cells using insertional mutagenesis in KBM7 cells, a chronic myeloid leukemia cell line that is haploid for all chromosomes except chromosome 81. However, this required individual clones to be isolated, expanded and used for DNA isolation to map gene-trap insertions by inverse-PCR and Sanger sequencing. Such screens are labor intensive, do not necessarily reach saturation and thus may not produce a reliable genome-wide overview of genes that contribute to phenotypes of interest.
To overcome these shortcomings, we report here an approach to interrogate millions of mutant alleles using deep sequencing as a means of assigning genes to phenotypes with high saturation and accuracy. Analogous to recent developments in high-density insertional mutagenesis in microorganisms3–5, this approach may enable the comparison of mutant phenotypes under different conditions. We first benchmarked a large population of mutagenized cells by examination of the mutation frequencies in individual genes by deep sequencing. We then used this benchmarked population of mutant cells for 12 independent phenotypic selections. As inferred from the number of independent insertions in genes of interest in a given screen, we achieved a high level of saturation and thus a genome-wide overview of the genes involved. Finally, we apply these technological advances to 4 comparative genome-wide screens, using toxins secreted by gram-negative bacteria as the selecting agents. We could distinguish amongst toxins produced by related pathogens that evolved to affect different anatomical sites in the human body. These comparative screens identify with high confidence 10 host factors not previously implicated in intoxication and furthermore provide evidence that these structurally and functionally related, yet distinct toxins evolved to exploit distinct sets of host factors to achieve intoxication of their hosts.
To obtain accurate genome-wide overviews of genes involved in particular phenotypes we increased the saturation of gene trap mutagenesis in two ways. First, for a given screen, we increased the total number of cells infected with a promotor-less retroviral gene trap vector to 100 million cells and improved the retroviral transduction rate to ~75%. Second, in an improvement over our earlier approach1, we aimed to retain insertion events also in genes that are silent or that show low or heterogeneous expression. Typically, mutagenesis by promoter trap vectors involves a selection step for insertions into active genes by tracking the reporter gene embedded in the gene trap. We omitted this step and characterized the mutagenized cell pool without selection, thereby extending the mutagenized cell population to all other types of gene-trap insertions: in silent genes, in lowly or heterogeneously expressed genes, opposite to direction of transcription, etc. To characterize the extent and type of insertions obtained in our mutagenized cell population we mapped the flanking sequences of ~900,000 independent insertion sites, using a Linear Amplification Mediated-PCR (LAM-PCR), followed by ssDNA linker ligation and massively parallel sequencing (Table S1).
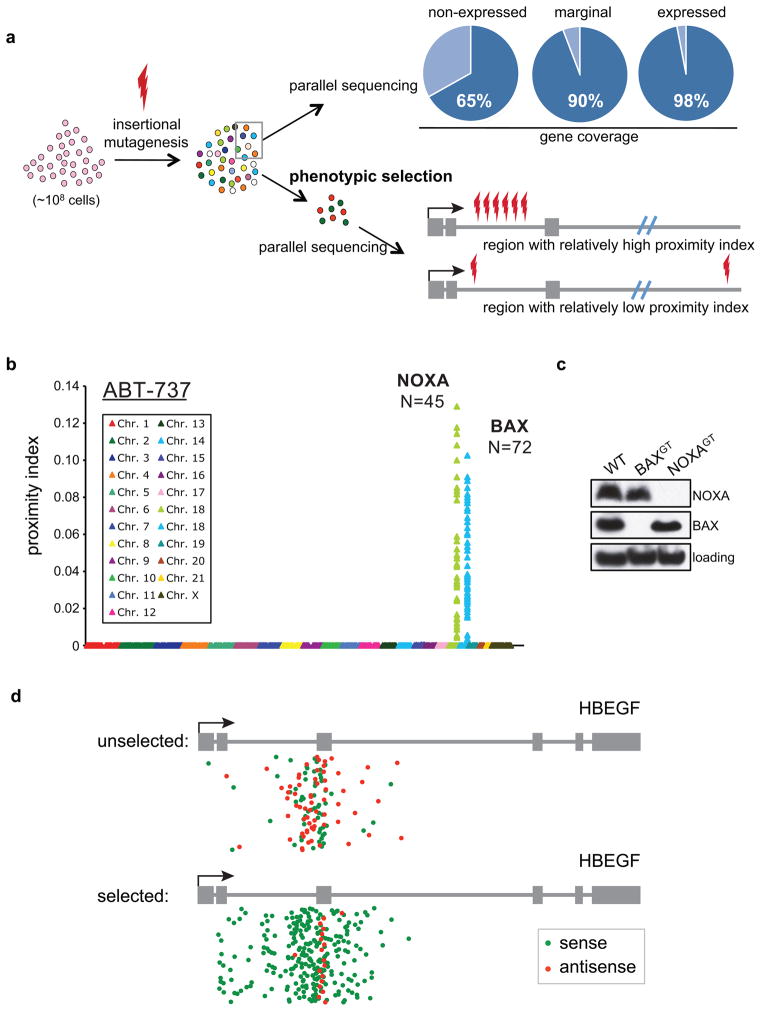
Insertion sites were distributed over all chromosomes but were biased towards genes, because ~49% of the insertions were present within Refseq annotated genes (Table S2). These insertions covered ~70% of all Refseq genes and each gene is hit with an average of ~30 insertions. Although we did not impose a selection a priori for active genes by using the selection embedded in the gene trap vector, it is known that gammaretroviral insertion sites have a preference for genomic regions near histone marks that are positively associated with transcription6. To assess the extent of mutagenesis obtained, we compared our mapped insertion database with expression data in KBM7 cells7. Ninety-eight percent of the genes classified as expressed based on KBM7 microarray data (Table S3) contain at least one gene trap insertion. These percentages decrease to 90% for marginally expressed genes and to 65% for genes classified as non-expressed. Given that we sequenced only ~1 % of the mutations present in the input cell population (0.9 million out of the 75 million mutants), we conclude that our full library contains many independent mutations in nearly all expressed genes, including those expressed at low levels and for the majority of genes that are heterogeneously expressed or silent under basal growth conditions. Phenotypic selection of mutant cells, followed by mapping of the mutations in the selected pool, should therefore yield a detailed genome-wide view of the genetic components associated with a particular phenotype. We termed this approach Phenotypic Interrogation via Tag Sequencing (PhITSeq)
As a first screening experiment, we exposed 100 million mutagenized cells to a recently developed antagonist of the anti-apoptotic BCL-2 family, the small molecule ABT-7378, which induces regression of solid tumours. We confirmed that, prior to selection, the unselected population of mutagenized cells contains mutations in all main components of the apoptotic machinery (Figure S1). After selection, cells were expanded and sequences flanking the insertion sites were amplified using an inverse PCR protocol, followed by massively parallel sequencing. These sequences were then mapped to the human genome and for each insertion the distances to its closest neighboring, independent insertions were determined, which allowed the calculation of a proximity index (PI) for each insertion. A high numerical value for this index represents a short distance to its neighbors (Figures 1A and 1B). Because passenger mutations will be randomly distributed over the genome, these should have a low PI, whereas driver mutations are expected to cluster closely in distinct genomic regions and should have a high PI. After ABT-737 selection of the mutant pool, we observed two regions on chromosome 18 and 19 with a high density of insertions (Figure 1B). These regions show a combined total of 117 independent mutations distributed over regions encoding the genes NOXA and BAX. Clonally derived cell lines that contain gene trap insertions in these genes show loss of expression of the corresponding gene (Figure 1C). Both pro-apoptotic proteins have been implicated in ABT-737 mediated induction of cell death9,10.
Figure 1. Phenotypic interrogation via tag sequencing (PhITSeq).
A. Approximately 100 million near-haploid KBM7 cells were infected with gene trap vectors and expanded without selection. Short DNA sequences flanking the inserted gene trap vectors were amplified and sequenced in parallel and aligned to the human genome. Insertion sites were identified in genes that were expressed and non-expressed in KBM7 cells. The population of 100 million cells was used to select several thousand clones for particular phenotypes. Selected clones were expanded and used for parallel sequencing for insertion sites. For each insertion site a proximity index was calculated. The proximity index corresponds to the calculated inverse of the average distance between a specific insertion and its immediate upstream and downstream insertions. B. Mutagenized cells were selected with ABT-737 and insertion sites were mapped in the selected populations. N indicates the number of insertions found in each gene. C. Immunoblot analysis of BAX and NOXA protein expression in clonally derived cell lines that contain gene trap insertions in corresponding genes. D. Insertions in the HBEGF locus in the unselected mutagenized pool and in a cell population that is selected using diphtheria toxin. Gene trap insertions in the same transcriptional orientation as the gene (sense) are depicted in green and in the antisense orientation are drawn in red. Note that selection against HBEGF function causes an enrichment for sense orientation insertions in introns but not in exons.
PhITSeq couples an abundance of independent insertional mutations to a phenotype. We do not select for inactivating mutations a priori: our mutant library contains insertions in introns in the sense (thereby inactivating the gene) and antisense (not likely to inactivate a gene when inserted in intronic sequences) orientation. As viral integration in the sense or antisense orientation is a random event, a skewed distribution of insertions in a particular gene is indicative of selection, and a functional consequence of inactivation. Indeed, when we compare the collections of insertions in cells that survive selection, there is strong enrichment of the inactivating sense mutations. As a graphic illustration of this point, we chose a gene for which the largest number of insertions was identified (Figure 1D). In our screen for resistance to diphtheria toxin, we disrupted the gene that encodes the entry receptor of diphtheria toxin receptor (HBEGF, 295 independent insertions). Whereas sense and antisense insertions are present in equal proportions in the unselected starting collection of mutant cells, the antisense insertions in introns are almost completely lost from the selected survivors. The remaining antisense insertions are located mainly within exonic sequences of the gene and are therefore likely to be mutagenic as well. Thus, PhITSeq accurately links inactivating mutations to a phenotype of interest and is not dependent on prior knowledge or gene annotation.
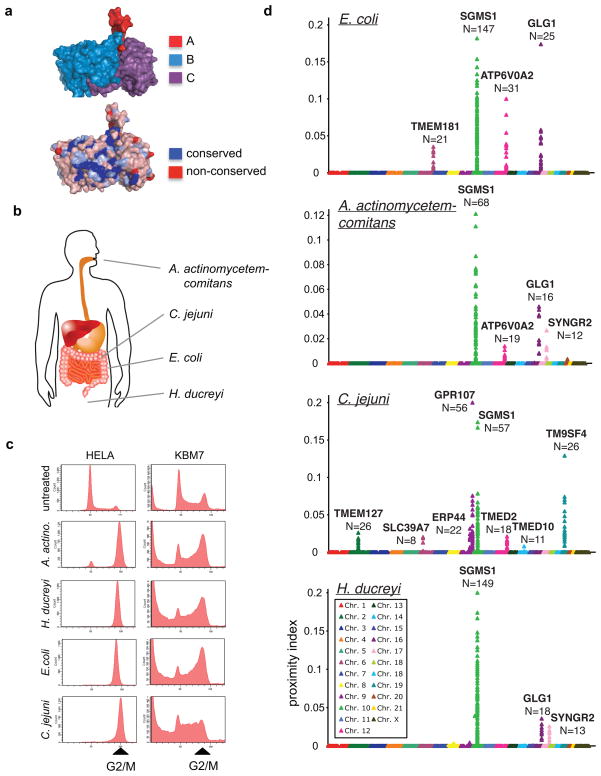
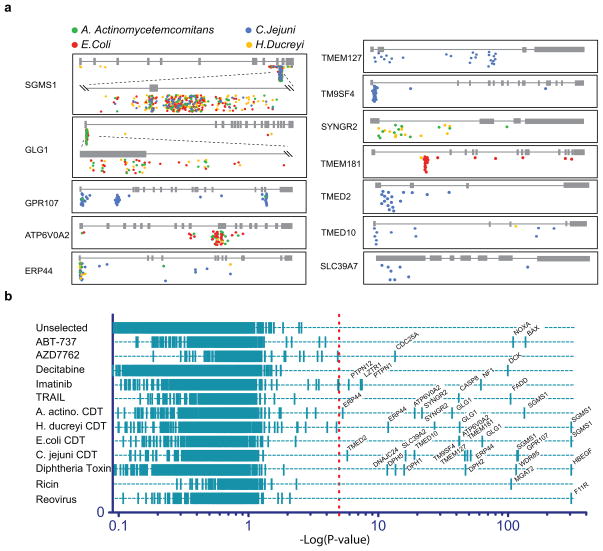
Our second screening experiment involved bacterial toxins. Cytolethal distending toxins (CDTs) are secreted by multiple bacterial species that cause disease2, including food borne diseases (Escherichia coli, Shigella dysenteriae, Campylobacter jejuni and Salmonella typhi), aggressive periodontitis (Aggregatibacter actinomycetemcomitans) and sexually transmitted disease (Haemophilus ducreyi). Their CDTs have been proposed to be virulence factors and are further suspected of having carcinogenic properties, because the catalytic subunit of these toxins displays DNAse I-like activity11. Exposure of human cells to these toxins causes cell cycle arrest and cytotoxicity by inducing DNA breaks11, 12. Virulence factors often interact closely with host cells at the site of infection to create an environment favorable to colonization. It is unclear whether CDTs that target different anatomical sites also use different host factors for cell intoxication. Notwithstanding similar overall structure, CDTs from different species diverge in sequence, with subunits A and C being the cell-binding component and the more conserved B subunit the catalytic domain with homology to DNAses11 (Figure 2A). These differences in sequence likely reflect the lifestyles and anatomical host niches targeted by the bacteria that produce them (Figure 2B). The lesser degree of sequence conservation of the cell binding subunits may reflect the use of different entry factors. We used PhITSeq to determine whether CDTs of diverse origin and divergent in structure use different pathways for entry and intoxication, or whether there are shared components as well. Four different bacterial toxins were produced, each of them causing the characteristic G2/M arrest in HeLa and KBM7 cells (Figure 2C). Four pools of 100 million library mutant cells were treated each with a different CDT, surviving cells were isolated 20 days after intoxication, and insertions were mapped as described above. The proximity plots show 12 host factors with a combined total of 743 mutations (Figure 2D and 3A). The observed enrichment scores, as calculated by comparing mutations in genes prior to and after selection, were highly significant (p-values between 4 × 10−6 and 1 × 10−307, Figure 3B). Among the genes identified were TMEM181 and SGMS1, already found in a haploid screen using E. coli CDT1. None of the 10 other host factors had been previously implicated in CDT intoxication. These host gene products are unlikely to act as generic entry factors for pathogens, because none of them were enriched in any of the other phenotypic screens performed to date, e.g. for diphtheria toxin, ricin or reovirus (Figure 3B and data not shown). Their role in intoxication or even the function of most of the newly identified factors is not known. With the exception of ERP44, a soluble oxidoreductase that resides in the ER lumen13, they all are integral membrane proteins as inferred from their predicted amino acid sequence. They reside either in the plasma membrane and/or in the endomembrane system, including Golgi (GLG1)14, ER lumen (ERP44) and transport vesicles (TMED2 and TMED10)15. CDTs of different origin require distinct sets of host factors, with sphingomyelin synthase 1 (SGMS1) being the only host factor common to all 4 screens. E. coli CDT requires the G-protein coupled receptor homolog TMEM181, sphingomyelin synthase 1 (SGMS1), Golgi glycoprotein 1 (GLG1) and the vacuolar ATPase subunit 2 (ATP6V0A2). A. actinomycetemcomitans CDT shows a similar pattern but does not require TMEM181 and is critically reliant on the presence of a synaptogyrin 2 (SYNGR2), an ubiquitously expressed integral membrane protein16. C. jejuni has evolved dependency on a different set of host factors compared to the other CDTs. Besides proteins involved in vesicular transport, three plasma membrane proteins were identified as essential: TMEM127, a familial tumor suppressor gene17, GPR107, an uncharacterized G-protein coupled receptor and TM9SF4, linked to resistance to pathogenic gram-negative bacteria by preventing phagocytosis in Drosophila18.
Figure 2. Host factors used by different CDTs.
A. CDTs are tripartite protein toxins that show the highest sequence conservation in the catalytic CdtB-subunit and lower conservation in the cell binding CdtA and CdtC subunits. Sequence conservation of the four CDTs used in this study is depicted using the H. ducreyi CTD crystal structure11. B. CDTs are secreted by pathogenic bacteria that infect and colonize the human body at different anatomical locations. C. CDTs from different bacterial species induce a G2/M cell cycle arrest in both HeLa cells and KBM7 cells. D. PhITSeq screens performed with CDTs secreted by different bacteria. The Y-axis represents the proximity index calculated for each insertion. The X-axis represents the chromosomes in which each insertion is located. N indicates the number of insertions found in each gene.
Figure 3. Genes linked to different phenotypes.
A. Gene trap insertions identified in loci essential for CDT intoxication. A color code distinguishes gene trap insertions that were enriched using distinct CDTs: A. actinomycetemcomitans in green, E. coli in red, H. ducreyi in yellow and C. jejuni in blue. B. Loci linked to 12 separate phenotypes. Cells were exposed to small molecule inhibitors of the Bcl2-family (ABT-737), chk1-kinase activity (AZD7762), Bcr-Abl-activity (imatinib), or DNA methylation (decitabine). Additional phenotypic selections were done using biological agents including TRAIL, CDTs, diphtheria toxin, ricin toxin and reovirus. Gene trap insertions in exonic sequences or in the sense orientation of genes were counted per individual selection. Enrichment p-value was calculated for each gene locus by comparing this number to the number of insertions identified in the same locus within the unselected cell population. Each screen resulted in a distinct set of between 1 and 8 genes with high significance. These include already known entry factors used by pathogens such as the entry receptor for diphtheria toxin (HBEGF), the reovirus receptor (F11R) and an enzyme involved in carbohydrate synthesis required for ricin entry (MGAT2). It also includes downstream effectors of kinases for example CDC25A in a screen with a Chk1 inhibitor or PTPN1 and PTPN12 identified by BCR-ABL inhibition using imatinib. Strong resistance against decitabine was observed in cells containing mutations in deoxycytidine kinase (DCK), the rate-limiting kinase for activation of several nucleoside analogs20.
These results are consistent with the notion that CDTs from different species have evolved partially overlapping yet distinct routes of intoxication. Remarkably, CDTs secreted by bacteria that colonize different anatomical sites may still show very similar host factor requirements (e.g. H. ducreyi and A. actinomycetemcomitans), whereas CDTs from bacteria that occupy similar niches (e.g. E. coli and C. jejuni) can have very distinct requirements. To determine if the genes identified here are also involved in cell intoxication by CDTs in other cell types we followed up on ATP6V0A2. Our screen suggests that this v-ATPase subunit is required for intoxication by E. coli CDT whereas intoxication by CDT derived from C. jejuni appears to be less dependent on this host factor. We treated HeLa cells with concanamycin A, a specific inhibitor of v-ATPase19 and exposed them to either E. coli CDT or C. jejuni CDT. Concanamycin treatment abolished the ability of E. coli CDT to induce cell cycle arrest at the G2/M phase of the cell cycle, whereas the action of C. jejuni CDT was not impaired, in agreement with the differential requirement for this host factor suggested by our screens (Figure S2). Thus, comparative profiling using PhITSeq identified 10 novel host factors required for a family of bacterial toxins and provides a solid genetic framework for further study of the molecular mechanisms of host-pathogen interactions. With the identity of these host factors revealed, it should now be possible to examine their involvement in tissue damage inflicted by CDTs in vivo, at the actual anatomical sites they target.
The PhITSeq approach is scalable and allows precise comparative analyses by using the same well-characterized library of mutants for multiple phenotypic selections. Here we present 12 examples of independent phenotypic screens, not only using different pathogens but also a collection of targeted cancer therapeutics (Figure 3B). Each screen yields a select number of hits. In the examples of TRAIL, ABT-737, decitabine, AZD7762, diphtheria toxin and reovirus, each of the hits correspond to established key regulators of the phenotype, including cell surface receptors (HBEGF, F11R), downstream effector molecules (FADD, CASP8) and a drug metabolizing enzyme (DCK). These findings suggest that these screens are unlikely to be confounded by large numbers of false positive results. In the case of CDTs, all significant hits are either transmembrane proteins or proteins involved in membrane trafficking events. These might act as toxin receptors or be involved in intracellular trafficking of receptor-toxin complexes. The clusters of insertion sites found in the various selected cell populations are located within genes and are predicted to disrupt gene function, based on their orientation and location. It is therefore likely that the gene-trap insertions directly affect the genes into which they insert, rather than perturb neighboring genes through action at a distance. Indeed, the retroviral vectors used for mutagenesis themselves lack strong promoter or enhancer sequences, disfavoring “long distance” effects. Our screens are unlikely to identify factors that are either essential for cell viability in the absence of selection or that show genetic redundancy (such as the TRAIL receptors). Indeed, essential genes can be recognized by a paucity of sense orientation gene-trap insertions in the mutagenized unselected cell population. A clear example of this is BCR-ABL (Figure S3). Genes whose disruption severely reduces cell fitness without outright cytolethal effects would be underrepresented in our screens and therefore may fail to reach levels of high significance, even when involved in the phenotype of interest. In contrast to RNA interference based screens, which can be applied to many cell types, our approach, for now, relies on the use of one particular human near-haploid cell line. Although many cellular phenotypes should be accessible in these cells the generation or isolation of additional haploid cell types, for example by reprogramming, would be useful.
Methods Online
Generation of mutagenized cells
Gene trap virus was produced by transfection of 293T cells in T175 dishes using turbofectin 8 (Origene) with a mixture of pGT-GFP, pGT-GFP+1 and pGT-GFP+21 (6.7 μg) combined with 1.7 μg pAdvantage, 2.6 μg CMV-VSVG and 4 μg Gag-pol. The virus-containing supernatant was concentrated using ultracentrifugation for 1.5 h at 25,000 r.p.m. in a Beckman SW28 rotor. Batches of mutant KBM7 cells (100 million cells in total) were prepared by transduction in 24-well tissue-culture dishes containing 1.5 million cells per well using spin infection for 45 minutes at 2,000 rpm in the presence of 8ug/ml protamine sulfate. Screens were started at least 6 days after gene trap infection.
Sequence analysis of the gene trap insertion sites in the unselected mutagenized cell population
Genomic DNA was isolated from ~40 million cells using QIAamp DNA mini kit (Qiagen) according to manufacturer’s protocol. After digestion with NlaIII or MseI, genomic DNA was used as template for a linear PCR (99 cycles) using a 5′-biotinylated primer (5′-GGTCTCCAAATCTCGGTGGAAC-3′) annealing to the GT-GFP gene trap vector. The resulting single stranded DNA (ssDNA) product contains the 5′LTR of the retroviral vector followed by the genomic DNA sequence flanking the insertion site ending at either an NlaIII or MseI restriction site. This product was purified using streptavidin-coated beads (Dynabeads M-280; Invitrogen) and a 5′-phosphorylated and 3′-modified (dideoxycytidine, ddC) ssDNA linker was ligated to the 3′end of the product using a ssDNA ligase (CircLigase II, Epicenter Biotechnologies). The linker (5′-TCGTATGCCGTCTTCTGCTTGACTCAGTAGTTGTGCGATGGATTGATG-3′) contains adaptor sequence II required for sequencing using the Genome Analyzer (Illumina). After ligation the product was purified and used as template for a PCR that adds adaptor sequence I with primers (5′-AATGATACGGCGACCACCGAGATCTGATGGTTCTCTAGCTTGCC-3′) and (5′-CAAGCAGAAGACGGCATACGA-3′). The PCR generates products of different lengths representing the abundance of individual insertion sites present in the sample, which visualizes as a smear on an agarose gel. After column purification (Qiaquick PCR purification kit, Qiagen), 8pM of the product was sequenced on one lane of an Illumina Model GA2X Genome Analyzer using a custom sequencing primer annealing to the extreme end of the 5′LTR (5′-CTAGCTTGCCAAACCTACAGGTGGGGTCTTTCA-3′) resulting in sequences directly flanking the site of insertion of the gene trap vector.
Analysis of gene trap insertions in the unselected mutagenized cell population
The 36 base pair (bp) sequences in the FASTQ data file were aligned to the human genome (hg18) using Bowtie alignment software21. We used stringent criteria to exclude ambiguous alignments by not allowing any mismatches in the full 36 bp sequence and by excluding all sequences that align non-uniquely to the human genome. Of the sequence reads 59% aligned uniquely over the full 36 bp sequence, 33% were excluded because they contained one or more mismatches and 8% were excluded because of non-unique alignment. Using these criteria, we obtained an insertion data table that contains ~900.000 independent insertion sites mapped to the human genome (Table S1). Based on their position on the human genome, insertion sites were identified as located in genomic regions annotated to contain genes. We further classified these insertions as being in the sense or antisense orientations compared to the gene. This was done by intersecting (based on chromosome intervals) the insertion database with a data table containing the chromosomal coordinates of Refseq22 annotated genomic regions retrieved from the UCSC genome table browser database23, using BEDTools software24. The resulting gene insertion data table contains ~450.000 insertions meeting these criteria (Table S2).
To determine the percentage of expressed genes that contain insertions we used gene expression data from KBM7 cells (GEO: GSE7114)7. The present/absent calls of 5 replicates were summarized, coupled to gene symbol and this table was joined to the gene insertion data table (Table S3). From this table we derive the percentage of expressed (P, present), marginally expressed (M, marginal) and non-expressed (A, absent) genes that contain insertions. Discrepancies of gene symbol annotation of the Affymetrix platform with the Refseq data table are indicated and excluded from the analysis.
Treatment of cells for phenotypic enrichments
For a phenotypic enrichment screen, 100 million cells were incubated with the selection agent. In a typical screen the resistant cells were expanded over the course of ~20 days. When the cells were expanded to 30 million cells, cell debris was removed by multiple wash steps with PBS and genomic DNA was isolated to map the insertion sites. In general the selection agent was present during the course of the experiment. Concentrations used were: diphtheria toxin (Sigma Aldrich) 400 ng/ml, ricin (Vector laboratories) 40 ng/ml, ABT-737 (Selleck Chemicals) 15 μM, AZD7762 (Selleck Chemicals) 250 nM, and decitabine (Tocris Bioscience) 15 μM. Imatinib was added at 2 μM for 4 days after which it was further diluted to 600 nM followed by a 20 day incubation. Recombinant TRAIL (Sigma Aldrich) was added at a concentration of 1 μg/ml for 7 days after which it was diluted two-fold and surviving cells were expanded. The cytolethal distending toxins were produced as described below. The following dilutions were used for the screen: CDT E. Coli 0.4 μl/ml, CDT C. jejuni 2.0 μl/ml, CDT H. ducreyi 0.05 μl/ml and A. actinomycetemcomitans 0.01 μl/ml.
Cloning and production of cytolethal distending toxins
E.coli CDT
The construct for the full operon of E.coli CDT was generously provided by Dr. James Kaper, University of Maryland School of Medicine, Baltimore, MD (pDS7.96)25. A starter culture of E. coli BL21 cells transformed with this plasmid was used to inoculate 500 ml of sterile M9 minimal media (SIGMA) supplemented with 1% (w/v) glucose. After 21 hrs at 37°C with vigorous shaking the supernatant was filter sterilized (0.22 μm pore size filter). The filtrate was concentrated using a Centricon-Plus 70 (Millipore) with a cut-off filter of 30kDa to a final volume of 5ml. The buffer was exchanged to PBS using a PD10 desalting column (GE Healthcare Life Sciences) and filter sterilized.
A. actinomycetemcomitans CDT
The operon sequence between subunits CdtA and CdtC was amplified from genomic DNA of Aggregatibacter actinomycetemcomitans (ATCC 700685D-5) using the following forward: 5′-ATCTAAGGAGAGGTACAATGAAA-3′ and reverse 5′-TTAGCTACCCTGATTTCTCC-3′ primers. The PCR product was purified from agarose gel using a QIAquick Gel Extraction Kit (Qiagen) and cloned into the pGemTeasy vector (Promega). A clone displaying the 5′ end in the orientation of the T7 promoter was used as template to introduce a Shine-Delgarno sequence upstream the initial methionine via Quickchange® II Site-Directed Mutagenesis (Stratagene) and the following primer: 5′-GGGCCCGACGTCGCTTAACTTTAAGAAGGAGCTCCGGCCGCCATGG-3′. A starter culture of E. coli BL21 (DE3) transformed with the resulting construct was used to inoculate 250ml M9 minimal media (SIGMA) supplemented with 1% (w/v) glucose. When the culture reached an optical density of ~0.4 at 600nm it was induced with (IPTG 0.5mM) and incubated for 5hr at 37°C with vigorous shaking. The culture was centrifuged at 10,000×g for 15min in a Sorvall RC6 PLUS centrifuge (SLA1500 rotor). The resultant supernatant was filtered and concentrated following the protocol described for E. coli CDT.
C. jejuni CDT
Individual toxin subunits were amplified from genomic DNA (ATCC 43432D-5), thereby removing secretion signal sequences, using the following primers: CdtA 5′-GCGCCATGGGATGTTCTTCTAAATTTG-3′ and 5′-CGCCTCGAGTCGTACCTCTCCTTGGCG-3′, CdtB 5′-GCGCCATGGGAAATTTAGAAAATTTTA-3′ and 5′-CGCCTCGAGAAATTTTCTAAAATTTAC-3′, CdtC 5′-GCGCCATGGGAACTCCTACTGGAGATT-3′ and 5′-CGCCTCGAGTTCTAAAGGGGTAGCAGC-3′, and 5′-CGCCTCGAGGCTACCCTGATTTCTTCG-3′. Each subunit was cloned into pET28(+) vector (Novagen) between the NcoI and XhoI restriction sites to equip each subunit with a C-terminal His6 tag. Individual subunits were expressed separately in E. coli BL21 grown in TB medium (Terrific Broth, Sigma) supplemented with 1 % glycerol at 37°C under agitation (220 rpm, Innova 5000 Gyrotory Tier Shaker, New Brunswick Scientific). Expression was induced in mid-log growth phase with 0.3 mM IPTG (Isopropyl-β-D-1-thiogalactopyranoside, Sigma). After an expression time of 5 hours, the three CDT subunit expression cultures were pooled, centrifuged (6000 × g, 15 minutes, Avanti J-E Centrifuge, Beckman Coulter) and pellets freeze-thawed. Proteins were solubilized under denaturing conditions at 37°C (8 M urea, 20 mM HEPES (N-[2-Hydroxyethyl]piperazine-N-[2-ethanesulphonic acid]) pH 7.5, 200 mM NaCl, 0.1% (v/v) TritonX-100, 2.5 mM dithiothreitol (DTT), 2 mM ethylenediaminetetraacetic acid (EDTA), purified at 4°C using nickel chelating affinity resin (Ni-NTA Agarose, Qiagen) and eluted with 0.3 M imidazole. CDT was co-refolded by stepwise dilution of 8M urea to 1 M urea with 20 mM HEPES pH 7.5, 200 mM NaCl, 2.5 mM DTT, 2 mM (EDTA), protease inhibitor (Complete Mini, Roche) buffer at 4°C on a stirring plate. Following a second nickel chelating affinity resin purification and concentration step (Amicon Ultra Centrifugal filter units, MWCO = 30 kDa, Millipore), the holotoxin was further purified with size exclusion chromatography (Äkta FPLC gel filtration system, HiLoad 16/60 Superdex 200 column, GE Healthcare). Protein-containing fractions were tested for toxicity by cell cycle analysis of intoxicated HeLa cells. Toxic fractions were pooled and concentrated.
H. ducreyi CDT
Individual toxin subunits were amplified from genomic DNA (ATCC 700724D-5), thereby removing secretion signal sequences, using the following primers: CdtA 5′-GCGCCATGGGATGTTCATCAAATCAAC-3′ and 5′-CGCCTCGAGATTAACCGCTGTTGCTTC-3′, CdtB 5′-GCGCCATGGGAAACTTGAGTGACTTCA-3′ and 5′-CGCCTCGAGGCGATCACGAACAAAACT-3′, CdtC 5′-GCGCCATGGGAAGTCATGCAGAATCAA-3′ and 5′-CGCCTCGAGGCTACCCTGATTTCTTCG-3′. Cloning and production of the toxin was performed similar to the protocol described for C. jejuni CDT.
Sequencing of gene trap insertion sites in phenotypically enriched populations
The lower complexity of selected cell populations allowed the insertion sites to be mapped by inverse PCR reaction. Genomic DNA was isolated from ~30 million cells using QIAamp DNA mini kit (Qiagen) according to manufacturer’s protocol. After digestion with MseI or NlaIII, DNA was ligated using T4 DNA ligase to circularize the fragments. After column purification (Qiaquick PCR purification kit, Qiagen), a PCR was performed using outward facing primers annealing internally in the gene trap vector thereby introducing the Illumina adaptor sequences I and II. The primers used for ligated DNA digested with NlaIII were (5′-AATGATACGGCGACCACCGAGATCTGATGGTTCTCTAGCTTGCC-3′ and 5′-CAAGCAGAAGACGGCATACGACCCAGGTTAAGATCAAGGTC) and for ligated DNA digested with MseI were (5′-AATGATACGGCGACCACCGAGATCTGATGGTTCTCTAGCTTGCC-3′ and 5′-CAAGCAGAAGACGGCATACGACGTTCTGTGTTGTCTCTGTCTG). After column purification, 8pM of the products was sequenced on one lane of an Illumina Model GA2X Genome Analyzer using a custom sequencing primer annealing to the extreme end of the 5′LTR (5′-CTAGCTTGCCAAACCTACAGGTGGGGTCTTTCA-3′).
Identification of unique insertion sites in the phenotypically enriched populations
The 36 base pair (bp) sequences in the FASTQ data file were mapped to the human genome (hg18) using Bowtie alignment software21. Because the complexity of insertions is significantly lower in the selected pools compared to the unselected pools and because PCR amplification is not unbiased, some individual insertion sites are sequenced in very high frequency. An unwanted consequence of this is the appearance of sequencing errors. Stringent criteria were used to identify unique insertion sites. No mismatches were allowed in the 36 bp sequence and the sequence should uniquely align to the human genome even when 1 or 2 mismatches are allowed. If two insertions align 1 or 2 base pairs apart only one is retained. For each selection a data table was produced containing these insertion sites (Tables S4–16). To distinguish genomic regions with a high density of insertions, we define the proximity index for a given insertion as the inverse value of the average distances with its neighboring insertion sites. The inverse value is calculated from the average distance (in base pairs) between the given insertion and the two neighboring upstream insertions and the two next downstream insertion sites. This method of analysis pinpoints insertion-rich regions in phenotypic screens and includes all insertions (sense and antisense). It provides a graphical illustration of insertion site clustering and thereby allows non-annotated elements also to be identified. An alternative method of analysis focuses on the insertions in a given screen that are present in genes and compares these to the unselected population. To create a control dataset, the FASTQ data file for the unselected population described above was analyzed using the same criteria as for the selected pools. The total number of insertions in genes and the number of insertions per individual gene were counted by intersecting the insertions with the data table containing chromosomal coordinates of Refseq annotated genes. If an insertion was located in a genomic region shared by multiple transcripts of the same gene, it only counted once. For a given screen, the number of inactivating mutations (=sense orientation or present in exon) per individual gene was counted as well as the total number of inactivating insertions for all genes. Enrichment of a particular gene in a particular screen was calculated by comparing how often that gene was mutated in the screen compared to how often the genes carries an insertion in the control dataset. For each gene a p-value (corrected for false discovery rate) was calculated using the one-sided Fisher exact test run in the R software environment. In some cases the p-value was lower than the R software could report. In these cases the numerical value was set to the smallest non-zero normalized floating-point number R could report (~1 × 10 −307).
Supplementary Material
Supplemental figure 1. Coverage of gene-trap mutagenesis. The number of gene trap insertions mapped in core members of the intrinsic and extrinsic cell death pathways was examined in the unselected mutagenized cell population to determine the gene coverage of our mutagenesis approach. The number of insertions identified is indicated above the gene name in red.
Supplemental figure 2. Differential requirement for the v-ATPase by E. coli CDT and C. jejuni CDT in HeLa cells. HeLa cells were treated with either E. coli or C. jejuni CDTs in the presence or absence of 50 nM concanamycin A to inhibit the v-ATPase. Subsequently the ability of these toxins to induce a G2/M cell cycle arrest was examined using flow cytometry.
Supplemental figure 3. Orientation bias of gene traps in essential genes in the unselected population of mutagenized cells. Gene trap insertions in BCR, encoding the N-terminal domain of the p210 bcr-abl fusion protein that is required for cell viability of the CML cell line KBM7. Gene trap insertions in the same transcriptional orientation as the gene (sense) are depicted in green and in the antisense orientation are drawn in red. Note the enrichment for antisense orientation insertions suggesting that cells containing sense insertions are lost during cell growth.
Acknowledgments
We would like to thank D. Sabatini and J. Roix for critical reading of this manuscript, S. Boulant, M. Nibert and R. Rooswinkel for providing reagents and T. DiCesare for graphics support. I.W. was supported by a PhD fellowship from the Boehringer Ingelheim Fonds. T. R. B. was supported by NIH grant R21-HG004938-01.
Footnotes
Author contributions
J.E.C., C.P.G., I.W., V.A.B., M.V., C.S., M.K.M., S.M.N. and T.R.B designed and performed experiments. B.Y., G.B., J.E.C., V.A.B. and T.R.B. were involved in the bioinformatics. J.E.C., C.P.G., I.W., V.A.B., H.L.P. and T.R.B. wrote the manuscript.
Competing financial interests
J. C. and T.R.B. are named inventors on a patent application on technology described in this manuscript. S.M.N. and T.R.B. are co-founders of an early-stage startup company involved in haploid genetic approaches.
References
- 1.Carette JE, et al. Haploid genetic screens in human cells identify host factors used by pathogens. Science. 2009;326:1231–1235. doi: 10.1126/science.1178955. [DOI] [PubMed] [Google Scholar]
- 2.Ge Z, Schauer DB, Fox JG. In vivo virulence properties of bacterial cytolethal-distending toxin. Cell Microbiol. 2008;10:1599–1607. doi: 10.1111/j.1462-5822.2008.01173.x. [DOI] [PubMed] [Google Scholar]
- 3.Mazurkiewicz P, Tang CM, Boone C, Holden DW. Signature-tagged mutagenesis: barcoding mutants for genome-wide screens. Nature Reviews Genetics. 2006;7:929–939. doi: 10.1038/nrg1984. [DOI] [PubMed] [Google Scholar]
- 4.Gawronski JD, Wong SMS, Giannoukos G, Ward DV, Akerley BJ. Tracking insertion mutants within libraries by deep sequencing and a genome-wide screen for Haemophilus genes required in the lung. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:16422–16427. doi: 10.1073/pnas.0906627106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Opijnen T, Bodi KL, Camilli A. Tn-seq: high-throughput parallel sequencing for fitness and genetic interaction studies in microorganisms. Nature Methods. 2009;6:767–721. doi: 10.1038/nmeth.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brady T, et al. Integration target site selection by a resurrected human endogenous retrovirus. Genes and Development. 2009;23:633–642. doi: 10.1101/gad.1762309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bao F, et al. Comparative gene expression analysis of a chronic myelogenous leukemia c-ell line resistant to cyclophosphamide using oligonucleotide arrays and response to tyrosine kinase inhibitors. Leukemia Research. 2007;31:1511–1520. doi: 10.1016/j.leukres.2007.03.002. [DOI] [PubMed] [Google Scholar]
- 8.Oltersdorf T, et al. An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature. 2005;435:677–681. doi: 10.1038/nature03579. [DOI] [PubMed] [Google Scholar]
- 9.van Delft MF, et al. The BH3 mimetic ABT-737 targets selective Bcl-2 proteins and efficiently induces apoptosis via Bak/Bax if Mcl-1 is neutralized. Cancer Cell. 2006;10:389–399. doi: 10.1016/j.ccr.2006.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hauck P, Chao BH, Litz J, Krystal GW. Alterations in the Noxa/Mcl-1 axis determine sensitivity of small cell lung cancer to the BH3 mimetic ABT-737. Molecular Cancer Therapeutics. 2009;8:883–892. doi: 10.1158/1535-7163.MCT-08-1118. [DOI] [PubMed] [Google Scholar]
- 11.Lara-Tejero M, Galan JE. A bacterial toxin that controls cell cycle progression as a deoxyribonuclease I-like protein. Science. 2000;290:354–357. doi: 10.1126/science.290.5490.354. [DOI] [PubMed] [Google Scholar]
- 12.Nesic D, Hsu Y, Stebbins CE. Assembly and function of a bacterial genotoxin. Nature. 2004;429:429–433. doi: 10.1038/nature02532. [DOI] [PubMed] [Google Scholar]
- 13.Higo T, et al. Subtype-specific and ER lumenal environment-dependent regulation of inositol 1,4,5-trisphosphate receptor type 1 by ERp44. Cell. 2005;120:85–98. doi: 10.1016/j.cell.2004.11.048. [DOI] [PubMed] [Google Scholar]
- 14.Steegmaier M, Borges E, Berger J, Schwarz H, Vestweber D. The E-selectin-ligand ESL-1 is located in the Golgi as well as on microvilli on the cell surface. Journal of Cell Science. 1997;110:687–694. doi: 10.1242/jcs.110.6.687. [DOI] [PubMed] [Google Scholar]
- 15.Gommel D, et al. p24 and p23, the major transmembrane proteins of COPI-coated transport vesicles, form hetero-oligomeric complexes and cycle between the organelles of the early secretory pathway. Febs Letters. 1999;447:179–185. doi: 10.1016/s0014-5793(99)00246-x. [DOI] [PubMed] [Google Scholar]
- 16.Janz R, Sudhof TC. Cellugyrin, a novel ubiquitous form of synaptogyrin that is phosphorylated by pp60(c-src) Journal of Biological Chemistry. 1998;273:2851–2857. doi: 10.1074/jbc.273.5.2851. [DOI] [PubMed] [Google Scholar]
- 17.Qin YJ, et al. Germline mutations in TMEM127 confer susceptibility to pheochromocytoma. Nature Genetics. 2010;42:229–U231. doi: 10.1038/ng.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bergeret E, et al. TM9SF4 is required for Drosophila cellular immunity via cell adhesion and phagocytosis. Journal of Cell Science. 2008;121:3325–3334. doi: 10.1242/jcs.030163. [DOI] [PubMed] [Google Scholar]
- 19.Dröse S, et al. Inhibitory effect of modified bafilomycins and concanamycins on P- and V-type adenosinetriphosphatases. Biochemistry. 1993;32:3902–3906. doi: 10.1021/bi00066a008. [DOI] [PubMed] [Google Scholar]
- 20.Momparler RL. Pharmacology of 5-Aza-2′-deoxycytidine (decitabine) Seminars in Hematology. 2005;42:S9–S16. doi: 10.1053/j.seminhematol.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 21.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biology. 2009;10 doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Research. 2007;35:D61–D65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rhead B, et al. The UCSC Genome Browser database: update 2010. Nucleic Acids Research. 2010;38:D613–D619. doi: 10.1093/nar/gkp939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scott DA, Kaper JB. Cloning and Sequencing of the Genes Encoding Escherichia-Coli Cytolethal Distending Toxin. Infection and Immunity. 1994;62:244–251. doi: 10.1128/iai.62.1.244-251.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental figure 1. Coverage of gene-trap mutagenesis. The number of gene trap insertions mapped in core members of the intrinsic and extrinsic cell death pathways was examined in the unselected mutagenized cell population to determine the gene coverage of our mutagenesis approach. The number of insertions identified is indicated above the gene name in red.
Supplemental figure 2. Differential requirement for the v-ATPase by E. coli CDT and C. jejuni CDT in HeLa cells. HeLa cells were treated with either E. coli or C. jejuni CDTs in the presence or absence of 50 nM concanamycin A to inhibit the v-ATPase. Subsequently the ability of these toxins to induce a G2/M cell cycle arrest was examined using flow cytometry.
Supplemental figure 3. Orientation bias of gene traps in essential genes in the unselected population of mutagenized cells. Gene trap insertions in BCR, encoding the N-terminal domain of the p210 bcr-abl fusion protein that is required for cell viability of the CML cell line KBM7. Gene trap insertions in the same transcriptional orientation as the gene (sense) are depicted in green and in the antisense orientation are drawn in red. Note the enrichment for antisense orientation insertions suggesting that cells containing sense insertions are lost during cell growth.