Abstract
In plants, plasmodesmata (PD) are intercellular channels that function in both metabolite exchange and the transport of proteins and RNAs. Currently, many of the PD structural and regulatory components remain to be elucidated. Receptor-like kinases (RLKs) belonging to a notably expanded protein family in plants compared to the animal kingdom have been shown to play important roles in plant growth, development, pathogen resistance, and cell death. In this study, cell biological approaches were used to identify potential PD-associated RLK proteins among proteins contained within cell walls isolated from rice callus cultured cells. A total of 15 rice RLKs were investigated to determine their subcellular localization, using an Agrobacterium-mediated transient expression system. Of these six PD-associated RLKs were identified based on their co-localization with a viral movement protein that served as a PD marker, plasmolysis experiments, and subcellular localization at points of wall contact between spongy mesophyll cells. These findings suggest potential PD functions in apoplasmic signaling in response to environmental stimuli and developmental inputs.
Electronic supplementary material
The online version of this article (doi:10.1007/s00709-010-0251-4) contains supplementary material, which is available to authorized users.
Keywords: Plasmodesmata, Receptor-like kinase, Symplasmic signaling, Apoplasmic signaling, Cell wall proteomics
Introduction
In plants, plasmodesmata (PD) are cytoplasmic channels that play a pivotal role in both the symplasmic exchange of metabolites and intercellular trafficking of information molecules, including proteins and RNA (Cilia and Jackson 2004; Heinlein and Epel 2004; Kim 2005; Maule 2008). The size exclusion limit (SEL) of PD probed by movement of micro-injected fluorescent dyes is on the order of 1 kDa (Robards and Lucas 1990; Lucas 1995). However, this intercellular movement can be regulated by modulating the size exclusion limit of the PD microchannels through the action of dilating proteins (Oparka 2004; Maule 2008; Lucas et al. 2009).
Movement proteins (MP) encoded by various plant viruses have the capacity to modulate PD SEL in order to facilitate the cell-to-cell movement of the viral RNA (Heinlein and Epel 2004; Lucas 2006). In addition, developmental stages and environmental stresses are also important factors that can modulate the SEL of diverse PD types (Crawford and Zambryski 2001; Roberts et al. 2001; Zambryski and Crawford 2000). For example, simple primary PD can undergo a significant change in structure, becoming highly branched, thereby greatly increasing the symplasmic pathway available for exchange of metabolites and/or signaling agents. Alternatively, during leaf development, the PD pathway can be downregulated to reduce the process of intercellular communication (Liarzi and Epel 2005; Oparka et al. 1999; Roberts et al. 2001). Finally, many abiotic and biotic stresses can reduce PD SEL through the accumulation of callose at the neck region of the PD (Chen and Kim 2009; Levy and Epel 2009; Turner et al. 1994). However, much still remains to be learned about the signaling machinery involved in both developmental and stress-induced regulation of PD function.
The intercellular transport of molecules through PD can occur by two mechanistically different processes, involving non-selective and selective movement. Evidence for the non-selective mode of movement has been gained through the use of fluorescent probes, including free green fluorescent protein (GFP); provided the molecular dimensions of these molecules are compatible with the physical void spaces within the PD microchannels, cell-to-cell movement can be driven by diffusion (Imlau et al. 1999; Oparka et al. 1997, 1999). Evidence for selective trafficking has been provided by studies on viral MP (Lucas 2006), as well as from studies of endogenous proteins, such as KNOTTED1 (Lucas et al. 1995; Kim et al. 2005) and other transcription factors (Gallagher et al. 2004; Gallagher and Benfey 2009; Kurata et al. 2005). Intercellular trafficking on the selective pathway involves an interaction between the non-cell-autonomous protein (NCAP) and components of the non-cell-autonomous protein pathway (Lee et al. 2003; Kragler et al. 2000; Taoka et al. 2007). Such interactions generally involve a protein-mediated increase in the PD SEL, a step that appears to be required for intercellular trafficking of this class of NCAPs.
In recent years, a number of studies have focused on identifying the molecular constituents of higher plant PD. A novel high-throughput screen was conducted using a cDNA library tagged with GFP (Escobar et al. 2003). However, further characterization of the identified putative PD proteins remains to be performed. A range of biochemical methods have also been developed to enrich for PD constituents from cell wall preparations. This strategy yielded a number of NCAP pathway components (Lee et al. 2003, 2005; Sagi et al. 2005; Levy et al. 2007a). Proteomic analyses of PD-enriched cell wall preparations from Arabidopsis, Chara corallina, and tobacco plants have also identified PD proteins (Bayer et al. 2006; Faulkner et al. 2005; Levy et al. 2007a).
Subcellular localization of these putative PD proteins has generally been confirmed using confocal laser scanning microscopy (CLSM) to detect the co-localization of the candidate protein, as a fusion with fluorescent reporters such as GFP, with a bona fide PD protein, such as a tagged viral MP (Levy et al. 2007a; Thomas et al. 2008). These CLSM analyses have also been complemented with transmission electron microscopy-based immunological experiments (Raffaele et al. 2009; Simpson et al. 2009). Currently, these biochemical and proteomic studies have identified some 30 PD-associated proteins from a range of plant species. Considering the likely complexity of the PD structure, which may well be comparable to the nuclear pore complex that is approximately 120 MDa in size and contains some 100 subunits (Lee et al. 2000), it would seem that many more PD proteins remain to be isolated and characterized.
A potential role for PD-localized proteins in apoplasmic signaling was recently raised based on the identification of a family of PD-located proteins (PDLP) belonging to a class of type I membrane receptor-like proteins (Thomas et al. 2008). This family contains two domains of unknown function (DUF26) that are located on the extracellular side of the plasma membrane. This general orientation is found in a wide array of receptor-like protein kinase (RLK) families. In plants, members of such RLKs have been shown to play important roles in plant growth, development, pathogen resistance, and cell death (Stone et al. 1998; Chen et al. 2003; Searle et al. 2003; Chinchilla et al. 2007; Wan et al. 2008). Typically, an RLK is comprised of an extracellular receptor domain that binds to a signal ligand, a trans-membrane domain that anchors the protein within the membrane, and a cytoplasmic kinase domain for signal transduction. It was reported that the Arabidopsis genome encodes for at least 610 RLKs, representing more than ten subfamilies (Chae et al. 2009).
Here, we report that six RLKs among 15 rice RLKs tested are associated with plasmodesmata based on their co-localization with a PD marker, the viral MP encoded by Turnip vein-clearing mosaic virus (TVCV). These findings are discussed in terms of potential signaling between the apoplasm and PD in terms of control over symplasmic signaling in response to environmental stimuli and developmental inputs.
Results
Subcellular localization of receptor-like protein kinases
Our analysis of the cell wall proteome of rice (unpublished) identified a number of membrane-associated proteins, including some RLKs. Since cell wall preparations can be substantially free of extraneous membrane, while at the same time retaining PD as membrane-rich wall components (Bayer et al. 2006), we decided to investigate whether some of these membrane proteins may reside in PD. A similar strategy, followed previously, identified the receptor-like molecule, PDLP, as a membrane-associated PD protein (Bayer et al. 2006; Thomas et al. 2008). The rice genome encodes more than 600 RLKs which are targeted by the secretory pathway to the plasma membrane and cell periphery. Using all the identified peptides from the proteomic analysis as preliminary evidence for the occurrence of RLKs (single and multiple hits) in the cell wall, we selected a subset of RLKs to study for their subcellular targeting to PD. The selection was also influenced by the availability of full length cDNAs for rapid and convenient cloning. On this basis, 15 RLKs were studied for targeting as fluorescent protein fusions with GFP using an Agrobacterium-based transient expression system. 15 RLK cDNAs were obtained from the rice PIPELINE database (http://cdna01.dna.affrc.go.jp/PIPE/; Yazaki et al. 2004). Table 1 presents the genomic information for these 15 RLK genes.
Table 1.
Candidate PD-associated receptor-like kinases identified based on a rice cell wall proteome
| Index | cDNA accession no. | Rice accession no. | RLK subfamilya | SPb | TMDc | Length | Subcellular localizationd |
|---|---|---|---|---|---|---|---|
| 1 | AK107192 | Os05g25540 | RLCK-X | Y | 1 | 478 | PM |
| 2 | AK101690 | Os03g12470 | WAK_LRK10L-1 | Y | 1 | 704 | PD |
| 3 | AK111508 | Os04g51050 | WAK | Y | 2 | 712 | PD |
| 4 | AK060765 | Os03g12150 | SD-2b | Y | 1 | 843 | PM, ER |
| 5 | AK065935 | Os04g01874 | L-LEC | Y | 0 | 731 | PD |
| 6 | AK060565 | Os07g35004 | SD1 | Y | 1 | 674 | PM, Nuc |
| 7 | AK111650 | Os07g35690 | DUF26 | Y | 1 | 695 | PM |
| 8 | AK100000 | Os07g41140 | LRR-XV | Y | 1 | 1,084 | PM |
| 9 | AK111558 | Os11g12530 | LRR-XI | Y | 1 | 987 | PM |
| 10 | AK110462 | Os06g47750 | LRR-Xb | Y | 1 | 1,066 | PD |
| 11 | AK111546 | Os02g05960 | LRR-Xb | Y | 1 | 1,051 | PD |
| 12 | AK111516 | Os10g25090 | LRR-V | Y | 1 | 719 | PM |
| 13 | AK103598 | Os07g48310 | LRR-III | Y | 1 | 640 | PM |
| 14 | AK106346 | Os09g02250 | LRR-III | Y | 1 | 794 | PD |
| 15 | AK100532 | Os08g10330 | LRR-VIII-2 | Y | 1 | 1,023 | PM |
SP signal peptides, TMD trans-membrane domains, PM plasma membrane, ER endoplasmic reticulum, Nuc nucleus, PD plasmodesmata
aSubfamilies for each candidate RLK were categorized according to Lehti-Shiu et al. (2009)
bSP were predicted by SignalP 3.0 (http://www.cbs.dtu.dk/services/SignalP/) with two algorithms (NN and HMM methods)
cTMD were predicted by a TMHMM v. 2.0 program (http://www.cbs.dtu.dk/services/TMHMM/)
dSubcellular localization for each RLK was determined by transient expression assays: PM, ER, Nuc, and PD
The SMART program (http://smart.embl-heidelberg.de/; Letunic et al. 2009) was employed to identify the conserved domains within each of these 15 candidate RLKs. Each protein appeared to contain a single N-terminally located signal peptide and a kinase domain housed in the C-terminal region (Fig. 1). Transmembrane domains (TMDs) were present in 14 proteins, but Os04g51050 had two, whereas Os04g01874 did not have a predicted TMD. These RLKs were assigned to the respective subfamilies, according to their conserved domains (Lehti-Shiu et al. 2009; Table 1). These 15 RLKs were members of 13 subfamilies and that they contained a broad array of additional domains, including epidermal growth factor (EGF), lectin, PAN, DUF26, and leucine-rich repeat (LRR) domains. Interestingly, eight of these 15 RLKs belonged to the LRR RLK family.
Fig. 1.
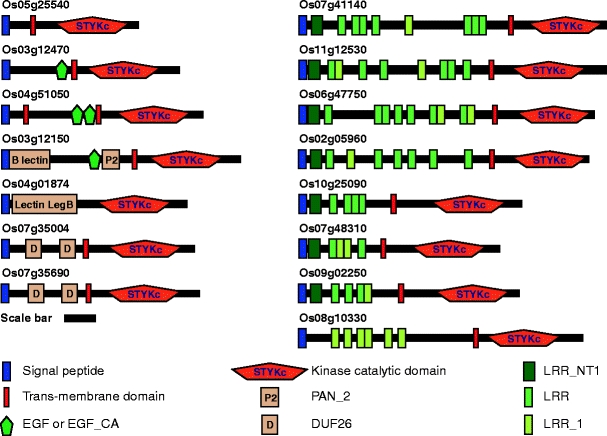
Predicted domain structures of 15 candidate PD-associated receptor-like kinases identified based on a rice cell wall proteome. The SMART program (http://smart.embl-heidelberg.de/) was employed to identify the domains for each RLK. Signal peptide, blue box; trans-membrane domain, red box; conserved kinase catalytic domain, STYKc; variable elongation growth factor domains (EGF and EGF_CA), green pentagon; B_lectin domain; Lectin_legB domain; plasminogen/hepatocyte growth factor family domain (PAN_2), P2 in orange box; domain of unknown function 26 (DUF26), D in orange box; N-terminal leucine-rich repeats (LRR) domain (LRRNT_1), deep green box; variable leucine-rich repeat domains (LRR and LRR_1), bright green and yellow-green box, respectively. Scale bar = 100 amino acid residues
To determine the subcellular localization of the RLKs, they were translationally fused to fluorescent proteins and expressed under control of the strong cauliflower mosaic virus 35S promoter upon agroinfiltration into Nicotiana benthamiana leaves. TVCV MP-red fluorescent protein (RFP) (Kim et al. 2003) was used as a PD marker for co-localization studies. First of all, to validate TVCV MP-RFP as a PD marker, co-localization experiments were performed with two bona fide PD markers, namely callose (Turner et al. 1994) and PLRV MP-GFP (Schmitz et al. 1997). Red fluorescence from TVCV MP-RFP overlapped with callose stained with aniline blue (Fig. 2a, yellow signal). In addition, TVCV MP-RFP was also found to be co-localized with PLRV MP-GFP (Fig. 2b). The observed co-localization between TVCV MP-RFP and two bona fide PD markers, callose and PLRV MP17, confirmed the utility of this tobamovirus MP as a PD marker in a tobacco transient expression system.
Fig. 2.
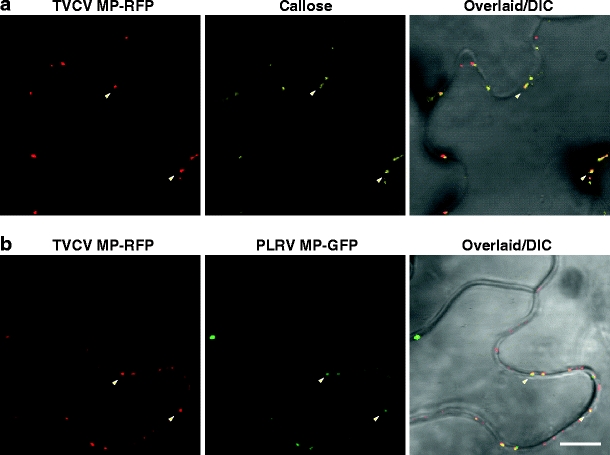
Validation of TVCV MP-RFP as a PD marker. Co-localization between TVCV MP-RFP and callose stained by aniline blue (a). Co-localization between TVCV MP-RFP and PLRV MP-GFP (b). Representative co-localized spots are indicated by arrowheads. Scale bar = 10 μm, common to all images
To identify the appropriate timeframe over which to conduct these subcellular localization screens, protein localization was assessed at 12, 24, 36, and 48 h post-agroinfiltration (Fig. 3). Based on these studies, 36 h post-agroinfiltration was optimal and selected for all localization studies.
Fig. 3.
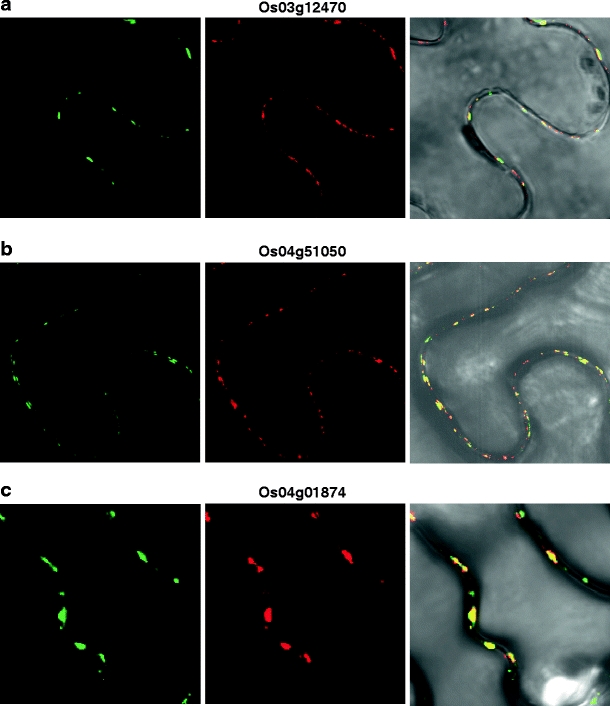
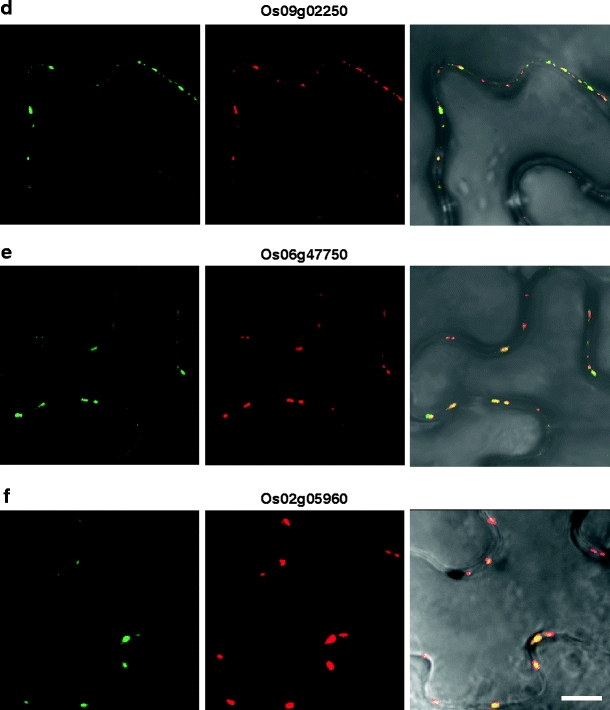
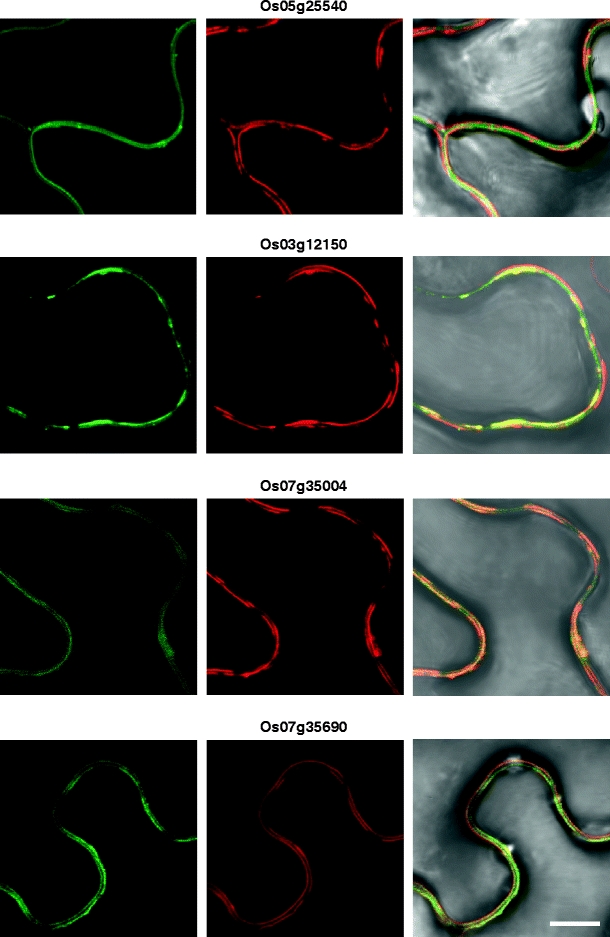
Six rice cell wall-associated RLKs are co-localized to PD. Indicated rice RLK-GFP fusion proteins were transiently co-expressed with a TVCV MP-RFP PD marker in N. benthamiana leaves. Punctate green fluorescent signals (left panels) were observed for 2 WAK-RLKs (a, b), a lectin RLK (c), and 3 LRR-RLKs (d–f). Red fluorescent signal (central panels) indicates the subcellular localization of the TVCV MP-RFP reporter. Merged images of GFP and RFP (right panels) confirm that these six RLKs were co-localized to PD. All CLSM images were collected 36 h post-agroinoculation. Scale bar = 10 μm, common to all images
Based on these experiments, six RLKs were identified as being targeted to PD (Fig. 3), and nine RLKs were found to be targeted to the PM (Fig. 4 and Supplemental Fig. 1). Here, it is important to note that images presented in Figs. 3 and 4 and Supplemental Fig. 1 are representative of three replicate experiments. We could not observe significant red fluorescences of six PDRLK-RFPs at cortical position or at the upper side of epidermis, at our optimized condition. Interestingly, two members of the LRR-Xb subfamily, Os06g47750 and its homolog, Os02g05960, were targeted to PD. In contrast, although Os07g48310 and Os09g02250 are homologous and belong to the LRR-III RLK family, Os09g02250 was targeted to PD, whereas Os07g48310 was localized to the PM. A number of these RLKs were also detected within the cytoplasm. For example, an SD-2b RLK (Os03g12150) localized to both the cytoplasm surrounding the nucleus, probably the endoplasmic reticulum (ER) and the PM (Supplemental Fig. 1c, d). An SD1 RLK (Os07g35004) was detected within the nucleus and the PM (Supplemental Fig. 1m, n).
Fig. 4.
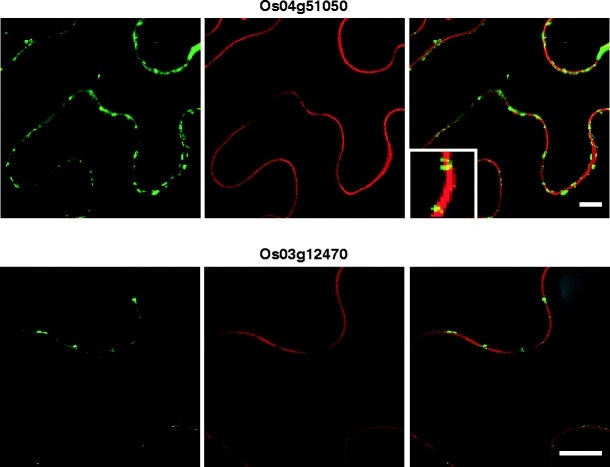
Subset of rice cell wall-associated RLKs are co-localized to the plasma membrane. Indicated rice RLK-GFP fusion proteins were transiently co-expressed with an RFP-tagged plasma membrane (PM) marker in N. benthamiana leaves. Green fluorescent signals associated with expression of Os05g25540-, Os03g12150-, Os07g35004-, and Os07g35690-GFP (left panels) were localized to the cell periphery. PM visualized using a PM-RFP reporter (central panels). Merged GFP and PM-RFP images establish that all four RLK proteins were co-localized to the PM (right panels). All images were collected 36 h post-agroinoculation. Scale bar = 10 μm, common to all images
The PM localization of the nine RLKs was further confirmed by co-expression with a known PM-RFP marker (Nelson et al. 2007). These experiments were performed with a subset of putative PM-targeted RLK-GFP constructs. As shown in Fig. 4, green fluorescent signals associated with all four RLKs were found to be highly co-localized with the red fluorescent signal from the PM marker.
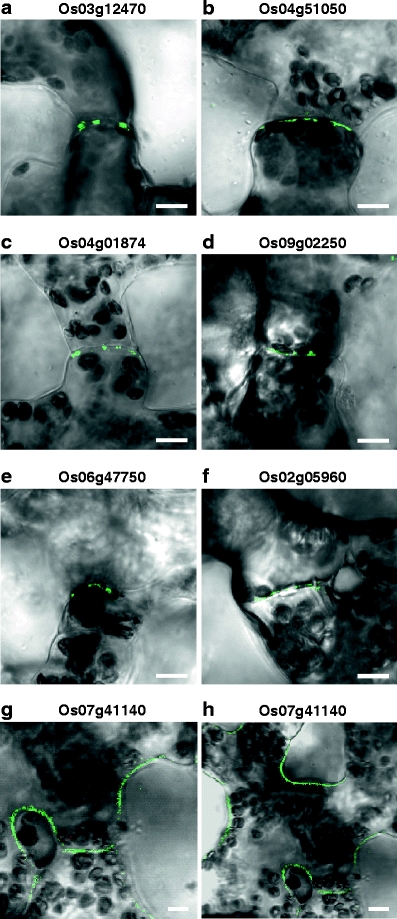
To also confirm that the six PD-targeted RLKs were localized within the region of the cell wall, leaves previously infiltrated with Agrobacterium expressing RLK-GFP constructs were incubated with propidium iodide (PI). As illustrated in Fig. 5, the Os04g51050-GFP and Os03g12470-GFP signals overlapped with the PI red fluorescence; the merged images clearly demonstrated that this PD-targeted RLK was localized within the cell wall (Fig. 5, right panels). Equivalent results were obtained for the other four PD-targeted RLKs.
Fig. 5.
PD-associated RLKs are located within the cell wall. Green fluorescent signal shows RLK-GFP expression (left panels) and red fluorescent signal (central panels) reflects propidium iodide staining within the wall of epidermal cells. Merged images of green and red fluorescent signals (right panels) confirm that Os04g51050 and Os3g12470 proteins are located within the wall. Inset, magnified image of wall GFP punctae. Scale bars = 10 μm
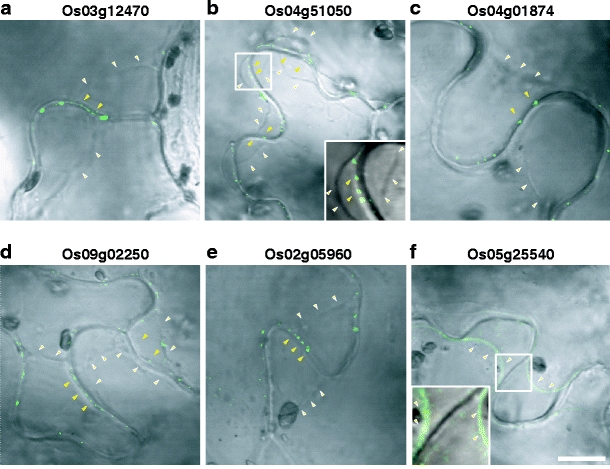
Mesophyll cells have a unique feature in that PD are only located in points of cellular contact. Hence, this system was next employed to test whether the candidate RLKs were targeted specifically to PD. Signal from all six PD-targeted RLK-GFP fusion proteins was detected only in the contact region between neighboring mesophyll cells (Fig. 6a–f). No signal was observed along the PM facing the intercellular air space. By contrast, the two PM-targeted RLKs that served as a control for these experiments were localized along the periphery of the mesophyll cell (Fig. 6g, h).
Fig. 6.
Rice PD-associated RLKs accumulate in the contact region between neighboring mesophyll cells. Indicated rice RLK-GFP fusion proteins were transiently expressed within N. benthamiana leaves. Punctate green fluorescent signals associated with the 6 PD-targeted RLKs were only detected within the contact walls of neighboring mesophyll cells (a–f). Control experiments performed with Os07g41140 (g, h) and yielded GFP signal along the PM in both contact regions and the wall facing the intercellular air space. All CLSM images were collected 36 h post-agroinoculation. Scale bars = 10 μm
Plasmolysis experiments confirm PD localization of RLKs
A routine test for proteins targeted to PD is whether the fluorescent signal associated with the tagged putative PD-localized protein is retained within the wall after plasmolysis (Tilney et al. 1991; Thomas et al. 2008). Such experiments were next performed using a 0.45 M mannitol solution. GFP punctate signals from PD-targeted RLKs were retained within the wall after plasmolysis (Fig. 7a–e), whereas GFP signal from PM-associated RLKs was displaced from the wall and remained localized over the retracted PM (Fig. 7f).
Fig. 7.
PD localized RLKs remain at the cell wall after plasmolysis. Yellow-colored arrowheads indicate putative PD-localized GFP signal within the cell wall; white-colored arrowheads indicate location of plasmolyzed PM. Scale bar = 10 μm, common to all images except inset image. Boxed images are shown in insets
Discussion
A range of experimental approaches have been developed to identify the molecular components of both algal and higher plant PD. A combination of cellular and proteomic tools has provided a high-throughput method to screen for PD proteins (Faulkner et al. 2005; Lee et al. 2005; Levy et al. 2007b; Bayer et al. 2008; Thomas et al. 2008). Although this approach has proven successful, it has certain drawbacks. For example, the protocols developed to isolate plasmodesmal enriched cell wall proteins (PECP) still yield protein populations containing a significant number of cell wall-specific proteins (Bayer et al. 2006). Furthermore, because of the unique structural features of PD that include various components of the cell wall, PM, and ER, it is a challenge to select potential PD proteins from these PECP preparations for in depth study.
The capacity to integrate cell wall proteomics with bioinformatics and cell biological tools, in order to use rational screens to filter the proteomic database, has proven successful in identifying bona fide PD proteins (Thomas et al. 2008; Simpson et al. 2009). In the current study, using a cell biological approach, we identified six PD-associated RLKs, with three belonging to the LRR-RLK family, the largest subfamily of RLKs comprising some 200 members in Arabidopsis (Lehti-Shiu et al. 2009).
A number of ways have been developed to determine subcellular localization of proteins targeted to PD. Although transient expression of proteins tagged with various fluorescence proteins, such as GFP, can provide information on potential subcellular targets, the use of strong promoters like the CaMV 35S must be used with caution. As found in the present study, protein overexpression can lead to mistargeting of a PD-associated protein. Time course experiments are essential to identify the sites within the cell first targeted by a candidate protein. As the capacity of the PD to accept newly synthesized proteins is likely to vary depending on the protein, the situation can be envisaged where the level of tagged protein is low and even on the margin of detection. In this case, overexpression would result in accumulation of signal along the PM, leading to a false negative. Thus, the possibility exists that some of the RLKs characterized as PM-targeted might well be PD-associated proteins. The use of endogenous promoters would avoid this complication.
A second method to detect PD-associated proteins involves the co-expression of the candidate protein along with a bona fide PD protein. Here, viral MPs that accumulate within PD have served as effective markers (Blackman et al. 1998; Chen et al. 2000; Roberts et al. 2001; Vogel et al. 2007). In our studies, we used the TVCV MP-RFP, which was co-localized with bona fide PD markers, PLRV MP17-GFP (Schmitz et al. 1997) and callose (Turner et al. 1994), to screen the 15 RLK-GFP proteins (Fig. 3). This system, combined with analyses performed on mesophyll cells, provided evidence in support of our conclusion that six RLKs are targeted to PD. Plasmolysis experiments (Fig. 7) further supported this assignment. Once transgenic rice plants are available for these newly identified PD-associated RLKs, high-resolution immunogold localization studies can be performed to identify the location occupied by these proteins within the PD.
The functions of more than 30 LRR-RLKs have been studied, and in Arabidopsis, individual members have been shown to play roles in a wide range of biological processes. For example, CLAVATA1 (CLV1) plays an important role in meristem and floral development (Clark et al. 1993). BRASSINOSTEROID-INSENSITIVE 1 (BR1) and BRI1-ASSOCIATED RECEPTOR KINASE 1 (BAK1) are involved in brassinosteroid (BR) signaling (Li and Chory 1997; Li et al. 2002). It will be of great interest to determine the roles played by the three rice PD-associated LRR-RLKs. As pointed out by Thomas et al. (2008), locating such membrane receptor-like proteins within the PD could well mediate in direct signaling between apoplasmic signaling ligands and PD constituents to regulate various aspects of the symplasmic pathway. Future studies will be performed to further test the hypothesis that these PD-associated RLKs function in mediating apoplasmic signaling to regulate cell-to-cell communication. An alternative hypothesis is that PD, as symplasmic bridges, might act as target sites to receive environmental or developmental apoplasmic signals.
A fourth PD-associated RLK, Os04g01874, a member of the Lectin LegB subfamily, is noteworthy as it does not contain a trans-membrane domain. This is interesting as our bioinformatic analysis also failed to detect a trans-membrane domain for PDLP1. Either this RLK has a trans-membrane domain similar to PDLP1 or it may be secreted and subsequently targeted/anchored to PD by a novel mechanism.
Two of the identified PD-associated RLKs (Os04g51050 and Os03g12470) are members of the WAK-RLK subfamily that contain EGF repeats in their extracellular domain (Anderson et al. 2001). The rice genome contains some 125 WAKs (Zhang et al. 2005), and based on studies with other species, these rice kinases will likely function in development as well as in various stress-related signaling modules (He et al. 1999; Kohorn et al. 2006; Wagner and Kohorn 2001). The role of these RLKs in stress response pathways is further supported by a recent gene expression analysis which demonstrated that, in Arabidopsis, RLK transcript levels were dramatically changed in response to various abiotic and biotic stresses, as well as by applied hormones (Chae et al. 2009). Future studies will be performed to probe the role of these two PD-targeted WAK-RLKs.
In summary, we have identified a new class of PD proteins, PDRLKs, belonging to the WAK, LEC, and LRR subfamilies. These rice PDRLKs now provide an opportunity to identify orthologs in other plant species. Finally, a functional characterization of these PDRLKs will provide important insights into the evolution of signaling between the apoplasm and PD in terms of control over symplasmic signaling in response to environmental stimuli and developmental inputs.
Materials and methods
Plant material
Seeds of N. benthamiana were sown in a vermiculite/soil mixture, and plants were grown in a growth chamber under a 16:8-h light/dark regime at a constant temperature of 25°C. Three-week-old plants were used for Agrobacterium-mediated transient expression studies.
Cloning of TVCV MP-mCherry fusion construct
The mCherry ORF was amplified by PCR from the pRSETB vector (Shaner et al. 2004) using the following primers: mCherry-Sal-d1 (AAATGTCGACATGGTGAGCAAGGGCG) and mCherry-SmBgl-r1 (CTAGCCTAGAT-CTGCCCGGGTGCCCTTGTACAGCTCGTCC). The amplified mCherry was then used to replace GFP in p35S-TVCV MP-GFP (a gift from David Jackson), using the SalI and SmaI enzyme sites, to yield pCCL133 containing p35S-TVCV MP-mCherry. The fragment carrying the p35S-TVCV MP-mCherry was obtained from the pCCL133 vector by enzyme digestion using HindIII and PvuI. Using blunt end ligation, the p35S-TVCV MP-mCherry fragment was inserted at the HindIII site of pCAMBIA1300 to produce pCCL162. In this study, we used RFP to describe our mCherry results as it is a modified member of the RFP family.
Plasmid construction
The cDNAs for the selected 15 RLKs were obtained from the rice PIPELINE (http://cdna01.dna.affrc.go.jp/PIPE/). Table 1 provides the accession numbers for each cDNA and protein. For the C-terminal GFP fusion constructs, all cDNAs without stop codons were again amplified by PCR using gene-specific primers. Detailed information for PCR primers is provided in Supplemental Table 1. Purified PCR products were cloned into the pDONR207 entry vector with gentamicin resistance by the BP reaction, according to the manufacturer’s instructions (Invitrogen, Darmstadt, Germany). Entry clones were transformed into Escherichia coli DH10B competent cells by electroporation, using a Gene Pulser Plus (Bio-Rad, Hercules, CA, USA) with settings of 2.5 kV, 200 Ω, and 25 μF. Cells were then grown in Luria–Bertani medium containing gentamicin (25 μg/ml). Plasmid DNAs were prepared using Solgent Miniprep kit, according to the manufacturer’s instructions (Solgent, Daejeon, South Korea), and all constructs were verified by sequencing (Solgent, Daejeon, South Korea). LR clonase reactions were performed to transfer each cDNA from the entry clone into a pMDC83 gateway compatible destination vector containing an N-terminal GFP-his6-tagged fusion driven by the 35S promoter, according to the manufacturer’s instructions (Invitrogen; Curtis and Grossniklaus 2003). The product of recombination reactions (LR reactions) was used to transform competent E. coli strain DH10B by electroporation. Purified plasmids were finally transformed into competent Agrobacterium strain GV3101 by electroporation. PM marker fused with RFP (Nelson et al. 2007) was obtained from the Arabidopsis Biological Resource Center.
Agrobacterium-mediated transient expression assays
Agrobacterium cells containing each RLK-GFP fusion construct were grown to an OD600 of 1.0 in liquid yeast extract peptone medium (10 g of peptone, 5 g of NaCl, and 10 g yeast extract in 1 l volume) supplemented with antibiotics (kanamycin 50 μg/ml and gentamicin 25 μg/ml) and 100 μM acetosyringone. Cultured cells were centrifuged at 4,000 rpm for 15 min at 4°C, and then the cell pellet was resuspended in infiltration buffer (10 mM MES, pH 5.6, 10 mM MgCl2, 200 μM acetosyringone). Cell density was adjusted with infiltration buffer to give an OD of approximately 1.0. Agrobacterium harboring the Tomato bushy stunt virus P19 silencing suppressor was also co-infiltrated in order to minimize the gene silencing effects on RLK-RFP expression. Aliquots (1 ml) of Agrobacterium cells containing a RLK-GFP fusion construct and P19 construct were mixed together, and then a syringe was used to infiltrate the mixture into the lower surface of N. benthamiana leaves. Transient expression was monitored from 12 to 48 h post infiltration, using 12 h increments.
Confocal microscopy
Aniline blue staining for callose detection and PI staining were performed just before microscopic observation using 0.1% and 10 μg/ml, respectively. Blue, green, and red fluorescent signals were observed using an Olympus (Tokyo, Japan) confocal laser scanning microscope (model FV1000). Excitation and emission wavelengths for GFP, RFP, aniline blue, and PI were 488/510–540, 543/587–625, 405/425–525, and 543/555–655 nm, respectively.
Plasmolysis experiments
Cut leaf sections from N. benthamiana leaves transiently expressing RLK-GFP were incubated in 0.45 M mannitol until epidermal cells were plasmolyzed. After a short incubation period, GFP signals were observed using a CLSM.
Electronic supplementary materials
Below is the link to the electronic supplementary material.
Supplemental Table 1 List of primers used to obtain the entry clones from cDNAs. (XLS 30 kb)
Supplemental Fig. 1 Subcellular localization of nine RLKs targeted to the plasma membrane. Each GFP fusion protein was transiently expressed in N. benthamiana leaves by agroinfiltration. Images shown were collected by CLSM 36 h post-agroinoculation. Scale bars = 20 μm. Arrows indicate nuclei. (JPG 265 kb)
Acknowledgments
We thank Sunseon Kim for technical assistance, David Jackson (Cold Spring Harbor Laboratory) and David Baulcombe (University of Cambridge) for the P19 clone, and Uwe Sonnewald (Institut für Pflanzengenetik und Kulturpflanzenforschung) and Roger Tsien (University of California, San Diego) for the mCherry clone. This work was supported by World Class University program (R33-10002) through the National Research Foundation of Korea, funded by the Ministry of Education, Science and Technology.
Conflict of interest
The authors declare that they have no conflict of interest.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Abbreviations
- PD
Plasmodesmata
- PM
Plasma membrane
- ER
Endoplasmic reticulum
- MP
Movement protein
- RLK
Receptor-like kinase
Footnotes
Yeonhwa Jo, Won Kyong Cho, and Yeonggil Rim contributed equally to this work.
References
- Anderson CM, Wagner TA, Perret M, He ZH, He D, Kohorn BD. WAKs: cell wall-associated kinases linking the cytoplasm to the extracellular matrix. Plant Mol Biol. 2001;47:197–206. doi: 10.1023/A:1010691701578. [DOI] [PubMed] [Google Scholar]
- Bayer EM, Bottrill AR, Walshaw J, Vigouroux M, Naldrett MJ, Thomas CL, Maule AJ. Arabidopsis cell wall proteome defined using multidimensional protein identification technology. Proteomics. 2006;6:301–311. doi: 10.1002/pmic.200500046. [DOI] [PubMed] [Google Scholar]
- Bayer E, Thomas C, Maule A. Symplastic domains in the Arabidopsis shoot apical meristem correlate with PDLP1 expression patterns. Plant Signal Behav. 2008;3:853–855. doi: 10.4161/psb.3.10.6020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackman LM, Boevink P, Cruz SS, Palukaitis P, Oparka KJ. The movement protein of Cucumber mosaic virus traffics into sieve elements in minor veins of Nicotiana clevelandii. Plant Cell. 1998;10:525–538. doi: 10.1105/tpc.10.4.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae L, Sudat S, Dudoit S, Zhu T, Luan S. Diverse transcriptional programs associated with environmental stress and hormones in the Arabidopsis receptor-like kinase gene family. Mol Plant. 2009;2:84–107. doi: 10.1093/mp/ssn083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen XY, Kim JY. Callose synthesis in higher plants. Plant Signal Behav. 2009;4:489–492. doi: 10.4161/psb.4.6.8359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen MH, Sheng J, Hind G, Handa AK, Citovsky V. Interaction between the tobacco mosaic virus movement protein and host cell pectin methylesterases is required for viral cell-to-cell movement. EMBO J. 2000;19:913–920. doi: 10.1093/emboj/19.5.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Du L, Chen Z. Sensitization of defense responses and activation of programmed cell death by a pathogen-induced receptor-like protein kinase in Arabidopsis. Plant Mol Biol. 2003;53:61–74. doi: 10.1023/B:PLAN.0000009265.72567.58. [DOI] [PubMed] [Google Scholar]
- Chinchilla D, Zipfel C, Robatzek S, Kemmerling B, Nürnberger T, Jones JD, Felix G, Boller T. A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature. 2007;448:497–500. doi: 10.1038/nature05999. [DOI] [PubMed] [Google Scholar]
- Cilia ML, Jackson D. Plasmodesmata form and function. Curr Opin Cell Biol. 2004;16:500–506. doi: 10.1016/j.ceb.2004.08.002. [DOI] [PubMed] [Google Scholar]
- Clark SE, Running MP, Meyerowitz EM. CLAVATA1, a regulator of meristem and flower development in Arabidopsis. Development. 1993;119:397–418. doi: 10.1242/dev.119.2.397. [DOI] [PubMed] [Google Scholar]
- Crawford KM, Zambryski PC. Non-targeted and targeted protein movement through plasmodesmata in leaves in different developmental and physiological states. Plant Physiol. 2001;125:1802–1812. doi: 10.1104/pp.125.4.1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U. A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 2003;133:462–469. doi: 10.1104/pp.103.027979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escobar NM, Haupt S, Thow G, Boevink P, Chapman S, Oparka K. High-throughput viral expression of cDNA-green fluorescent protein fusions reveals novel subcellular addresses and identifies unique proteins that interact with plasmodesmata. Plant Cell. 2003;15:1507–1523. doi: 10.1105/tpc.013284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulkner CR, Blackman LM, Cordwell SJ, Overall RL. Proteomic identification of putative plasmodesmatal proteins from Chara corallina. Proteomics. 2005;5:2866–2875. doi: 10.1002/pmic.200401186. [DOI] [PubMed] [Google Scholar]
- Gallagher KL, Benfey PN. Both the conserved GRAS domain and nuclear localization are required for SHORT-ROOT movement. Plant J. 2009;57:785–797. doi: 10.1111/j.1365-313X.2008.03735.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher KL, Paquette AJ, Nakajima K, Benfey PN. Mechanisms regulating SHORT-ROOT intercellular movement. Curr Biol. 2004;14:1847–1851. doi: 10.1016/j.cub.2004.09.081. [DOI] [PubMed] [Google Scholar]
- He ZH, Cheeseman I, He D, Kohorn BD. A cluster of five cell wall-associated receptor kinase genes, Wak1-5, are expressed in specific organs of Arabidopsis. Plant Mol Biol. 1999;39:1189–1196. doi: 10.1023/A:1006197318246. [DOI] [PubMed] [Google Scholar]
- Heinlein M, Epel BL. Macromolecular transport and signaling through plasmodesmata. Int Rev Cytol. 2004;235:93–164. doi: 10.1016/S0074-7696(04)35003-5. [DOI] [PubMed] [Google Scholar]
- Imlau A, Truernit E, Sauer N. Cell-to-cell and long-distance trafficking of the green fluorescent protein in the phloem and symplastic unloading of the protein into sink tissues. Plant Cell. 1999;11:309–322. doi: 10.1105/tpc.11.3.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JY. Regulation of short-distance transport of RNA and protein. Curr Opin Plant Biol. 2005;8:45–52. doi: 10.1016/j.pbi.2004.11.005. [DOI] [PubMed] [Google Scholar]
- Kim JY, Yuan Z, Jackson D. Developmental regulation and significance of KNOX protein trafficking in Arabidopsis. Development. 2003;130:4351–4362. doi: 10.1242/dev.00618. [DOI] [PubMed] [Google Scholar]
- Kim JY, Rim Y, Wang J, Jackson D. A novel cell-to-cell trafficking assay indicates that the KNOX homeodomain is necessary and sufficient for intercellular protein and mRNA trafficking. Genes Dev. 2005;19:788–793. doi: 10.1101/gad.332805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohorn BD, Kobayashi M, Johansen S, Riese J, Huang LF, Koch K, Fu S, Dotson A, Byers N. An Arabidopsis cell wall-associated kinase required for invertase activity and cell growth. Plant J. 2006;46:307–316. doi: 10.1111/j.1365-313X.2006.02695.x. [DOI] [PubMed] [Google Scholar]
- Kragler F, Monzer J, Xoconostle-Cázares B, Lucas WJ. Peptide antagonists of the plasmodesmal macromolecular trafficking pathway. EMBO J. 2000;19:2856–2868. doi: 10.1093/emboj/19.12.2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurata T, Ishida T, Kawabata-Awai C, Noguchi M, Hattori S, Sano R, Nagasaka R, Tominaga R, Koshino-Kimura Y, Kato T, Sato S, Tabata S, Okada K, Wada T. Cell-to-cell movement of the CAPRICE protein in Arabidopsis root epidermal cell differentiation. Development. 2005;132:387–398. doi: 10.1242/dev.02139. [DOI] [PubMed] [Google Scholar]
- Lee JY, Yoo BC, Lucas WJ. Parallels between nuclear-pore and plasmodesmal trafficking of information molecules. Planta. 2000;210:177–187. doi: 10.1007/PL00008124. [DOI] [PubMed] [Google Scholar]
- Lee JY, Yoo BC, Rojas MR, Gomez-Ospina N, Staehelin LA, Lucas WJ. Selective trafficking of non-cell-autonomous proteins mediated by NtNCAPP1. Science. 2003;299:392–396. doi: 10.1126/science.1077813. [DOI] [PubMed] [Google Scholar]
- Lee JY, Taoka K, Yoo BC, Ben-Nissan G, Kim DJ, Lucas WJ. Plasmodesmal-associated protein kinase in tobacco and Arabidopsis recognizes a subset of non-cell-autonomous proteins. Plant Cell. 2005;17:2817–2831. doi: 10.1105/tpc.105.034330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehti-Shiu MD, Zou C, Hanada K, Shiu SH. Evolutionary history and stress regulation of plant receptor-like kinase/pelle genes. Plant Physiol. 2009;150:12–26. doi: 10.1104/pp.108.134353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Doerks T, Bork P. SMART 6: recent updates and new developments. Nucleic Acids Res. 2009;37:D229–D232. doi: 10.1093/nar/gkn808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy A, Epel BL. Cytology of the (1–3)-beta-glucan (callose) in plasmodesmata and sieve plate pores. In: Bacic A, Fincher GB, Stone BA, editors. Chemistry, biochemistry and biology of (1, 3)-beta glucans and related polysaccharides. San Diego: Academic; 2009. pp. 439–463. [Google Scholar]
- Levy A, Erlanger M, Rosenthal M, Epel BL. A plasmodesmata-associated beta-1, 3-glucanase in Arabidopsis. Plant J. 2007;49:669–682. doi: 10.1111/j.1365-313X.2006.02986.x. [DOI] [PubMed] [Google Scholar]
- Levy A, Guenoune-Gelbart D, Epel BL. beta-1, 3-Glucanases: plasmodesmal gate keepers for intercellular communication. Plant Signal Behav. 2007;2:404–407. doi: 10.4161/psb.2.5.4334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Chory J. A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell. 1997;90:929–938. doi: 10.1016/S0092-8674(00)80357-8. [DOI] [PubMed] [Google Scholar]
- Li J, Wen J, Lease KA, Doke JT, Tax FE, Walker JC. BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell. 2002;110:213–222. doi: 10.1016/S0092-8674(02)00812-7. [DOI] [PubMed] [Google Scholar]
- Liarzi O, Epel BL. Development of a quantitative tool for measuring changes in the coefficient of conductivity of plasmodesmata induced by developmental, biotic, and abiotic signals. Protoplasma. 2005;225:67–76. doi: 10.1007/s00709-004-0079-x. [DOI] [PubMed] [Google Scholar]
- Lucas WJ. Plasmodesmata: intercellular channels for macromolecular transport in plants. Curr Opin Cell Biol. 1995;7:673–680. doi: 10.1016/0955-0674(95)80109-X. [DOI] [PubMed] [Google Scholar]
- Lucas WJ. Plant viral movement proteins: agents for cell-to-cell trafficking of viral genomes. Virology. 2006;344:169–184. doi: 10.1016/j.virol.2005.09.026. [DOI] [PubMed] [Google Scholar]
- Lucas WJ, Bouche-Pillon S, Jackson DP, Nguyen L, Baker L, Ding B, Hake S. Selective trafficking of KNOTTED1 homeodomain protein and its mRNA through plasmodesmata. Science. 1995;270:1980–1983. doi: 10.1126/science.270.5244.1980. [DOI] [PubMed] [Google Scholar]
- Lucas WJ, Ham BK, Kim JY. Plasmodesmata—bridging the gap between neighboring plant cells. Trends Cell Biol. 2009;19:495–503. doi: 10.1016/j.tcb.2009.07.003. [DOI] [PubMed] [Google Scholar]
- Maule AJ. Plasmodesmata: structure, function and biogenesis. Curr Opin Plant Biol. 2008;11:680–686. doi: 10.1016/j.pbi.2008.08.002. [DOI] [PubMed] [Google Scholar]
- Nelson BK, Cai X, Nebenfuhr A. A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 2007;51:1126–1136. doi: 10.1111/j.1365-313X.2007.03212.x. [DOI] [PubMed] [Google Scholar]
- Oparka KJ. Getting the message across: how do plant cells exchange macromolecular complexes? Trends Plant Sci. 2004;9:33–41. doi: 10.1016/j.tplants.2003.11.001. [DOI] [PubMed] [Google Scholar]
- Oparka KJ, Prior DA, Santa Cruz S, Padgett HS, Beachy RN. Gating of epidermal plasmodesmata is restricted to the leading edge of expanding infection sites of tobacco mosaic virus (TMV) Plant J. 1997;12:781–789. doi: 10.1046/j.1365-313X.1997.12040781.x. [DOI] [PubMed] [Google Scholar]
- Oparka KJ, Roberts AG, Boevink P, Santa Cruz S, Roberts I, Pradel KS, Imlau A, Kotlizky G, Sauer N, Epel B. Simple, but not branched, plasmodesmata allow the nonspecific trafficking of proteins in developing tobacco leaves. Cell. 1999;97:743–754. doi: 10.1016/S0092-8674(00)80786-2. [DOI] [PubMed] [Google Scholar]
- Raffaele S, Bayer E, Lafarge D, Cluzet S, German Retana S, Boubekeur T, Leborgne-Castel N, Carde JP, Lherminier J, Noirot E, Satiat-Jeunemaitre B, Laroche-Traineau J, Moreau P, Ott T, Maule AJ, Reymond P, Simon-Plas F, Farmer EE, Bessoule JJ, Mongrand S. Remorin, a Solanaceae protein resident in membrane rafts and plasmodesmata, impairs potato virus X movement. Plant Cell. 2009;21:1541–1555. doi: 10.1105/tpc.108.064279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robards AW, Lucas WJ. Plasmodesmata. Annu Rev Plant Physiol Plant Mol Biol. 1990;41:369–419. doi: 10.1146/annurev.pp.41.060190.002101. [DOI] [Google Scholar]
- Roberts IM, Boevink P, Roberts AG, Sauer N, Reichel C, Oparka KJ. Dynamic changes in the frequency and architecture of plasmodesmata during the sink-source transition in tobacco leaves. Protoplasma. 2001;218:31–44. doi: 10.1007/BF01288358. [DOI] [PubMed] [Google Scholar]
- Sagi G, Katz A, Guenoune-Gelbart D, Epel BL. Class 1 reversibly glycosylated polypeptides are plasmodesmal-associated proteins delivered to plasmodesmata via the Golgi apparatus. Plant Cell. 2005;17:1788–1800. doi: 10.1105/tpc.105.031823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz J, Stussi-Garaud C, Tacke E, Prüfer D, Rohde W, Rohfritsch O. Long-distance signaling in nodulation directed by a CLAVATA1-like receptor kinase. Science. 2003;299:109–112. doi: 10.1126/science.1077937. [DOI] [PubMed] [Google Scholar]
- Searle IR, Men AE, Laniya TS, Buzas DM, Iturbe-Ormaetxe I, Carroll BJ, Gresshoff PM. Long-distance signaling in nodulation directed by a CLAVATA1-like receptor kinase. Science. 2003;299:109–112. doi: 10.1126/science.1077937. [DOI] [PubMed] [Google Scholar]
- Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE, Tsien RY. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- Simpson C, Thomas C, Findlay K, Bayer E, Maule AJ. An Arabidopsis GPI-anchor plasmodesmal neck protein with callose binding activity and potential to regulate cell-to-cell trafficking. Plant Cell. 2009;21:581–594. doi: 10.1105/tpc.108.060145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone JM, Trotochaud AE, Walker JC, Clark SE. Control of meristem development by CLAVATA1 receptor kinase and kinase-associated protein phosphatase interactions. Plant Physiol. 1998;117:1217–1225. doi: 10.1104/pp.117.4.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taoka K, Ham BK, Xoconostle-Cázares B, Rojas MR, Lucas WJ. Reciprocal phosphorylation and glycosylation recognition motifs control NCAPP1 interaction with pumpkin phloem proteins and their cell-to-cell movement. Plant Cell. 2007;19:1866–1884. doi: 10.1105/tpc.107.052522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas CL, Bayer EM, Ritzenthaler C, Fernandez-Calvino L, Maule AJ. Specific targeting of a plasmodesmal protein affecting cell-to-cell communication. PLoS Biol. 2008;6:e7. doi: 10.1371/journal.pbio.0060007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilney LG, Cooke TJ, Connelly PS, Tilney MS. The structure of plasmodesmata as revealed by plasmolysis, detergent extraction, and protease digestion. J Cell Biol. 1991;112:739–747. doi: 10.1083/jcb.112.4.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner A, Wells B, Roberts K. Plasmodesmata of maize root tips: structure and composition. J Cell Sci. 1994;107:3351–3361. doi: 10.1242/jcs.107.12.3351. [DOI] [PubMed] [Google Scholar]
- Vogel F, Hofius D, Sonnewald U. Intracellular trafficking of Potato leafroll virus movement protein in transgenic Arabidopsis. Traffic. 2007;8:1205–1214. doi: 10.1111/j.1600-0854.2007.00608.x. [DOI] [PubMed] [Google Scholar]
- Wagner TA, Kohorn BD. Wall-associated kinases are expressed throughout plant development and are required for cell expansion. Plant Cell. 2001;13:303–318. doi: 10.1105/tpc.13.2.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan J, Zhang XC, Neece D, Ramonell KM, Clough S, Kim SY, Stacey MG, Stacey G. A LysM receptor-like kinase plays a critical role in chitin signaling and fungal resistance in Arabidopsis. Plant Cell. 2008;20:471–481. doi: 10.1105/tpc.107.056754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazaki J, Kojima K, Suzuki K, Kishimoto N, Kikuchi S. The rice PIPELINE: a unification tool for plant functional genomics. Nucleic Acids Res. 2004;32:D383–D387. doi: 10.1093/nar/gkh001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambryski P, Crawford K. Plasmodesmata: gatekeepers for cell-to-cell transport of developmental signals in plants. Annu Rev Cell Dev Biol. 2000;16:393–421. doi: 10.1146/annurev.cellbio.16.1.393. [DOI] [PubMed] [Google Scholar]
- Zhang S, Chen C, Li L, Meng L, Singh J, Jiang N, Deng XW, He ZH, Lemaux PG. Evolutionary expansion, gene structure, and expression of the rice wall-associated kinase gene family. Plant Physiol. 2005;139:1107–1124. doi: 10.1104/pp.105.069005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table 1 List of primers used to obtain the entry clones from cDNAs. (XLS 30 kb)
Supplemental Fig. 1 Subcellular localization of nine RLKs targeted to the plasma membrane. Each GFP fusion protein was transiently expressed in N. benthamiana leaves by agroinfiltration. Images shown were collected by CLSM 36 h post-agroinoculation. Scale bars = 20 μm. Arrows indicate nuclei. (JPG 265 kb)


