Abstract
Muscle wasting in catabolic patients is in part mediated by glucocorticoids and is associated with increased expression and activity of the transcription factor C/EBPβ. It is not known, however, if C/EBPβ is causally linked to glucocorticoid-induced muscle atrophy. We used dexamethasone-treated L6 myoblasts and myotubes to test the role of C/EBPβ in glucocorticoid-induced expression of the muscle-specific ubiquitin ligases atrogin-1 and MuRF1, protein degradation, and muscle atrophy by transfecting cells with C/EBPβ siRNA. In myoblasts, silencing C/EBPβ expression with siRNA inhibited dexamethasone-induced increase in protein degradation, atrogin-1 and MuRF1 expression, and muscle cell atrophy. Similar effects of C/EBPβ siRNA were seen in myotubes except that the dexamethasone-induced increase in MuRF1 expression was not affected by C/EBPβ siRNA in myotubes. In additional experiments, overexpressing C/EBPβ did not influence atrogin-1 or MuRF1 expression in myoblasts or myotubes. Taken together, our observations suggest that glucocorticoid-induced muscle wasting is at least in part regulated by C/EBPβ. Increased C/EBPβ expression alone, however, is not sufficient to upregulate atrogin-1 and MuRF1 expression.
Keywords: Skeletal muscle, Muscle wasting, Glucocorticoids, Transcription factors, Ubiquitin ligases, Proteolysis
Introduction
Loss of muscle mass is commonly seen in patients with sepsis, severe injury, and cancer (Hasselgren et al, 2005; Lecker et al, 2006). Muscle wasting in these conditions is mainly caused by accelerated ubiquitin-proteasome-dependent protein breakdown (Attaix et al, 2008; Murton et al, 2008), although inhibited protein synthesis may contribute as well (Lang et al, 2007). Studies suggest that increased expression of the ubiquitin ligases atrogin-1 and MuRF1 may be particularly important for the development of muscle atrophy in various catabolic conditions (Bodine et al, 2001; Gomes et al, 2001; Wray et al, 2003). Because stimulated transcription of the genes for atrogin-1 and MuRF1 as well as for multiple other components of the ubiquitin-proteasome pathway is essential for loss of muscle mass (Khal et al, 2005; Tiao et al, 1994), it is likely that factors regulating gene transcription are involved in muscle wasting. Indeed, there is evidence suggesting that expression and activity of the transcription factors NF-kB (Cai et al, 2004; Penner et al, 2001), AP-1 (Moore-Carrasco et al, 2006; Penner et al, 2001), and FOXO1 and 3a (Kamei et al, 2004; Sandri et al, 2004; Smith et al, 2010; Stitt et al, 2004), and of the nuclear cofactors p300 (Alamdari et al, 2010; Yang et al, 2005b, 2007) and PGC-1α and β (Arany et al, 2007; Menconi et al, 2010; Sandri et al, 2006), play important roles in the regulation of muscle mass in various catabolic conditions.
Previous studies suggest that the transcription factors C/EBPβ and δ may also be involved in muscle wasting (Penner et al, 2002; Yang et al, 2005a). In experiments in our laboratory, sepsis in rats resulted in increased expression and activity of C/EBPβ and δ in skeletal muscle and evidence was reported that the promoter regions of several genes in the ubiquitin-proteasome pathway contain multiple putative C/EBP binding sites (Penner et al, 2002). In additional experiments in the same study, treatment of rats with the glucocorticoid receptor antagonist RU38486 attenuated the sepsis-induced activation of C/EBPβ and δ, suggesting that sepsis-induced activation of these transcription factors was, at least in part, glucocorticoid-dependent (Penner et al, 2002).
Further support for a role of glucocorticoids in the upregulation of C/EBP transcription factors in atrophying skeletal muscle was found in experiments in which treatment of cultured muscle cells in vitro or rats in vivo with dexamethasone resulted in increased expression and DNA binding activity of C/EBPβ and δ (Yang et al, 2005a). Those observations are significant because other studies suggest that glucocorticoids are important regulators of ubiquitin-proteasome-dependent muscle proteolysis (Hasselgren, 1999; Tiao et al, 1996) and that dexamethasone-treated muscle cells can be used as a valid in vitro model of muscle wasting (Menconi et al, 2008). Indeed, in previous reports, dexamethasone-induced atrogin-1 and MuRF1 expression and protein degradation resulted in atrophy of cultured muscle cells (Bodine et al, 2001; Gomes et al, 2001; Menconi et al, 2008).
Although previous experiments provided evidence for an association between activation of C/EBP transcription factors and ubiquitin-proteasome-dependent protein degradation in dexamethasone-treated muscle cells (Menconi et al, 2008; Yang et al, 2005a), it is not known from those experiments whether there is a causative link between C/EBP transcription factors and muscle wasting. Cells in which a transcription factor is inactivated represent an important tool to determine the role played by the factor in the transcriptional control of candidate target genes and in the metabolic consequences of certain treatments. Here we used siRNA technique to test the role of C/EBPβ in dexamethasone-induced upregulation of atrogin-1 and MuRF1 expression, protein degradation, and atrophy in cultured L6 myoblasts and myotubes. Among the C/EBP transcription factors, we focused on C/EBPβ because the DNA binding activity, as determined by EMSA and supershift analysis, was upregulated to a greater extent for C/EBPβ than for C/EBPδ (Yang et al, 2005a) and the nuclear cofactor p300 interacted with C/EBPβ but not C/EBPδ in dexamethasone-treated myotubes (Yang et al, 2005b). Results in the present study suggest that dexamethasone-induced atrogin-1 expression, protein degradation, and atrophy in cultured muscle cells are at least in part regulated by C/EBPβ. Interestingly, C/EBPβ siRNA prevented the dexamethasone-induced increase in MuRF1 expression only in myoblasts, suggesting that the role of C/EBPβ in glucocorticoid-induced MuRF1 expression may be different in undifferentiated and differentiated muscle cells. Because results observed in differentiated myotubes are probably more relevant for skeletal muscle in vivo than results observed in myoblasts, the present observations suggest that MuRF1 may not be essential for glucocorticoid-induced C/EBPβ-regulated muscle wasting.
Materials and methods
Cell Culture
Experiments were performed in cultured L6 myoblasts and myotubes. L6 muscle cells, a rat skeletal muscle cell line (American Type Culture Collection, Manassas, VA), were maintained and cultured as described in detail recently (Menconi et al, 2008). In short, cells were grown in Dulbeco’s modified Eagle’s medium (DMEM; Mediatech Inc., Herndon, VA) supplemented with 10% fetal bovine serum (FBS; Sigma, St. Louis, MO) in 10% CO2 at 37°C. At approximately 80% confluence, cells were washed with phosphate buffered saline (PBS), pH 7.4, and removed with 0.25% trypsin in PBS. Cells were suspended in DMEM supplemented with 10% FBS and seeded in 6 or 12 well culture plates. Myoblasts were grown to approximately 60% confluence whereafter they were transfected with C/EBPβ siRNA or C/EBPβ expression plasmids as described below. When experiments were performed in myotubes, the myoblasts were grown in DMEM with 10% FBS until they reached approximately 80% confluence. The medium was then replaced with DMEM containing 2% FBS for induction of myotube formation. Formation of myotubes was observed after 5–9 days and after treatment with cytosine arabinoside (10 µM) for 24 h to remove any remaining myoblasts, the myotubes were transfected with C/EBPβ siRNA as described below.
Cell transfections
L6 myoblasts or myotubes were transfected with C/EBPβ siRNA or non-targeting (scrambled) siRNA (Dharmacon RNA Technologies, Lake Placid, NY) utilizing the transfection reagent Lipofectamine RNAiMAX (Invitrogen, Grand Island, NY). The siRNA constructs were added to the culture medium at a concentration of 165 nM in combination with Lipofectamine according to the manufacturer’s instructions. After 5 h, the medium was changed to fresh DMEM containing 10% FBS (when myoblasts were studied) or 2% FBS (when myotubes were studied). After 48 h, cell cultures were exposed to dexamethasone (1 µM) or corresponding concentration of solvent (0.1 % ethanol) for 24 h during which time protein degradation was measured as described below. In other experiments, myoblasts or myotubes were harvested after 24 h dexamethasone treatment for measurement of C/EBPβ and δ, atrogin-1 and MuRF1 mRNA levels by real-time PCR and C/EBPβ, atrogin-1, and MuRF1 protein levels by Western blotting. In other cell cultures, cell size was assessed as described below.
In additional experiments, L6 myoblasts were transfected with a C/EBPβ expression plasmid in order to test whether increased expression of C/EBPβ alone may be sufficient to upregulate atrogin-1 and MuRF1 expression. Transient transfections were performed using Lipofectamine 2000 (Invitrogen, Grand Island, NY) according to the manufacturer’s instructions. L6 myoblasts were seeded in 12-well plates at a density of 0.5–2 × 10(5) cells/well in DMEM containing 10% FBS and incubated at 37°C. Myoblasts were grown up to 70–80% confluence whereafter pcDNA3.1-C/EBPβ vector or pcDNA3.1-empty vector (1 µg/ml; kindly supplied by Dr. Daniel Tennen, Harvard Medical School) was added together with Lipofectamine 2000 for 6 h, followed by incubation in DMEM containing 10% FBS for 24 h. In other experiments, the medium was replaced with DMEM containing 2% FBS for induction of myotube differentiation and myotubes were studied after 5 days.
Promoter binding sites
Examination of potential C/EBP binding sites in the atrogin-1 and MuRF1 promoters was performed using Explain (Biobase, Beverly, MA). Promoter regions for rat atrogin-1 and rat MuRF1 were generated by extracting an interval of [−1000/+100] nucleotides around the canonical RefSeq transcription start site (UCSC Genome Browser, build rn4) (Karolchik et al, 2003). The transcription factor binding site search was limited to TRANSFAC motifs (Matys et al, 2006) matching the generic C/EBP family or the specific C/EBPβ binding sites, keeping putative binding sites with core match and site match scores ≥ 0.95.
Protein degradation
Protein degradation rates were determined by measuring the release of trichloroacetic acid (TCA)-soluble radioactivity during 24 h from proteins prelabeled with [3H]-tyrosine as described in detail previously (Menconi et al, 2008; Wang et al, 1998). Proteins in myoblasts or myotubes that had been transfected with non-targeting or C/EBPβ siRNA were labeled for 48 h with 1.0 µCi/ml of L-[3,5-3H]-tyrosine in DMEM containing 10% FBS (when myoblasts were studied) or 2% FBS (when myotubes were studied). After labeling of the proteins, the [3H]-tyrosine containing medium was removed, the cell cultures were rinsed, and fresh DMEM containing 10% or 2% FBS and 2 mM unlabeled tyrosine was added. The cell cultures were then treated for 24 h with 1 µM dexamethasone or corresponding concentration of solvent (0.1% ethanol). Protein degradation rates during the 24 h dexamethasone treatment were calculated as described in detail previously (Menconi et al, 2008; Wang et al, 1998) and expressed as %/24 h.
Real-time PCR
Messenger RNA levels for atrogin-1, MuRF1, C/EBPβ, and C/EBPδ were determined by real-time PCR performed as described in detail recently (Menconi et al, 2008; Yang et al, 2005a). In short, RNA was extracted by the acid guanidinium thiocyanate-phenol-chloroform method (Chomczynski and Sacchi, 1987) using TRI REAGENT (Molecular Research Center, Cincinnati, OH). TaqMan analysis using One Step PCR Master Mix Reagents Kit and subsequent calculations were performed with an ABI PRISM 7700 Sequence Detection System (Applied Biosystems). The primers and probes for rat atrogin-1, MuRF-1, C/EBPβ and C/EBPδ were designed by the use of Primer Express 1.5 design software (ABI, Foster City, CA) and were synthesized by Biosearch Technologies, Inc. (Novato, CA). The sequences for the forward, reverse, and double-labeled primer oligonucleotides for atrogin-1 were forward, 5'-CTT TCA ACA GAC TGG ACT TCT CGA-3', reverse, 5'- CAG CTC CAA CAG CCT TAC TAC GT-3', and double-labeled TaqMan probe, FAM-5'-TCG CAT CCT GGA TTC CAG AAG ATT CAA C-3'-TAMRA. The corresponding sequences for MuRF1 were forward, 5'-CGA CTC CTG CCG AGT GAC C-3', reverse, 5'-GCG TCA AAC TTG TGG CTC AG-3', and double-labeled Taq-Man probe, FAM-5'-AGG AAA ACA GCC ACC AGG TGA AGG AGG-3'-TAMRA. The corresponding sequences for C/EBPβ were forward, 5'-AAG ATG CGC AAC CTG GAG AC-3', reverse, 5'-CCT TCT TCT GCA GCC GCT C-3', and double-labeled TaqMan probe, FAM-5'-CAC AAG GTG CTG GAG CTG ACG GC-3'-TAMRA. The corresponding sequences for C/EBPδ were forward, 5'-AGA ACG AGA AGC TGC ATC AGC-3', reverse, 5'-TTG AAG AAC TGC CGG AGG C-3', and double-labeled TaqMan probe, FAM-5'-CAG CTC ACC CGG GAC CTG GC-3'-TAMRA.
Western blotting
Western blotting was performed as described in detail recently (Menconi et al, 2008; Yang et al, 2005a). For determination of C/EBPβ levels, a mouse monoclonal anti-rat C/EBPβ antibody was used as primary antibody at a dilution of 1:166 and a goat anti-mouse IgG horseradish peroxidase-conjugated antibody was used as secondary antibody at a dilution of 1:10,000 (both antibodies from Santa Cruz Biotechnology, Santa Cruz, CA). For determination of atrogin-1 protein levels, a rabbit polyclonal anti-mouse atrogin-1 antibody was used (1:1,000; kindly supplied by Dr. Stewart Lecker, Harvard Medical School). For determination of MuRF1 protein levels, a rabbit polyclonal anti-mouse MuRF1 antibody was used (1:1,000; kindly supplied by Regeneron Pharmaceuticals to Dr. Alfred Goldberg, Harvard Medical School, and used in the present experiments after permission from Regeneron Pharmaceuticals). In order to confirm equal loading, α-tubulin levels were determined by using a rabbit monoclonal anti-rat α-tubulin antibody at a dilution of 1:5000 (Sigma, St. Louis, MO) and a goat anti-rabbit IgG horseradish peroxidase-conjugated secondary antibody at a dilution of 1:5000 (Santa Cruz Biotechnology). Immunoreactive protein bands were visualized by chemiluminescence using the Western Lighting Kit (Perkin Elmer Life Sciences Inc., Boston, MA) followed by exposure to Kodak X-Omat blue film (Eastman Kodak, Rochester, NY). Molecular weights of protein bands were determined by Dual Precision molecular weight standards (BioRad, Hercules, CA). The protein bands were quantified by densitometry.
Cell size
After treating myoblasts for 24 h and myotubes for 48 h with 1 µM dexamethasone or corresponding concentration of solvent (0.1% ethanol), the cell cultures were photographed with a Nikon TE-DH100W phase contrast microscope (Nikon Instrument, Melville, NY) at 100X magnification. The cell size (area) was measured for at least 60 myoblasts from 10 random fields from each photograph using Image J software (NIH, Frederick, MD). Myotube diameters were measured for at least 60 myotubes from 10 random fields form each photograph using Image J software (NIH, Frederick, MD). The measurements were conducted in a “blinded” fashion with the investigator being unaware from which experimental group the cultures originated. Myoblast size was expressed as µm2 and myotube diameter as µm and the distribution of myoblasts with different size was depicted in histograms.
Statistics
Results are reported as means ± SEM. All experiments were repeated two or three times to ensure reproducibility. Statistical analysis was performed by Student’s t-test or ANOVA followed by Holm-Sidak’s test. p<0.05 was considered statistically significant.
Results
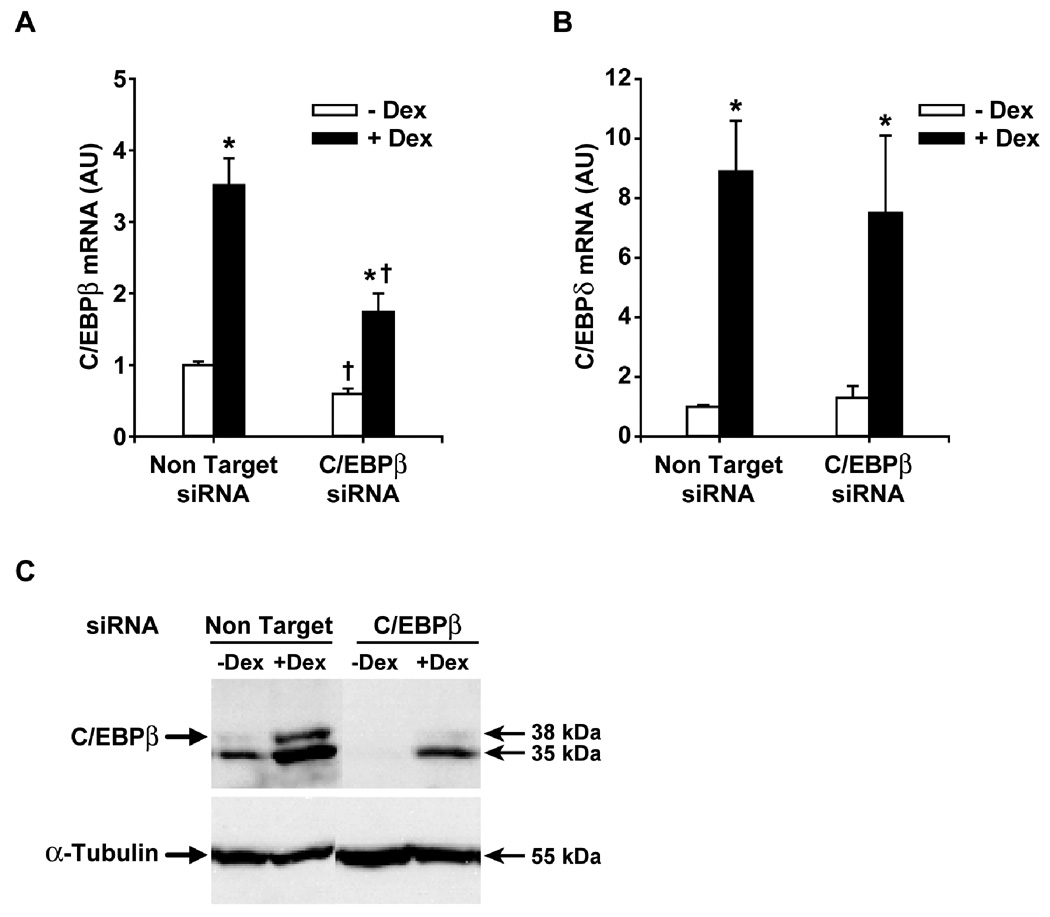
In initial experiments, we tested the effectiveness of C/EBPβ siRNA to downregulate the expression of C/EBPβ in control and dexamethasone-treated myoblasts. Transfection of the myoblasts with C/EBPβ siRNA resulted in an approximately 50% reduction of basal C/EBPβ mRNA levels (Fig 1A) but no reduction of C/EBPδ mRNA levels (Fig 1B). The effect of C/EBPβ siRNA on basal C/EBPβ mRNA levels was accompanied by a reduction of C/EBPβ protein to almost undetectable levels (Fig 1C). Treatment of the myoblasts with 1 µM dexamethasone for 24 h resulted in a 3.5-fold increase in C/EBPβ mRNA levels and a substantial increase in C/EBPβ protein levels (Fig 1A and C). Similar to previous studies in cultured muscle cells (Wei et al, 2007; Yang et al, 2005a), the 35 kDa isoform was the predominant isoform of C/EBPβ and treatment of the muscle cells with dexamethasone increased the expression of both the 35 and 38 kDa C/EBPβ isoforms. The effects of dexamethasone on C/EBPβ mRNA and protein expression were significantly blunted in myoblasts treated with C/EBPβ siRNA. In contrast, C/EBPδ mRNA levels in dexamethasone-treated myoblasts were not influenced by C/EBPβ siRNA (Fig 1B). Taken together, the results in Fig 1 suggest that transfecting L6 myoblasts with C/EBPβ siRNA resulted in a robust and specific downregulation of C/EBPβ mRNA and protein expression and blunted the dexamethasone-induced increase in C/EBPβ mRNA and protein levels. Thus, C/EBPβ siRNA-treated myoblasts provide a tool to study the role of C/EBPβ in glucocorticoid-induced muscle wasting-related changes.
Fig 1.
C/EBPβ siRNA reduces C/EBPβ expression and inhibits dexamethasone-induced C/EBPβ mRNA and protein levels in L6 myoblasts. (A) C/EBPβ mRNA levels in L6 myoblasts transfected with C/EBPβ siRNA or non-targeting siRNA and treated for 24 h with or without 1 µM dexamethasone. C/EBPβ mRNA in myoblasts transfected with non-targeting siRNA and cultured for 24 h in the absence of dexamethasone was arbitrarily set at 1.0. Results are means ± SEM with n=6 per group. *p<0.05 vs –Dex; +p<0.05 vs corresponding non-targeting group by ANOVA. (B) C/EBPδ mRNA levels in the same groups of myoblasts as depicted in panel A. Results are means ± SEM with n=6 per group. *p<0.05 vs –Dex. (C) C/EBPβ protein levels determined by Western blotting in L6 myoblasts transfected with C/EBPβ siRNA or non-targeting siRNA and treated for 24 h in the absence or presence of 1 µM dexamethasone. α-Tubulin levels were determined as loading control. Molecular weights on the right side of the blots were based on molecular weight markers used in the Western blots. Almost identical results were observed in three repeated experiments.
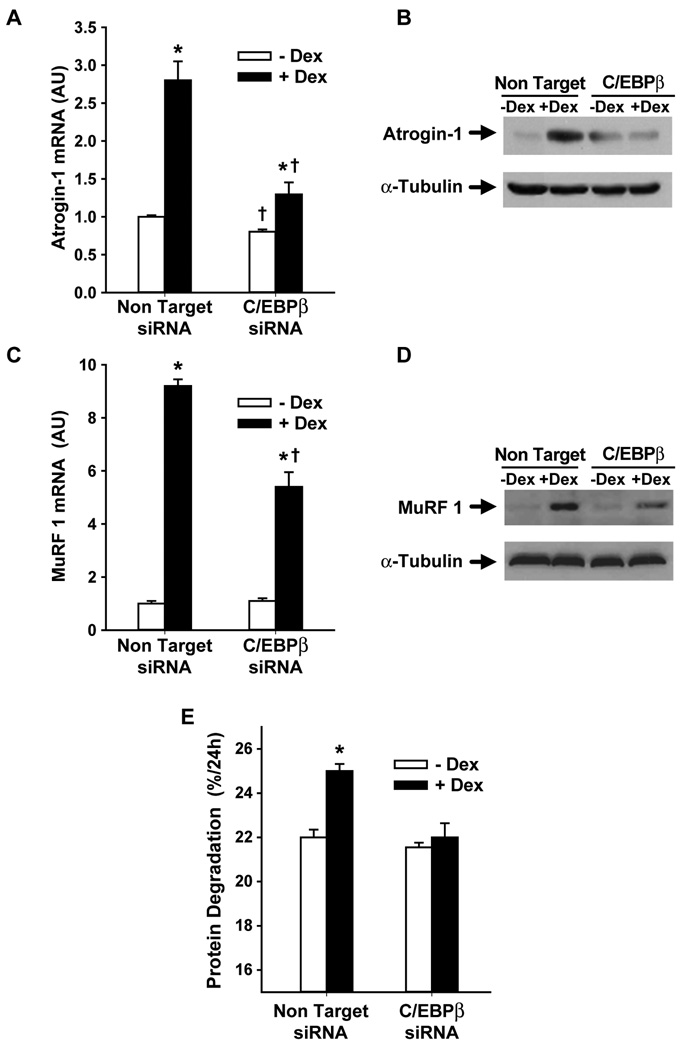
We next used C/EBPβ siRNA-treated myoblasts to test the role of C/EBPβ in dexamethasone-induced upregulation of atrogin-1 and MuRF1. Treatment of L6 myoblasts with 1 µM dexamethasone for 24 h resulted in a 2.5- to 3-fold increase in atrogin-1 mRNA levels (Fig 2A), similar to previous reports from our and other laboratories (Bodine et al, 2001; Gomes et al, 2001; Menconi et al, 2008). The increase in atrogin-1 mRNA levels was associated with a 2.3-fold increase (determined by densitometry of the Western blot) in atrogin-1 protein levels (Fig 2B). Importantly, the dexamethasone-induced increase in atrogin-1 mRNA and protein levels was substantially reduced in C/EBPβ siRNA-treated myoblasts. Of note, basal atrogin-1 mRNA levels, i.e., atrogin-1 mRNA levels in myoblasts that had not been treated with dexamethasone, were reduced by approximately 20% by C/EBPβ siRNA (Fig 2A).
Fig 2.
Silencing of C/EBPβ with C/EBPβ siRNA inhibits dexamethasone-induced increase in (A) atrogin-1 mRNA and (B) protein levels, (C) MuRF1 mRNA and (D) protein levels, and (E) protein degradation in cultured L6 myoblasts. L6 myoblasts were transfected with C/EBPβ siRNA or non-targeting siRNA followed by treatment for 24 h with or without 1 µM dexamethasone. Atrogin-1 and MuRF1 mRNA levels were determined by real-time PCR, atrogin-1 and MuRF1 protein levels were determined by Western blotting, and protein degradation rates were determined by measuring the release of TCA-soluble radioactivity from proteins labeled with [3H]-tyrsoine as described in Methods. Results are means ± SEM with n=6 per group. *p<0.05 vs –Dex; +p<0.05 vs corresponding non-targeting group by ANOVA.
Treatment of the myoblasts with dexamethasone resulted in an approximately 9-fold increase in MuRF1 mRNA levels accompanied by a 1.7-fold increase in MuRF1 protein levels (Fig 2C and D). These effects of dexamethasone were also inhibited in myoblasts transfected with C/EBPβ siRNA (Fig 2C and D). Basal levels of MuRF1 mRNA and protein were not influenced by C/EBPβ siRNA. Taken together, the results in Fig 2A–D suggest that C/EBPβ plays a role in glucocorticoid-induced upregulation of atrogin-1 and MuRF1 in muscle cells.
Although the present results provide strong evidence that glucocorticoid-induced upregulation of atrogin-1 and MuRF1 is C/EBPβ-dependent, it is not known at present whether the role of C/EBPβ reflected a direct or indirect effect. To shed some light on that question, we analyzed the rat atrogin-1 and rat MuRF1 promoters for putative C/EBP binding sites. The analysis showed 3 binding sites within the evolutionary conserved binding regions for atrogin-1 present on chromosome 7 (genomic coordinates: 9494264-74, 94942856-66, and 94942863-73) and 5 binding sites for MuRF1 on chromosome 5 (genomic coordinates: 153059398-08, 153059350-60, 153059351-61, 153059365-75, and 153059398-08). These sites exhibited high evolutionary conservation in 9 vertebrates (Sieple et al, 2005) suggesting that the predicted target sites may be functional. Although these observations suggest that C/EBPβ may bind directly to the atrogin-1 and MuRF1 promoters, it is important to point out that the present results of C/EBPβ dependency may also reflect an indirect effect, for example regulation by C/EBPβ of other gene(s), the products of which in turn may regulate the atrogin-1 and MuRF1 genes.
Because atrogin-1 and MuRF1 play important roles in regulating muscle proteolysis, we next tested the role of C/EBPβ in dexamethasone-induced protein degradation in cultured L6 myoblasts. Treatment of the myoblasts with dexamethasone resulted in an approximately 15% increase in protein degradation and this effect of dexamethasone was abolished in myoblasts that had been transfected with C/EBPβ siRNA (Fig 2E).
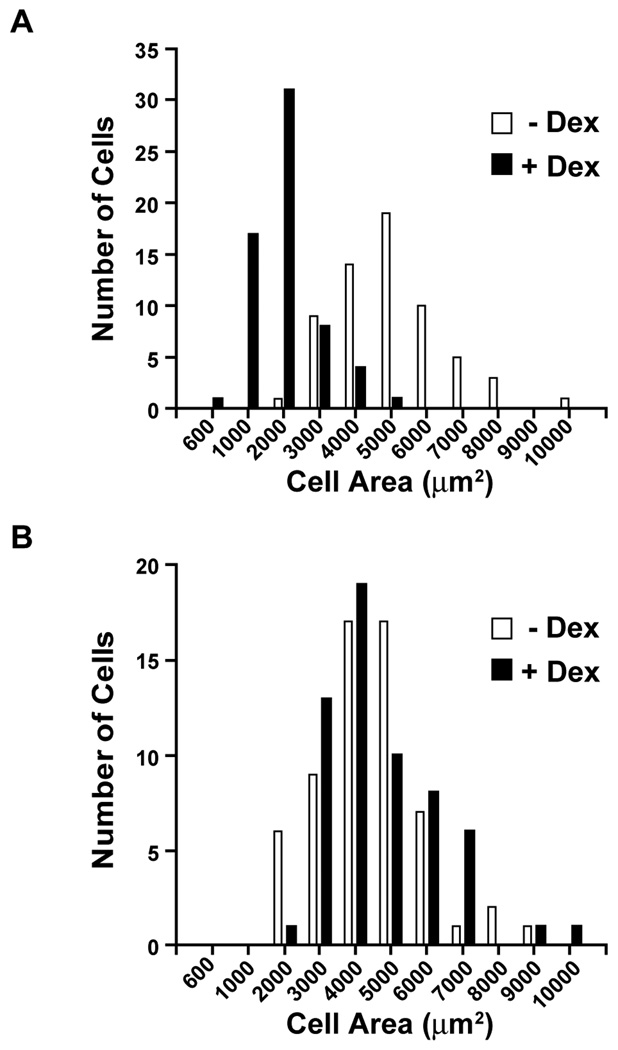
In order to test whether silencing of C/EBPβ would influence dexamethasone-induced changes in muscle cell size, we next examined the size of cultured L6 myoblasts that had been transfected with non-targeting or C/EBPβ siRNA. Treatment of non-targeting siRNA-transfected myoblasts with 1 µM dexamethasone for 24 h resulted in a substantial reduction of cell size (Fig 3A). Importantly, the dexamethsone-induced atrophy was abolished in myoblasts transfected with C/EBPβ siRNA (Fig 3B).
Fig 3.
Silencing of C/EBPβ with C/EBPβ siRNA prevents dexamethasone-induced atrophy of cultured L6 myoblasts. Distribution of L6 myoblasts with different size (cell area) and transfected with (A) non-targeting siRNA or (B) C/EBPβ siRNA and then treated for 24 h with or without 1 µM dexamethasone. The dexamethasone-induced decrease in cell size (illustrated by left-shifted filled bars in panel A) was prevented by C/EBPβ siRNA (as illustrated in panel B). The cell areas in panel A were 5,430 ± 191 µm2 and 2,471 ± 123 µm2 in control and dexamethasone-treated myoblasts, respectively (p<0.001 by Student’s t-test). The corresponding figures for panel B were 5,236 ± 215 µm2 and 4,924 ± 183 µm2 (N.S.).
The experiments described above in Fig 1–3 were performed in L6 myoblasts. We chose to study myoblasts in our initial experiments because efficient transfection with siRNA constructs may be more easily achieved in myoblasts than in myotubes (Balci and Dincer, 2009). Because the role of C/EBPβ in the response to dexamethasone may not be identical in myoblasts and myotubes and because changes observed in myotubes may be more relevant for the situation in adult muscle than changes observed in myoblasts, it was important to test whether the role of C/EBPβ in dexamethasone-induced changes was similar in differentiated myotubes as described above for myoblasts.
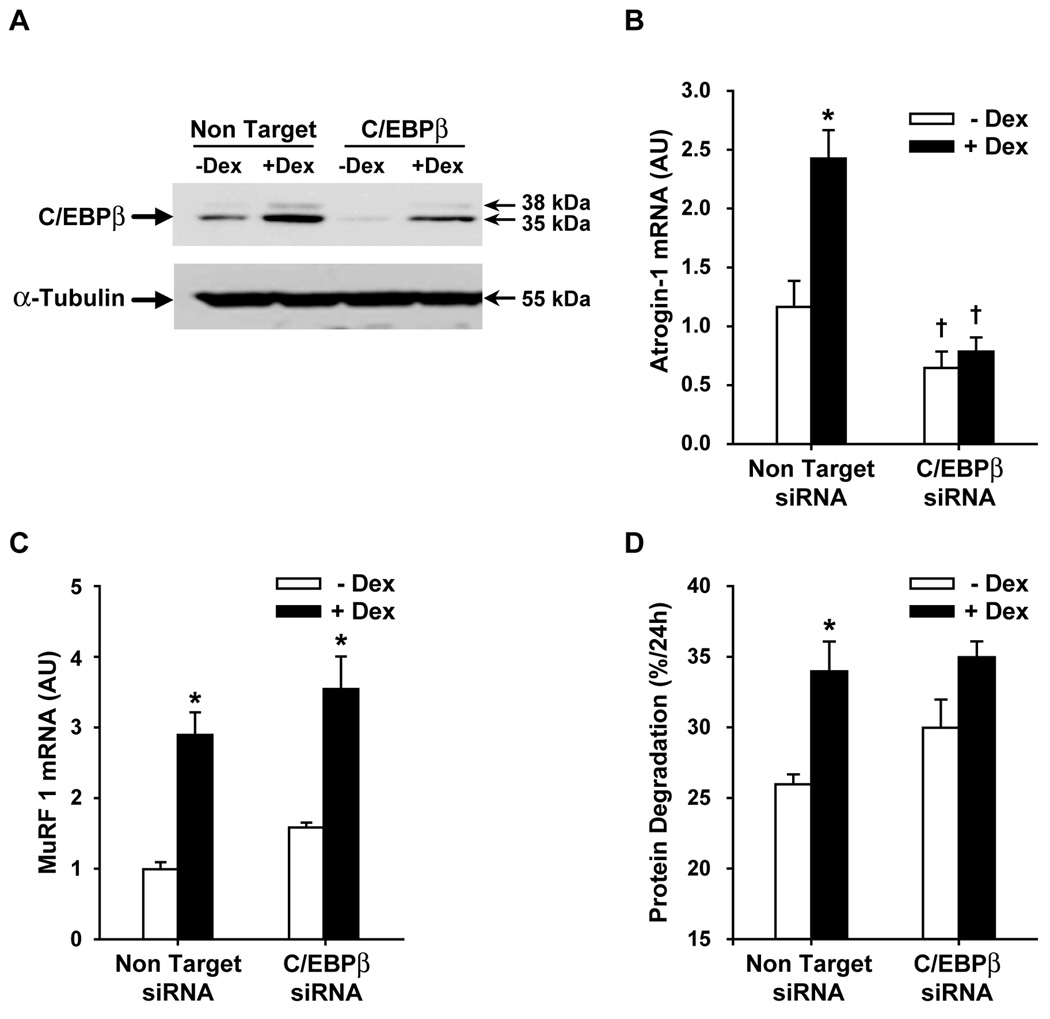
When L6 myotubes were transfected with C/EBPβ siRNA, C/EBPβ protein expression was reduced in both control and dexamethasone-treated myotubes (Fig 4A), similar to the findings in myoblasts (compare with Fig 1C). This result suggests that C/EBPβ siRNA transfection was efficient in both L6 myotubes and myoblasts. Treatment of the myotubes with dexamethasone resulted in a 2.5- and 3-fold increase in atrogin-1 and MuRF1 expression, respectively (Fig 4B and C). Silencing of C/EBPβ blocked the effect of dexamethasone on atrogin-1 expression in myotubes (similar to the effect of C/EBPβ siRNA in myoblasts; see Fig 2A). In contrast, downregulation of C/EBPβ did not influence the dexamethasone-induced upregulation of MuRF1 in myotubes which differs from the results observed in myoblasts (Fig 4C and compare with Fig 2C). In order to test whether the lack of effect of C/EBPβ siRNA on MuRF1 expression in myotubes was due to insufficient silencing of the C/EBPβ gene, we performed an additional experiment in which we increased the length of transfection with C/EBPβ siRNA from 24 to 48 h; the dexamethasone-induced increase in MuRF1 expression was unaffected by C/EBPβ siRNA in that experiment as well (data not shown). Thus, the role of C/EBPβ in glucocorticoid-induced expression of MuRF1 may be different in undifferentiated and differentiated muscle cells. Of note, basal atrogin-1 mRNA levels were reduced in C/EBPβ siRNA-treated myotubes (Fig 4B), similar to the finding in myoblasts (see Fig 2A). In contrast, basal MuRF1 mRNA levels were unaffected by C/EBPβ siRNA in both myotubes (Fig 4C) and myoblasts (see Fig 2C).
Fig 4.
Silencing of C/EBPβ with C/EBPβ siRNA inhibits dexamethasone-induced C/EBPβ and atrogin-1 expression and protein degradation in cultured L6 myotubes. L6 myotubes were transfected with non-targeting or C/EBPβ siRNA followed by treatment with 1 µM dexamethasone for 24 h and determination of (A) C/EBPβ protein levels, (B) atrogin-1 mRNA levels, (C) MuRF1 mRNA levels, and (D) protein degradation rates. Results are means ± SEM with n≥6 per group. *p<0.05 vs –Dex; +p<0.05 vs corresponding non-targeting group by ANOVA.
Protein degradation was increased by approximately 25% in dexamethasone-treated myotubes transfected with non-targeting C/EBPβ siRNA (Fig 4D). In contrast, dexamethasone did not significantly affect protein degradation in C/EBPβ siRNA-treated myotubes, similar to the observation in myoblasts.
Similar to the effect of dexamethasone observed in myoblasts, treatment of myotubes with dexamethasone resulted in cellular atrophy. The effect of dexamethasone on myotube size, however, was somewhat less pronounced than in myoblasts and required a longer time of treatment. Thus, treatment with 1 µM dexamethasone for 24 h did not influence myotube size (not shown) whereas treatment with dexamethasone for 48 h resulted in myotube atrophy (Fig 5A). We recently reported a similar more delayed atrophy in dexamethasone-treated L6 myotubes (Menconi et al, 2008). Importantly, the dexamethasone-induced atrophy of L6 myotubes was prevented by silencing the C/EBPβ gene (Fig 5B). Taken together, our results suggest that the role of C/EBPβ in dexamethasone-induced protein degradation and atrophy is similar, but not identical, in L6 myotubes and myoblasts (the main difference being inhibition of dexamethasone-induced MuRF1 expression in C/EBPβ siRNA-treated myoblasts but not in myotubes).
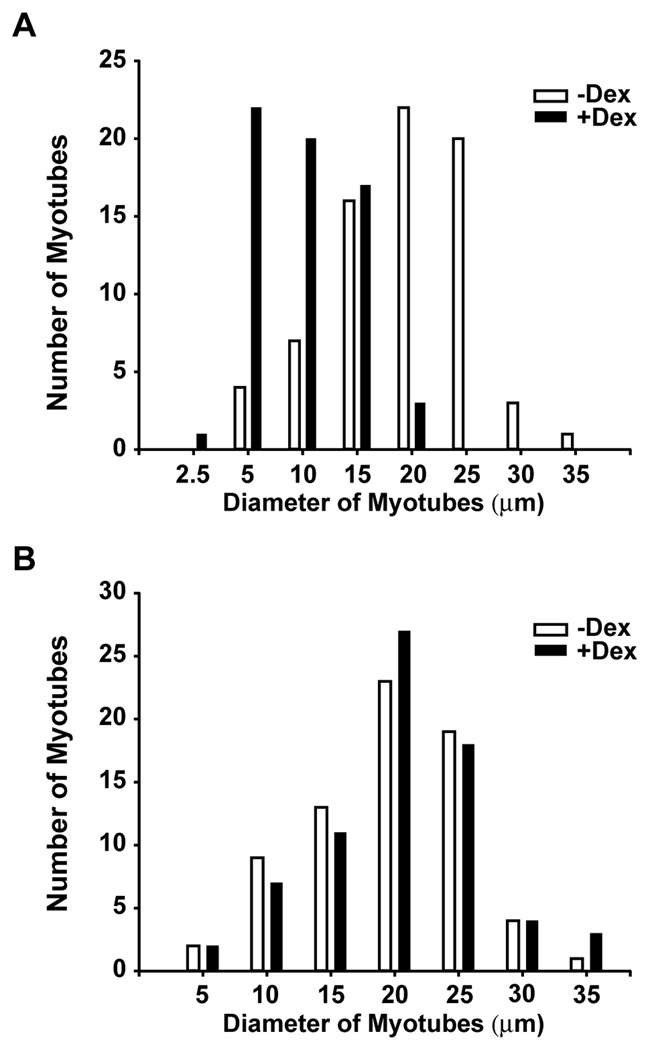
Fig 5.
Silencing of C/EBPβ with C/EBPβ siRNA prevents dexamethasone-induced atrophy of cultured L6 myotubes. Distribution of L6 myotubes with different diameters and transfected with (A) non-targeting siRNA or (B) C/EBPβ siRNA and then treated for 48 h with or without 1 µM dexamethasone. The dexamethasone-induced decrease in cell size (illustrated by left-shifted filled bars in panel A) was prevented by C/EBPβ siRNA (as illustrated in panel B). The myotube diameters in panel A were 19 ± 0.8 µm and 10 ± 0.5 µm in control and dexamethasone-treated myotubes, respectively (p<0.001 by Student’s t-test). The corresponding figures for panel B were 19 ± 0.7 µm and 20 ± 0.8 µm (N.S.).
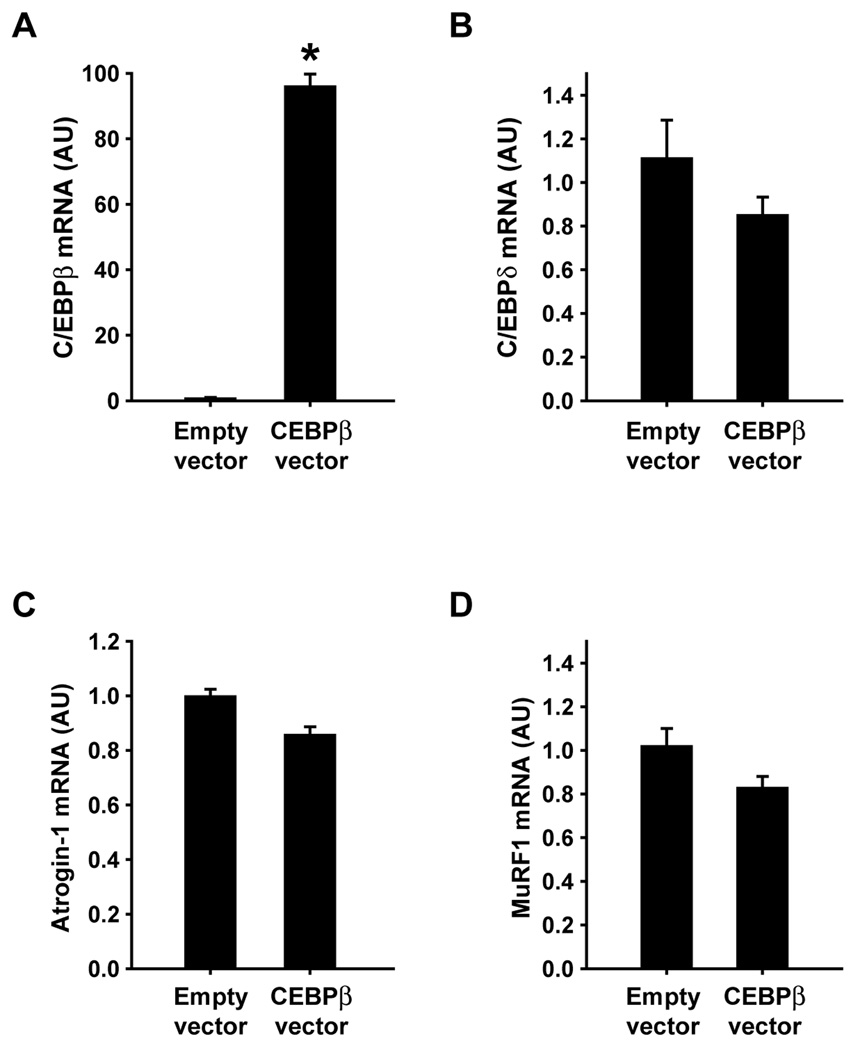
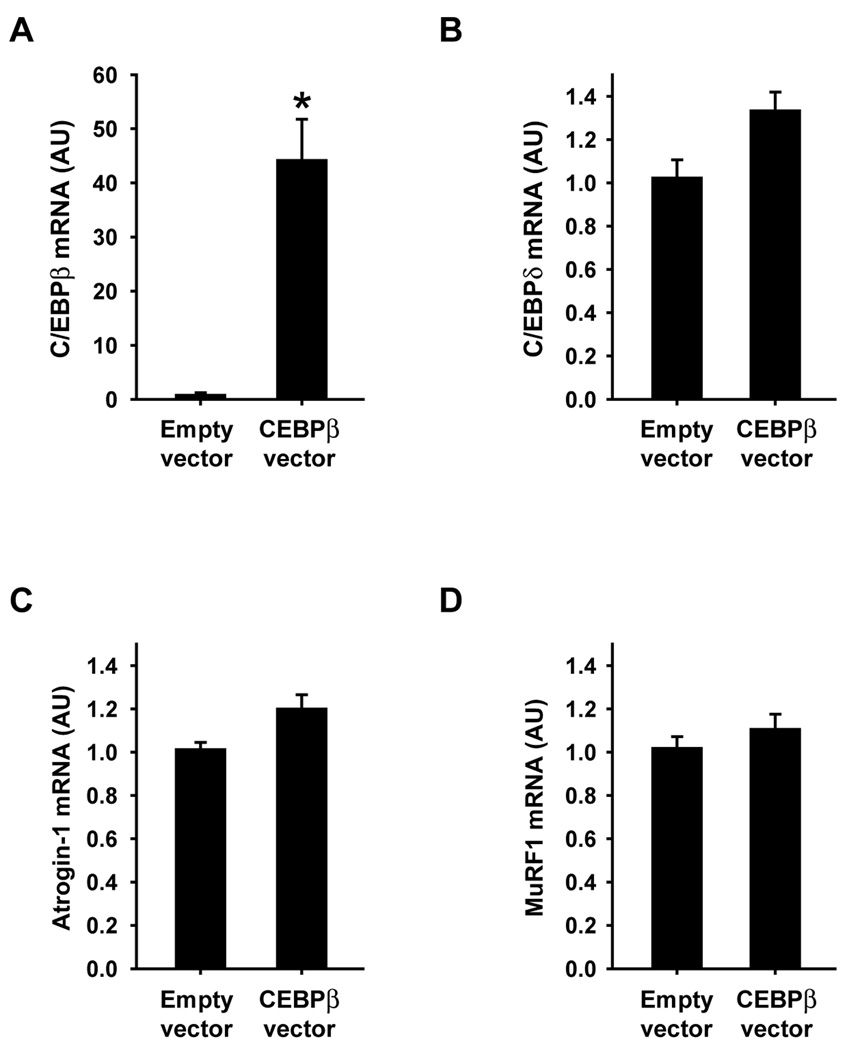
Although the results described above suggest that increased C/EBPβ expression is involved in dexamethasone-induced expression of atrogin-1 and MuRF1, it is not known if C/EBPβ alone is sufficient to increase the expression of atrogin-1 and MuRF1. In order to address that question, L6 myoblasts were transfected with a C/EBPβ expression plasmid. The transfection resulted in an approximately 90-fold increase in C/EBPβ mRNA levels but did not influence C/EBPδ mRNA levels, illustrating the effectiveness and specificity of the transfection (Fig 6A and B). Atrogin-1 and MuRF1 mRNA levels were unaffected by the increased expression of C/EBPβ (Fig 6C and D). In an additional experiment, L6 myoblasts that had been transfected with the C/EBPβ expression plasmid were allowed to differentiate into myotubes. Assessed from microscopic examinations, the differentiation of the myoblasts to myotubes was not affected by C/EBPβ overexpression (not shown). High levels of C/EBPβ mRNA were maintained in the myotubes although to a lesser degree than observed in the myoblasts (approximately 40-fold increase in the myotubes vs approximately 90-fold increase in the myoblasts). Similar to the finding in myoblasts, overexpression of C/EBPβ did not influence the expression of C/EBPδ, atrogin-1, or MuRF1 in myotubes (Fig 7B–D).
Fig 6.
Overexpression of C/EBPβ does not influence atrogin-1 or MuRF1 expression in L6 myoblasts. Cultured L6 myoblasts were transfected with C/EBPβ or empty vector followed by determination by real-time PCR of mRNA levels for (A) C/EBPβ, (B) C/EBPδ, (C) atrogin-1, and (D) MuRF1. Results are means ± SEM with n=6 per group. *p<0.05 vs empty vector by Student’s t-test.
Fig 7.
Overexpression of C/EBPβ does not influence atrogin-1 or MuRF1 expression in L6 myotubes. Cultured L6 myoblasts were transfected with C/EBPβ or empty vector whereafter the myoblasts were allowed to differentiate into myotubes. This was followed by determination by real-time PCR of mRNA levels for (A) C/EBPβ, (B) C/EBPδ, (C) atrogin-1, and (D) MuRF1. Results are means ± SEM with n=6 per group. *p<0.05 vs empty vector by Student’s t-test.
Discussion
In the present study, silencing of the C/EBPβ gene by siRNA inhibited dexamethasone-induced ubiquitin ligase expression, protein degradation, and atrophy in cultured L6 myoblasts and myotubes. The results are consistent with the concept that glucocorticoid-induced muscle wasting is, at least in part, regulated by C/EBPβ. In previous studies, we found that the expression and activity of C/EBPβ were increased in skeletal muscle of septic and dexamethasone-treated rats (Penner et al, 2002; Yang et al, 2005a), suggesting a close correlation between upregulated C/EBPβ expression and loss of muscle mass. The present study is important because it provides the first evidence of a link between C/EBPβ and the catabolic response to glucocorticoids in skeletal muscle.
Of note, the effects of C/EBPβ siRNA on dexamethasone-induced changes were similar in myoblasts and myotubes with the exception of upregulation of MuRF1 expression that was inhibited by C/EBPβ siRNA in myoblasts but not in myotubes. Importantly, the differential role of C/EBPβ in dexamethasone-induced upregulation of MuRF1 expression in myoblasts and myotubes suggests that C/EBPβ may not be involved in glucocorticoid-induced upregulation of MuRF1 in adult skeletal muscle. The present finding that the dexamethasone-induced expression of atrogin-1, but not MuRF1, was regulated by C/EBPβ in myotubes is in line with other studies suggesting that atrogin-1 and MuRF1 may be differentially regulated by other transcription factors as well. For example, atrogin-1 gene expression is mainly regulated by FOXO transcription factors in atrophying muscle (Sandri et al, 2004) whereas MuRF1 is mainly regulated by NF-kB (Cai et al, 2004).
Of note, a recent study by Waddell et al (2008) provided a detailed analysis of potential transcription factor binding sites in the MuRF1 promoter and the response to dexamethasone. Interestingly, a 5,000 base pair (bp) sequence of the MuRF1 promoter contained four C/EBP putative binding sites in addition to one glucocorticoid receptor (GR), four FOXO, and three NF-kB binding sites. The GR binding site and two of the FOXO binding sites were located within a 500 bp sequence immediately upstream of the MuRF1 transcription start site and this sequence was used for determination of the functional roles of FOXO transcription factors and the GR in dexamethasone-induced transcription of the MuRF1 gene. The functional roles of C/EBP transcription factors and NF-kB were not examined. When cultured HepG2 cells were transfected with expression plasmids for the GR or FOXO transcription factors, evidence was found that both FOXO transcription factors and the GR regulate MuRF1 gene transcription. Interestingly, FOXO1 (but not FOXO3a or FOXO4) showed a strong (20- to 40-fold) synergistic activation with the GR of the 500 bp MuRF1 promoter construct.
In additional experiments, Waddell et al (2008) used the GRdim mouse strain that expresses a GR with a point mutation in the DNA binding domain preventing binding of the GR to GR response elements (Reichardt et al, 1998) to further examine the role of MuRF1 in glucocorticoid-induced muscle atrophy. As expected, the upregulation of MuRF1 in skeletal muscle of dexamethasone-treated wild-type mice was reduced (but not abolished) in the GRdim mice. Despite this, however, the dexamethasone-induced muscle atrophy was identical in wild-type and GRdim mice. This observation was interpreted as indicating that the residual level of MuRF1 expression in the GRdim mice was sufficient to support full dexamethasone-induced muscle atrophy. An alternative interpretation that was also proposed was that MuRF1 may not be essential for dexamethasone-induced muscle atrophy. This interpretation is supported by the observation in the present study that silencing of C/EBPβ prevented dexamethasone-induced atrophy of differentiated myotubes despite the fact that MuRF1 expression was not reduced. In addition, a study by Hirner et al (2008) showed that transgenic overexpression of MuRF1 was not sufficient to induce muscle atrophy. These observations, of course, do not rule out the possibility that MuRF1 is essential for the regulation of muscle mass in certain conditions resulting in atrophy, such as denervation (Bodine et al, 2001).
The lack of atrogin-1 and MuRF1 upregulation in myoblasts and myotubes in which C/EBPβ was overexpressed does not necessarily contradict involvement of C/EBPβ in glucocorticoid-induced muscle atrophy but merely suggests that high levels of C/EBPβ alone are not sufficient to increase the expression of atrogin-1 and MuRF1. The transcriptional activation of most genes is regulated by multiple transcription factors that can interact with each other and with other nuclear proteins, including nuclear cofactors. It is possible that the involvement of C/EBPβ in dexamethasone-induced upregulation of atrogin-1 and MuRF1 reflects an interaction between C/EBPβ and other regulatory proteins. Interestingly, previous studies suggest that C/EBPβ can interact with the nuclear cofactor p300 (Mink et al, 1997). In recent studies from our laboratory, treatment of cultured myotubes with dexamethasone increased the expression and activity of p300 and its protein-protein interaction with C/EBPβ (Yang et al, 2005b, 2007). Thus, it is possible that C/EBPβ participates in glucocortiocid-induced muscle wasting at least in part secondary to its interaction with p300 (and perhaps other nuclear proteins as well) and that both C/EBPβ and interacting proteins need to be upregulated for C/EBPβ to play a role in the regulation of atrogin-1 and MuRF1.
Dexamethasone-treated cultured muscle cells have been used in several previous studies both from our and other laboratories as an in vitro model of muscle wasting (Bodine et al, 2001; Gomes et al, 2001; Hong and Forsberg, 1995; Menconi et al, 2008; Wang et al, 1998). The model is particularly relevant for sepsis-induced muscle wasting since the catabolic response to sepsis is to a great extent mediated by glucocorticoids (Hasselgren, 1999; Hasselgren et al, 2010; Tiao et al, 1996). It should be noted, however, that other mediators as well are probably involved in muscle wasting during sepsis, in particular the proinflammatory cytokines TNFα and IL-1 (Li et al, 2005; Pajak et al, 2008). C/EBPβ is involved in the response to TNF and IL-1 in other cell types (Gorgoni et al, 2001; Poli, 1998; Ramji and Foka, 2002; Ranjan and Boss, 2006) but it remains to be determined if these mediators regulate muscle mass via C/EBPβ.
Although the present results suggest that C/EBPβ plays an essential role in glucocorticoid-induced, and possibly sepsis-induced, muscle wasting, there is evidence that other transcription factors as well are important for the loss of muscle mass in various catabolic conditions. Thus, previous studies suggest that NF-kB (Cai et al, 2004; Penner et al, 2001; Van Gammeren et al, 2009) and FOXO transcription factors (Kamei et al, 2004; Sandri et al, 2004; Smith et al, 2010; Stitt et al, 2004) are involved in the regulation of muscle mass. This, of course, is not surprising since most genes are regulated by multiple transcription factors and the different transcription factors may interact with each other in a complex fashion. For example, C/EBPβ can form heterodimers with other members of the C/EBP family or with other transcription factors, including NF-kB (Kunsch et al, 1994; Poli, 1998; Ramji and Foka, 2002).
It should be emphasized again that although our results strongly suggest that dexamethasone-induced upregulation of atrogin-1 and MuRF1 was C/EBPβ-dependent in myoblasts and that dexamethasone-induced atrogin-1 expression was C/EBPβ-dependent in myotubes, the results do not necessarily imply that C/EBPβ regulates the atrogin-1 and MuRF1 genes through a direct effect (by binding to the atrogin-1 and MuRF1 promoters) but may regulate the atrogin-1 and MuRF1 genes indirectly, for example by stimulating other gene(s) the products of which then in turn regulate the atrogin-1 and MuRF1 genes. Although the atrogin-1 and MuRF1 promoters contain several putative C/EBP binding sites, additional experiments will be needed to determine whether C/EBPβ binds to the atrogin-1 and MuRF1 promoters under basal and dexamethasone-stimulated conditions. It should also be noted that it is possible that the inhibition of dexamethasone-induced protein degradation in C/EBPβ siRNA-treated muscle cells reflected a role of C/EBPβ in the regulation of genes in additional proteolytic pathways, other than the ubiquitin-proteasome pathway, involved in muscle wasting. For example, there is evidence that cathepsin L and autophagosome-dependent protein degradation is involved in loss of muscle mass (Deval et al, 2001; Mammucari et al, 2007; Zhao et al, 2007). The potential role of C/EBPβ in the regulation of genes in these proteolyitc pathways remains to be determined.
Although the present results suggest that glucocorticoid-induced muscle wasting can be prevented or reduced by decreasing the abundance of C/EBPβ, it should be pointed out that the activity of C/EBPβ is regulated not only by its abundance but by posttranslational modifications as well, in particular phosphorylation (Piwien-Pilipuk et al, 2001, 2002) and acetylation (Cesena et al, 2007). Our recent observation that C/EBPβ interacts with the nuclear cofactor p300 in dexamethasone-treated myotubes suggests that C/EBPβ may be acetylated in glucocorticoid-induced muscle atrophy (Yang et al, 2005b). The role of C/EBPβ acetylation (and phosphorylation) in glucocorticoid-regulated muscle wasting remains to be examined. It is possible that modulating both the overall abundance (as in the present study) and posttranslational modifications of C/EBPβ may prove important in the prevention of muscle wasting.
Acknowledgements
This work was supported in part by NIH grants R01 DK37908 (POH) and R01 NR08545 (POH). ZA was supported by the Department of Clinical Medicine, “Sapienza”, University of Rome, Rome, Italy. Dr. Oliver Hofmann, Department of Biostatistics, Harvard School of Public Health, provided assistance in examining the atrogin-1 and MuRF1 promoter regions for potential C/EBP binding sites.
References
- Alamdari N, Smith IJ, Aversa Z, Hasselgren PO. Sepsis and glucocorticoids upregulate p300 and downregulate HDAC6 expression and activity in skeletal muscle. Am J Physiol Regul Integr Comp Physiol. 2010;299:R509–R520. doi: 10.1152/ajpregu.00858.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arany Z, Lebrasseur N, Morris C, Smith E, Yang W, Ma Y, Chin S, Spiegelman M. The transcriptional coactivator PGC-1β drives the formation of oxidative type IIX fibers in skeletal muscle. Cell Metab. 2007;5:35–46. doi: 10.1016/j.cmet.2006.12.003. [DOI] [PubMed] [Google Scholar]
- Attaix D, Combaret L, Bechet D, Taillandier D. Role of the ubiquitin-proteasome pathway in muscle atrophy in cachexia. Curr Opin Support Palliat Care. 2008;2:262–266. doi: 10.1097/spc.0b013e3283196ac2. [DOI] [PubMed] [Google Scholar]
- Balci B, Dincer P. Efficient transfection of mouse-derived C2C12 myoblasts using a matrigel basement membrane matrix. Biotechnol J. 2009;4:1042–1045. doi: 10.1002/biot.200800269. [DOI] [PubMed] [Google Scholar]
- Bodine SC, Latres E, Baumheuter S, Lai VK, Nunez L, Clarke BA, Poueymirou WT, Panaro FJ, Na E, Dharmarajan K, Pan ZQ, Valenzuela DM, DeChiara TM, Stitt TN, Yancopoulos GD, Glass DJ. Identification of ubquitin ligases required for muscle atrophy. Science. 2001;294:1704–1708. doi: 10.1126/science.1065874. [DOI] [PubMed] [Google Scholar]
- Cai D, Frantz JD, Tawa NE, Melendez PA, Oh BC, Lidov HG, Hasselgren PO, Frontera WR, Lee J, Glass DJ, Shoelson SE. IKKβ/NF-kB activation causes severe muscle wasting in mice. Cell. 2004;119:285–298. doi: 10.1016/j.cell.2004.09.027. [DOI] [PubMed] [Google Scholar]
- Cesena TI, Cardinaux JR, Kwok R, Schwartz J. CCAAT/enhancer binding protein (C/EBP) β is acetylated at multiple lysines: acetylation of C/EBPβ at lysine 39 modulates its ability to activate transcription. J Biol Chem. 2007;282:956–967. doi: 10.1074/jbc.M511451200. [DOI] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Deval C, Mordier S, Obled C, Bechet D, Combaret L, Attaix D, Ferrara M. Identification of cathepsin L as a differentially expressed message associated with skeletal muscle wasting. Biochem J. 2001;360:143–150. doi: 10.1042/0264-6021:3600143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes MD, Lecker SH, Jagoe RT, Navon A, Goldberg AL. Atrogin-1, a muscle specific F-box protein highly expressed during muscle atrophy. Proc Natl Acad Sci USA. 2001;98:14440–14445. doi: 10.1073/pnas.251541198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorgoni B, Caivano M, Arizmendi C, Poli V. The transcription factor C/EBPβ is essential for inducible expression of the cox-2 gene in macrophages but not in fibroblasts. J Biol Chem. 2001;276:40769–40777. doi: 10.1074/jbc.M106865200. [DOI] [PubMed] [Google Scholar]
- Hasselgren PO. Glucocorticoids and muscle catabolism. Curr Opin Nutr Metab Care. 1999;2:201–205. doi: 10.1097/00075197-199905000-00002. [DOI] [PubMed] [Google Scholar]
- Hasselgren PO, Alamdari N, Aversa Z, Gonnella P, Smith IJ, Tizio S. Corticosteroids and muscle wasting – role of transcription factors, nuclear cofactors, and hyperacetylation. Curr Opin Clin Nutr Metabol Care. 2010;13:423–428. doi: 10.1097/MCO.0b013e32833a5107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasselgren PO, Menconi MJ, Fareed MU, Yang H, Wei W, Evenson A. Novel aspects on the regulation of muscle wasting in sepsis. Int J Biochem Cell Biol. 2005;37:2156–2168. doi: 10.1016/j.biocel.2005.01.017. [DOI] [PubMed] [Google Scholar]
- Hirner S, Krohne C, Schuster A, Hoffman S, Witt S, Erber R, Sticht C, Gasch A, Labeit S, Labeit D. MuRF1-dependent regulation of systemic carbohydrate metabolism as revealed from transgenic mouse studies. J Mol Biol. 2008;379:666–677. doi: 10.1016/j.jmb.2008.03.049. [DOI] [PubMed] [Google Scholar]
- Hong DH, Forsberg NE. Effects of dexamethasone on protein degradation and protease gene expression in rat L8 myotube cultures. Mol Cell Endocrinol. 1995;108:199–209. doi: 10.1016/0303-7207(95)03476-n. [DOI] [PubMed] [Google Scholar]
- Kamei Y, Miura S, Suzuki M, Kai Y, Mizukami J, Taniguchi T, Mochida K, Hata T, Matsuda J, Aburatani H, Nishino I, Ezaki O. Skeletal muscle FOXO1 (FKHR) transgenic mice have less skeletal muscle mass, down-regulated type 1 (slow twitch/red muscle) fiber genes, and impaired glycemic control. J Biol Chem. 2004;279:41114–41123. doi: 10.1074/jbc.M400674200. [DOI] [PubMed] [Google Scholar]
- Karolchik D, Baertsch R, Diekhans M, Furey TS, Hinrichs A, Lu YT, Roskin KM, Schwartz M, Sugnet CW, Thomas DJ, Weber RJ, Haussler D, Kent WJ. The UCSC genome browser database. Nucleic Acids Res. 2003;31:51–54. doi: 10.1093/nar/gkg129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khal J, Hine AV, Fearon KC, Dejong CH, Tisdale MJ. Increased expression of proteasome subunits in skeletal muscle of cancer patients with weight loss. Int J Biochem Cell Biol. 2005;37:2196–2206. doi: 10.1016/j.biocel.2004.10.017. [DOI] [PubMed] [Google Scholar]
- Kunsch C, Lang RK, Rosen CA, Shannon MF. Synergistic transcriptional activation of the IL-8 gene by NF-kB p65 (RelA) and NF-IL-6. J Immunol. 1994;153:153–164. [PubMed] [Google Scholar]
- Lang CH, Frost RA, Vary TC. Regulation of muscle protein synthesis during sepsis and inflammation. Am J Physiol. 2007;293:E453–E459. doi: 10.1152/ajpendo.00204.2007. [DOI] [PubMed] [Google Scholar]
- Lecker SH, Goldberg AL, Mitch WE. Protein degradation by the ubiquitin-proteasome pathway in normal and disease states. J Am Soc Nephrol. 2006;17:1807–1819. doi: 10.1681/ASN.2006010083. [DOI] [PubMed] [Google Scholar]
- Li YP, Chen Y, John J, Moylan J, Jin B, Mann DL, Reid MB. TNF-α acts via p38 MAPK to stimulate expression of the ubiquitin ligase atrogin1/MAFbx in skeletal muscle. FASEB J. 2005;19:362–370. doi: 10.1096/fj.04-2364com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mammucari C, Milan G, Romanello V, Masiero E, Rudolf R, Del Piccolo P, Burden SJ, DiLisi R, Sandri C, Zhao J, Goldberg AL, Schiaffino S, Sandri M. FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metab. 2007;6:458–471. doi: 10.1016/j.cmet.2007.11.001. [DOI] [PubMed] [Google Scholar]
- Matys V, Kel-Margoulis OV, Fricke E, Liebich I, Land S, Barre-Dirrie A, Reuter I, Chekmenev D, Krull M, Hornischer K, Voss N, Stegmaier P, Lewicki-Potapov B, Saxel H, Kel AE, Wingender E. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006;34:D108–D110. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menconi MJ, Arany Z, Alamdari N, Aversa Z, Gonnella P, O’Neal P, Smith IJ, Tizio S, Hasselgren PO. Sepsis and glucocorticoids downregulate the expression of the nuclear cofactor PGC-1β in skeletal muscle. Am J Physiol Endocrinol Metab. 2010;299:E533–E543. doi: 10.1152/ajpendo.00596.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menconi M, Gonnella P, Petkova V, Lecker SH, Hasselgren PO. Dexamethasone and corticosterone induce similar, but not identical, muscle wasting responses in cultured L6 and C2C12 myotubes. J Cell Biochem. 2008;105:353–364. doi: 10.1002/jcb.21833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mink S, Haenig B, Klempnauer KH. Interaction and functional collaboration of p300 and C/EBPβ. Mol Cell Biol. 1997;17:6609–6617. doi: 10.1128/mcb.17.11.6609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore-Carrasco R, Garcia-Martinez C, Busquets S, Ametller E, Barreiro E, Lopez-Soriano FJ, Argiles JM. The AP-1/CJUN signaling cascade is involved in muscle differentiation: implications in muscle wasting during cancer cachexia. FEBS Lett. 2006;580:691–696. doi: 10.1016/j.febslet.2005.12.084. [DOI] [PubMed] [Google Scholar]
- Murton AJ, Constantin D, Greenhaff PL. The involvement of the ubiquitin proteasome system in human skeletal muscle remodeling and atrophy. Biochim Biophys Acta. 2008;1782:730–743. doi: 10.1016/j.bbadis.2008.10.011. [DOI] [PubMed] [Google Scholar]
- Pajak B, Orzechowska S, Pijet B, Pijet M, Pogorzelska A, Gajkowska B, Orzechowski A. Crossroads of cytokine signaling – the chase to stop muscle cachexia. J Physiol Pharmacol. 2008;59:251–264. [PubMed] [Google Scholar]
- Penner G, Gang G, Sun X, Wray C, Hasselgren PO. C/EBP DNA-binding activity is upregulated by a glucocorticoid-dependent mechanism in septic muscle. Am J Physiol. 2002;282:R439–R444. doi: 10.1152/ajpregu.00512.2001. [DOI] [PubMed] [Google Scholar]
- Penner CG, Gang G, Wray C, Fischer JE, Hasselgren PO. The transcription factors NF-kB and AP-1 are differentially regulated in skeletal muscle during sepsis. Biochem Biophys Res Commun. 2001;281:1331–1336. doi: 10.1006/bbrc.2001.4497. [DOI] [PubMed] [Google Scholar]
- Piwien-Pilipuk G, MacDougald O, Schwartz J. Dual regulation of phosphorylation and dephosphorylation of C/EBPβ modulate its transcriptional activation and DNA binding in response to growth hormone. J Biol Chem. 2002;277:44557–44565. doi: 10.1074/jbc.M206886200. [DOI] [PubMed] [Google Scholar]
- Piwien-Pilipuk G, Van Mater D, Ross SE, MacDougald OA, Schwartz J. Growth hormone regulates phosphorylation and function of CCAAT/enhancer-binding protein β by modulating Akt and glycogen synthase kinase-3. J. Biol. Chem. 2001;276:19664–19671. doi: 10.1074/jbc.M010193200. [DOI] [PubMed] [Google Scholar]
- Poli V. The role of C/EBPβ isoforms in the control of inflammatory and native immunity functions. J Biol Chem. 1998;273:29279–29282. doi: 10.1074/jbc.273.45.29279. [DOI] [PubMed] [Google Scholar]
- Ramji DP, Foka P. CCAAT/enhancer-binding proteins: structure, function, and regulation. Biochem J. 2002;365:561–575. doi: 10.1042/BJ20020508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjan P, Boss JM. C/EBPβ regulates TNF induced MnSOD expression and protection against apoptosis. Apoptosis. 2006;11:1837–1849. doi: 10.1007/s10495-006-9530-0. [DOI] [PubMed] [Google Scholar]
- Reichardt HM, Kaestner KH, Tuckermann J, Kretz O, Wessely O, Bock R, Gass P, Scmid W, Herrlich P, Angel P, Schutz G. DNA binding of the glucocorticoid receptor is not essential for survival. Cell. 1998;93:531–541. doi: 10.1016/s0092-8674(00)81183-6. [DOI] [PubMed] [Google Scholar]
- Sandri M, Lin J, Handschin C, Yang W, Arany ZP, Lecker SH, Goldberg AL, Spiegelman BM. PGC-1α protects skeletal muscle from atrophy by suppressing FoxO3 action and atrophy-specific gene transcription. Proc Natl Acad Sci USA. 2006;103:16260–16265. doi: 10.1073/pnas.0607795103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandri M, Sandri C, Gilbert A, Skurk C, Calabria E, Picard A, Walsh K, Schiaffino S, Lecker SH, Goldberg AL. Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell. 2004;117:399–412. doi: 10.1016/s0092-8674(04)00400-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz C, Beck K, Mink S, Schmolke M, Budde B, Wenning D, Klempnauer KH. Recruitment of p300 by C/EBPβ triggers phosphorylation of p300 and modulates coactivator activity. EMBO J. 2003;22:882–892. doi: 10.1093/emboj/cdg076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieple A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S, Weinstock GM, Wilson RK, Gibbs RA, Kent WJ, Miller W, Haussler D. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith IJ, Alamdari N, O’Neal P, Gonnella P, Aversa Z, Hasselgren PO. Sepsis increases the expression and activity of the transcription factor Forkhead Box O 1 (FOXO1) in skeletal muscle by a glucocorticoid-dependent mechanism. Int J Biochem Cell Biol. 2010;42:701–711. doi: 10.1016/j.biocel.2010.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stitt TN, Drujan D, Clarke BA, Panaro F, Timofeyva Y, Kline WO, Gonzalez M, Yancopoulos GD, Glass DJ. The IGF-1/PI3K/Akt pathway prevents expression of muscle atrophy-induced ubiquitin ligases by inhibiting Foxo transcription factors. Mol Cell. 2004;14:395–403. doi: 10.1016/s1097-2765(04)00211-4. [DOI] [PubMed] [Google Scholar]
- Tiao G, Fagan J, Roegner V, Lieberman M, Wang JJ, Fischer JE, Hasselgren PO. Energy-ubiquitin-dependent proteolysis during sepsis in rats is regulated by glucocorticoids. J Clin Invest. 1996;97:339–348. doi: 10.1172/JCI118421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiao G, Fagan JM, Samules N, James JH, Hudson K, Lieberman M, Fischer JE, Hasselgren PO. Sepsis stimulates nonlysosomal, energy-dependent proteolysis and increases ubiquitin mRNA levels in rat skeletal muscle. J Clin Invest. 1994;94:2255–2264. doi: 10.1172/JCI117588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Gammeren D, Damrauer JS, Jackman RW, Kandarian S. The IkappaB kinases IKKα and IKKβ are necessary and sufficient for skeletal muscle atrophy. FASEB J. 2009;23:362–370. doi: 10.1096/fj.08-114249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddell DS, Baehr LM, van den Brandt J, Johnsen SA, Reichardt HM, Furlow JD, Bodine SC. The glucocorticoid receptor and FOXO1 synergistically activate the skeletal muscle atrophy-associated MuRF1 gene. Am J Physiol Endocrinol Metab. 2008;295:E785–E797. doi: 10.1152/ajpendo.00646.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Luo GJ, Wang JJ, Hasselgren PO. Dexamethasone stimulates proteasome- and calcium-dependent proteolysis in cultured L6 myotubes. Shock. 1998;10:298–306. doi: 10.1097/00024382-199810000-00011. [DOI] [PubMed] [Google Scholar]
- Wang LH, Yang XY, Zhang X, Farrar WL. Inhibition of adhesive interaction between multiple myeloma and bone marrow stromal cells by PPARγ cross talk with NF-kB and C/EBPβ. Blood. 2007;110:4373–4384. doi: 10.1182/blood-2006-07-038026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W, Yang H, Menconi M, Cao P, Chamberlain C, Hasselgren PO. Treatment of cultured myotubes with the proteasome inhibitor β-lactone increases the expression of the transcription factor C/EBPβ. Am J Physiol. 2007;292:C216–C226. doi: 10.1152/ajpcell.00282.2006. [DOI] [PubMed] [Google Scholar]
- Wray CJ, Mammen JM, Hershko DD, Hasselgren PO. Sepsis upregualtes the expression of multiple ubiquitin ligases in skeletal muscle. Int J Biochem Cell Bio. 2003;35:698–705. doi: 10.1016/s1357-2725(02)00341-2. [DOI] [PubMed] [Google Scholar]
- Yang H, Mammen J, Wei W, Menconi M, Evenson A, Fareed M, Petkova V, Hasselgren PO. Expression and activity of C/EBPβ and δ are upregulated by dexamethasone in skeletal muscle. J Cell Physiol. 2005a;204:219–226. doi: 10.1002/jcp.20278. [DOI] [PubMed] [Google Scholar]
- Yang H, Menconi M, Wei W, Petkova V, Hasselgren PO. Dexamethasone upregulates the expression of the nuclear cofactor p300 and its interaction with C/EBPβ in cultured myotubes. J Cell Biochem. 2005b;94:1058–1067. doi: 10.1002/jcb.20371. [DOI] [PubMed] [Google Scholar]
- Yang H, Wei W, Menconi M, Hasselgren PO. Dexamethasone-induced protein degradation in cultured myotubes is p300/HAT dependent. Am J Physiol. 2007;292:R337–R344. doi: 10.1152/ajpregu.00230.2006. [DOI] [PubMed] [Google Scholar]
- Zhao J, Brault JJ, Schild A, Cao P, Sandri M, Schiaffino S, Lecker SH, Goldberg AL. FoxO3 coordinately activates protein degradation by the autophagic/lysosomal pathways in atrophying muscle. Cell Metab. 2007;6:472–483. doi: 10.1016/j.cmet.2007.11.004. [DOI] [PubMed] [Google Scholar]