Abstract
The A loop is an essential RNA component of the ribosome peptidyltransferase center that directly interacts with aminoacyl (A)-site tRNA. The A loop is highly conserved and contains a ubiquitous 2′-O-methyl ribose modification at position U2552. Here, we present the solution structure of a modified and unmodified A-loop RNA to define both the A-loop fold and the structural impact of the U2552 modification. Solution data reveal that the A-loop RNA has a compact structure that includes a noncanonical base pair between C2556 and U2552. NMR evidence is presented that the N3 position of C2556 has a shifted pKa and that protonation at C2556-N3 changes the C-U pair geometry. Our data indicate that U2552 methylation modifies the A-loop fold, in particular the dynamics and position of residues C2556 and U2555. We compare our structural data with the structure of the A loop observed in a recent 50S crystal structure [Ban, N., Nissen, P., Hansen, J., Moore, P. B. & Steitz, T. A. (2000) Science 289, 905–920; Nissen, P., Hansen, J., Ban, N., Moore, P. B. & Steitz, T. A. (2000) Science 289, 920–930]. The solution and crystal structures of the A loop are dramatically different, suggesting that a structural rearrangement of the A loop must occur on docking into the peptidyltransferase center. Possible roles of this docking event, the shifted pKa of C2556 and the U2552 2′-O-methylation in the mechanism of translation, are discussed.
Precise alignment of A-(aminoacyl) site and P-(peptidyl) site tRNA at the peptidyltransferase center is essential to ribosome function. A recent crystal structure of the isolated large ribosomal subunit (50S) revealed that the peptidyltransferase center is a complex tertiary arrangement of RNA elements principally composed of domain V of 23S rRNA (1). The crystal structure confirms much of the previous biochemical and genetic work showing that highly conserved nucleotides of 23S rRNA position tRNA substrates at the active site of the ribosome (2, 3). The interactions that orient tRNA within the ribosome include Watson–Crick base pairing between rRNA and universally conserved nucleotides at the 3′ termini of both A- and P-site tRNA. In Escherichia coli, G2553, within a hairpin loop implicated in A-site tRNA binding (A loop), base pairs with C75 of A-site tRNA, and G2252, within a hairpin loop implicated in P-site tRNA binding (P loop), base pairs with C74 of P-site tRNA. These interactions are essential to peptidyltransferase activity of the ribosome (3, 4).
In the 50S subunit crystal structure, the A and P loops are juxtaposed at the active-site pocket above the peptide exit channel by docking against residues of the central loop of domain V and the domain IV ridge. Peptidyltransferase substrates, soaked into the 50S crystals, base paired with the A and P loops, as predicted from previous biochemical and genetic studies.
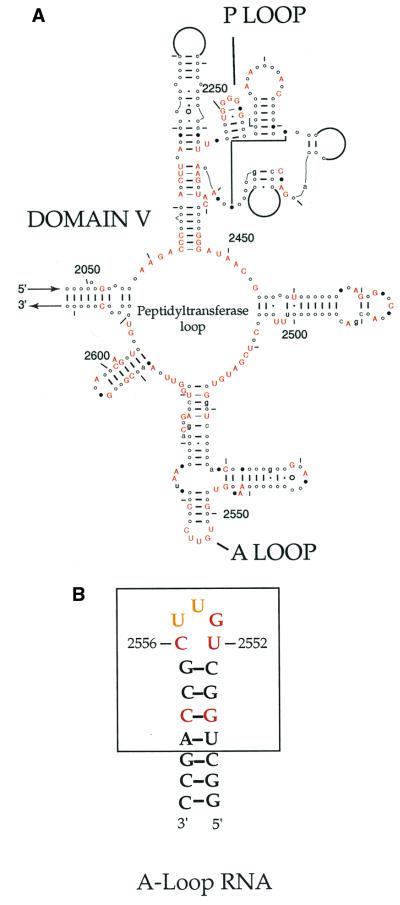
The P and A loops are found in domain V of 23S rRNA and in all 23S-like rRNA thus far sequenced (5) (Fig. 1a). All five A-loop residues are phylogenetically highly conserved; only transversions at positions U2554 and U2555 to cytidine occur within organisms of the Archea kingdom.
Figure 1.
(a) A portion of Domain V of 23S ribosomal RNA, highlighting conserved structural elements and nucleotides present in all kingdoms, including chloroplasts and mitochondria. Conservation data are superimposed on the E. coli secondary structural map and annotated with E. coli numbering. The A and P loops, flanking the peptidyltransferase (central) loop of Domain V, are indicated. AUCG residues shown in red are >95% conserved in all organisms; AUCG shown in lowercase are >90% conserved in all organisms; closed circles are 80–90% conserved, and open circles exist relative to E. coli >95% of the time. Arcs denote regions in nucleotide sequence highly variable in length and composition among rRNA. This figure has been modified from that found at the Comparative RNA web site (http://www.rna.icmb.utexas.edu/cite.html). (b) The A-loop RNA oligonucleotide designed to recapitulate the secondary structure within the E. coli ribosome. The boxed region indicates those nucleotides that exist in E. coli 23S rRNA. The three Watson–Crick base pairs extending the stem portion of the A loop were added to facilitate handling and increase thermodynamic stability. Nucleotides in red denote invariable residues within rRNA, and gold denotes >95% conservation; U2554 and U2555 infrequently vary to cytidine in Archeal organisms.
Binding of tRNA at the A loop may be critical to the mechanism of tRNA selection by the ribosome. Accommodation of aminoacyl-tRNA into the A site, a kinetically slow step in tRNA selection, follows aminoacyl-tRNA release from elongation factor-Tu and precedes rapid peptide-bond formation (6, 7). The accommodation step likely involves recognition of aminoacyl-tRNA by the A loop and movement of the A-site substrate into proper position at the active site before peptidyl transfer can occur. The exact nature of the rearrangements that occur during accommodation of A-site tRNA into the peptidyltransferase center remains unclear.
Mutations of residues within the A loop or the 3′ terminus of tRNA reduces translational fidelity and the efficiency of ribosome-catalyzed peptide-bond formation; these mutations likely affect the accommodation step of tRNA selection. Mutation of A76 and C75 of A-site tRNA or G2553, G2550, or G2557 of the A loop reduces the efficiency of ribosome-catalyzed peptide-bond formation in vitro (3, 8–11). Mutation of U2555 of the A loop to adenosine increases frameshifting errors and miscoding by the ribosome (12). These mutational data support a model in which binding of tRNA at the A loop is coupled to tRNA selection and accommodation. Accommodation rates for these mutants must be measured to test this hypothesis directly.
Posttranscriptional modification of rRNA at the peptidyltransferase center may play an important role in substrate binding, selection, or accommodation at the ribosomal A site. In all organisms thus far investigated, residues G2251 of the P loop and U2552 of the A loop are 2′-O-methylated, suggesting that 2′-O-methylations play an important role in modulating A- and P-loop function (13, 14). Only limited structural data are available on the impact of ribose modifications in RNA structure.
We present here the solution structure of the A loop (nucleotides 2548–2560 of E. coli 23S rRNA), with and without the U2552 2′-O-methyl modification, determined by NMR spectroscopy. In solution, the A loop adopts a compact fold in which conserved nucleotides form noncanonical base pairs that modulate loop architecture. Comparison of modified and unmodified A-loop structures suggests that U2552 2′-O-methylation plays a role in modifying the A-loop fold. Our data indicate a distinct structure from the A-loop fold observed within the 50S subunit crystal structure. These differences are carefully discussed.
Methods
Sample Preparation.
For the unmodified A loop, RNA was prepared by transcription from DNA templates by using phage T7 RNA polymerase and purified by using PAGE (15). Unlabeled and 13C,15N-labeled RNAs were prepared (16). RNAs were electroeluted from the gel and subsequently dialyzed against buffer in either 5% D2O/95% H2O or 99.9% D2O. NMR experiments were performed in Shigemi NMR tubes (5 mm OD, sample volume 250 μl) at RNA concentrations ranging from 1.0 to 3.0 mM. The 2′-O-methyl A-loop RNA was chemically synthesized by either the Yale oligonucleotide synthesis facility or Dharmacon Research, Boulder, CO. Following each facility's suggested deprotection protocol, modified A-loop RNA was purified and prepared for analysis exactly as described for unmodified A-loop RNA.
A-loop RNA molecules with and without the U2552 2′-O-methylation were investigated in a variety of solution conditions, including potassium (1–100 mM), sodium (5–100 mM), and magnesium (0–20 mM) salts, as well as in the presence of trifluoroethanol (0–10% vol/vol). All data used in the structure determination of both modified and unmodified A-loop RNA molecules were obtained in 10 mM sodium phosphate buffer at pH 6.2.
NMR Spectral Analyses.
NMR data were acquired at either 15 or 25°C on Varian Inova 500- and 800-MHz NMR spectrometers equipped with triple-resonance x-, y-, or z-axis gradient probes. Data for rapidly exchanging protons in the loop region were obtained at lower temperatures (2 and 5°C). 1H, 13C, 15N, and 31P assignments were obtained by using standard homonuclear and heteronuclear methods optimized for RNA structure determination (RnaPack, Varian User Library). Constant-time heteronuclear single quantum coherence spectroscopy, three-dimensional (3D) HCCH-total correlation spectroscopy (TOCSY), 3D HCCH-correlated spectroscopy (COSY), and two-dimensional (2D) HCCH-RELAY experiments were used to assign sugar-spin systems, and through-backbone assignments were made with HCP and HP-COSY experiments. Base-exchangeable protons were assigned by correlation to nonexchangeable base protons by using hetero-TOCSY methods. Intranucleotide H1′ to base proton correlations were obtained by using a 2D MQ-HCN experiment (17). Nuclear Overhauser effect (NOE) distance restraints were obtained from 2D-NOE spectroscopy (NOESY) (99.9% D2O) and WATERGATE-NOESY experiments (4% D2O) at 50, 100, and 150 ms mixing time. Base pairing was confirmed for the stem region of the A-loop RNA by using a HNN-COSY experiment (18). Generally, NOE restraints were characterized as strong (0–3.0 Å), medium (0–4.0 Å), weak (0–5 Å), and very weak (0–6 Å). In several instances, very weak NOE restraints observed in the loop region were loosened to 0–7 Å. Restraints to the 2′-O-methyl group were not included in the structure calculations because of difficulties determining NOE volume. Dihedral torsion restraints were obtained from TOCSY, double-quantum-filtered-COSY, 3D heteronuclear multiple quantum coherence-TOCSY, HP-COSY, and HCP experiments. Dihedral restraints were implemented as previously described (19). Spectra were analyzed with Varian vnmr software.
Structure Calculation.
Structures were calculated by using a simulated annealing protocol within BioSym Technologies (San Diego) INSIGHT II software. A random array of atoms corresponding to the sequence of either modified and unmodified A-loop RNA molecules was heated to 1,000 K, and bonding, distance and dihedral restraints and repulsive quartic potential were gradually increased to full value (distance: 50 kcal mol−1Å−2 and torsion: 50 kcal mol−1 rad−2) over 40 ps of molecular dynamics. The molecules were then cooled during 10 ps to 300 K and subjected to a final minimization step that included an attractive Lennard–Jones potential. An electrostatic term was not included in the target function at this stage of the calculation procedure. During this initial round of de novo calculations, Watson–Crick hydrogen-bonding restraints at 250 kcal mol−1 Å−2 were set for those stem residues experimentally determined to be canonically paired.
A total of 100 structures were generated in the first round of calculations. Low-energy structures (40 unmodified and 46 modified A-loop structures), cut off at three standard deviations above the low-energy average, were then subjected to additional rounds of refinement and minimization by using the X-PLOR 3.1 package. Refinement through 500 steps of restrained energy minimization (rMD) at 1,000 K while increasing the torsion angle force constant from 5 to 50 kcal mol−1 rad−2 during the protocol was completed by rMD while cooling to 300 K. Calculations were completed through 1,000 cycles of energy minimization, which included a Lennard–Jones potential and electrostatic terms. The final structures, shown in Fig. 2, had the lowest total energy and restraint violation energies. Nonconverged structures were at least one standard deviation higher in total and restraint violation energy.
Figure 2.
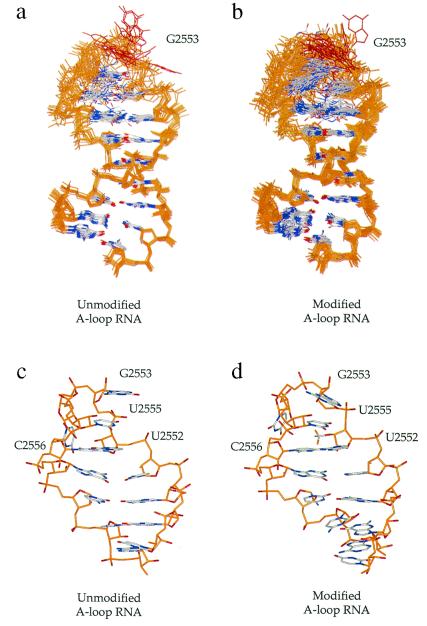
(a and b) Superpositions of (a) unmodified A-loop RNA and (b) modified A-loop RNA structures calculated from NMR data (superposing residues in the A-form stem). Bases are shown in gray, with nitrogen and oxygen atoms in blue and red, respectively. All other atoms are shown in amber. The G2553 base is highlighted in red to illustrate its disorder in the ensemble of structures. (c and d) Representative structures of the ensemble highlight the A-loop fold of both (c) unmodified and (d) modified A-loop RNA molecules. Colors are as indicated in the superposition of structures, except both phosphate oxygens and 2′-hydroxyl oxygens are shown in red.
Results
NMR Spectroscopy.
The A-loop structures with and without the U2552 2′-O-methyl modification were investigated by NMR by using a 19-nt RNA construct. The A-loop RNA bearing the U2552 modification was prepared synthetically, whereas the unmodified RNA was synthesized by transcription (Fig. 1b) (see Methods). Both A-loop RNA constructs contain a 5-nt loop closing a 7-bp A-form helix (Tm = 92°C ± 1.5° for both modified and unmodified A-loop RNA molecules; data not shown). Imino proton spectra, gel filtration, and UV melting studies demonstrated that the A-loop RNA is monomeric and forms the same secondary structure as found in the ribosome. Homonuclear and heteronuclear (13C, 15N, 31P) multidimensional NMR techniques enabled complete 1H resonance assignment of both modified and unmodified A-loop RNA oligonucleotides. Isotope labeling of the unmodified A-loop RNA facilitated complete 13C resonance assignment and partial 15N and 31P resonance assignment of the unmodified A-loop RNA (published as supplemental data on the PNAS web site; see www.pnas.org). Two- and three-dimensional NOE and through-bond correlation-based experiments provided 215 NOE distance restraints and 71 dihedral angle restraints (Table 1, second column) that were used in simulated annealing structure calculations (see Methods).
Table 1.
Structural statistics and atomic rms deviations for A-loop and U2552 2′-O-methyl A-loop RNA oligonucleotides
| A loop 〈SA〉1 | 2′-O-methyl A loop 〈SA〉2 | |
|---|---|---|
| Total experimental distance restraints | 215 | 212 |
| Total experimental dihedral restraints | 71 | 54 |
| Final forcing energies distance and dihedral restraints, kcal⋅mol−1 | 1.61 ± 0.41 | 1.28 ± 0.37 |
| rms deviation from experimental distance restraints, Å | 0.009 ± 0.003 | 0.008 ± 0.005 |
| rms deviation from experimental dihedral restraints, degrees | 0.54 ± 0.197 | 0.460 ± 0.341 |
| Deviations from idealized geometry | ||
| Bonds, Å | 0.004 ± 0.0001 | 0.006 ± 0.008 |
| Angle, degrees | 0.87 ± 0.01 | 0.88 ± 0.001 |
| Impropers, degrees | 0.20 ± 0.03 | 0.20 ± 0.05 |
| Heavy-atom rms deviation, Å | 〈SA〉 vs. SA | 〈SA〉 vs. SA |
| All A-loop RNA | 1.36 | |
| All 2′-O-methyl A-loop RNA | 1.69 |
〈SA〉1 refers to the final 15 simulated annealing structures. SA to the average structure are obtained by taking the average coordinates of the 15 simulated annealing structures best fitted to one another. 〈SA〉2 refers to the final 21 simulated annealing structures. SA to the average structure are obtained by taking the average coordinates of the 21 simulated annealing structures best fitted to one another. The final structures did not contain distance violations of >0.15 Å or dihedral violations of >5°.
The structure of the A-loop RNA bearing a U2552 2′-O-methylation was solved from 212 distance restraints and 54 dihedral restraints by using data obtained from homonuclear multidimensional experiments (Table 1, third column). Forty unmodified and forty-six modified structures (each of 100) converged to low energy in initial de novo structure calculations. These structures were subjected to refinement, and a final round of minimization and the lowest energy structures (15 unmodified and 21 modified) were superimposed. The overall rms deviation in heavy-atom position of the superimposed structures is 1.35 and 1.69 Å for unmodified and modified A-loop RNA, respectively (Fig. 2 a and b).
Both modified and unmodified A-loop RNA molecules adopt qualitatively similar folds, each having the highest degree of disorder within loop residues G2553 through C2556. In agreement, qualitative relaxation measurements of aromatic 13C-resonances and proton inversion recovery experiments indicate that G2553, U2554, and C2556 are the most dynamic residues within the unmodified A-loop RNA molecule (data not shown).
General Features of the A-Loop Fold.
Five conserved nucleotides cap an A-form helix in the A-loop structure (Fig. 2 c and d). The loop is closed by an unusual pyrimidine–pyrimidine base pair between universally conserved C2556 and U2552 nucleotides. This pyrimidine–pyrimidine base pair narrows the crosshelix backbone distance by 2 Å from A-form geometry to 11.5 Å (O4′-O4′).
The RNA backbone inverts at residue U2555 immediately 3′ of C2556. This polarity reversal creates a distinct kink in the loop backbone that directs the backbone of U2554 and G2553 toward the minor groove side of the helical axis. The NOEs defining backbone inversion at U2555 include U2555 H1′ to C2556 H5/H6, C2556 H5/H6 to U2555 H4′, and U2555 H1′ to G2557 H8/H1′.
Narrow pyrimidine–pyrimidine pairing and subsequent backbone reversal position the U2555 base directly above the hydrogen-bonding interface of the C-U pair. In this configuration, the 4′-ribose oxygen of U2555 is situated above the C2556 aromatic ring, and the base-pairing face of U2555 is oriented toward the major groove. Its position directly on the helical axis permits cross-strand stacking with G2553.
As seen in the ensemble of both modified and unmodified A-loop RNA structures, G2553 is highly mobile (Fig. 2 a and b). However, G2553 is predominantly (≈55% frequency in both ensembles) stacked above U2555, with its base-pairing face toward the major groove. Its position above U2555 extrudes U2554 into the minor groove side of the A loop. Nevertheless, the ensemble of calculated structures indicates that G2553 can move freely from major to minor groove with little energetic penalty.
In both modified and unmodified RNA molecules, G2553 has a predominantly C2′-endo ribose conformation but is highly dynamic and undergoes rapid syn-anti conformational switching. Syn-anti equilibria are often observed for purine residues having a C2′-endo sugar conformation, because the activation energy barrier for χ torsion angle rotation is on the order of kT (20). Syn-anti exchange of the G2553 base supports the observation that G2553 has a predominantly C2′-endo sugar conformation. Syn-anti exchange is evidenced by very strong H8/H1′ NOE volumes at short mixing time (50 ms) and interproton NOEs from the amino group of G2553 to the sugar protons of U2552 (H3′/H4′ and H5′/5′) (data not shown). Neither the sugar conformation of G2553 nor the NOEs defining the syn-anti equilibrium were included in either structure calculation. The methyl proton resonance (3.42 ppm) in the modified A-loop RNA provided seven clearly identifiable NOEs to loop protons. Although these NOEs were consistent with the calculated structure (data not shown), they were not included in structure calculations.
The A-loop fold is accommodated by unusual torsion angles within loop nucleotides. Both U2554 and U2555 possess a C2′-endo sugar conformation, which extends the phosphate–phosphate distance between residues to ≈7 Å from ≈5.8 Å found in canonical A-form C3′-endo geometry. Because U2555 inversion pushes the loop backbone into the minor groove, these two C2′-endo sugars may be necessary to span the additional distance across the helical stem. In three-dimensional HCP experiments performed on uniformly 13C- and 15N-labeled unmodified A-loop RNA, trans values were observed for the ɛ dihedral angles of residues C2556, U2555, and U2554. These unusual backbone angles facilitate formation of the 3-nt turn that bridges the two sides of the A loop.
The backbone torsion angles of C2556 are dynamic. C2556 possesses a mixed sugar-pucker conformation that interconverts between C2′-endo and C3′-endo on the timescale of the NMR experiment in both modified and unmodified A-loop RNA molecules (data not shown). However, it appears that U2552 2′-O-methylation shifts the C2556 sugar pucker toward a C2′-endo sugar pucker (data not shown). Conformational flexibility at the C2556 position is likely to impact the dynamic properties and position of neighboring nucleotides within the loop structure.
The C2556-U2552 Base Pair Changes with pH.
Chemical shifts of protons within the C2556-U2552 base pair and those proximal to it are sensitive to solution pH (chemical shifts at various pH values can be found in supplemental data, www.pnas.org). C2556 NH2, H5, U2552 H3, and C2556 H6 proton resonances show the largest changes (monitored over a range of two pH units, 5.5–7.5). Although one-dimensional NMR experiments at low temperature and low pH failed to reveal additional proton resonances, the chemical-shift data as a function of pH could be fit to a base titration curve whose estimated pKa is ≈6.4 (data not shown).
The modified and unmodified A-loop RNAs respond differently to pH changes. C2556 aromatic protons are unusually downfield shifted in both modified and unmodified A-loop RNA molecules. On lowering the pH from 7.5 to 5.5, both aromatic protons of C2556 shift further downfield (from pH 7.5 to 5.5, the ΔH5 shift is ≈0.12 ppm and the ΔH6 shift is ≈0.1 ppm) (Fig. 3a). These chemical-shift changes correspond to those that accompany acidification of cytidine monophosphate (pKa of N3 position is ≈4.5; data not shown). For the unmodified A-loop RNA, chemical-shift changes greater than those expected for N3 protonation alone are observed at residue C2556. Moreover, the chemical-shift changes observed throughout the loop are greater in the unmodified RNA. These data suggest that C2556 N3 protonation in the unmodified RNA is accompanied by additional conformational changes in the vicinity of the C2556-U2552 pair.
Figure 3.
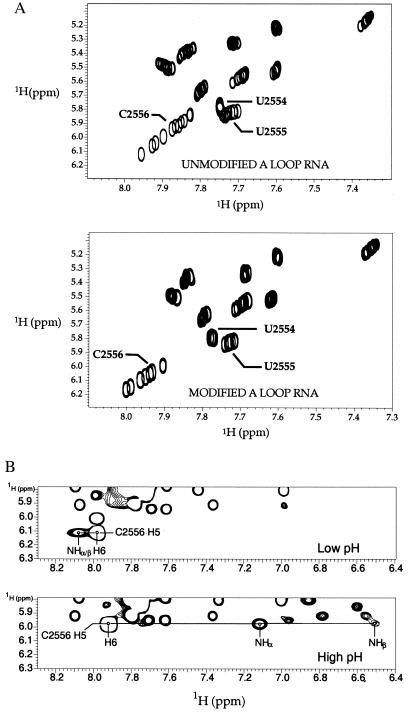
(a) pH dependence of selected A-loop RNA NMR spectra. Aromatic–aromatic region of wet-TOCSY spectra of unmodified (Upper) and modified (Lower) A-loop RNA molecules taken as a function of pH (5.5–7.5). Residues C2556, U2555, and U2554 are indicated. On decreasing solution pH by slow dialysis into isotonic 10 mM Na2PO4 buffer, C2556 moves downfield, U2555 moves upfield, and U2554 remains unchanged in both molecules. (b) WATERGATE-NOESY spectra of modified A-loop RNA taken at low (pH 5.5, Upper) and high (pH 7.5, Lower) pH. NOEs from C2556 H5 to its exocyclic amino protons are indicated. These experiments were performed at 500 MHz, 15°C, and a mixing time of 100 ms.
Protonation of the C2556 N3 changes the C-U pair geometry. Direct observation of a pH-induced C-U pair conformational change was achieved by monitoring the exocyclic amino proton resonances of C2556 at low and high pH. For both modified and unmodified RNA molecules, these proton resonances switch from being two peaks at ≈7.2 and ≈6.5 ppm in high pH solution (7.5) to a single resonance at ≈8.3 ppm in low pH solution (5.5) (Fig. 3b). The amino protons of C2556 probably experience two distinct chemical environments because of hydrogen bonding at high pH and one environment that is in fast rotational exchange at low pH (21).
The C2556-U2552 pair can adopt one of two hydrogen-bonding configurations, depending on the protonation state of the N3 position of C2556. On protonation, C2556 shifts toward the major groove and is unstacked from G2557 (Fig. 4). Change of the C-U pair geometry is probably accommodated by sugar-pucker and backbone torsion-angle changes around the C2556 nucleotide. Phosphorous-decoupled proton–proton COSY experiments (double-quantum-filtered-COSY) indicate that the sugar pucker of C2556 is affected by pH (data not shown). Specifically, H1′/H2′ and H3′/H4′ coupling data suggest that the sugar pucker of C2556 either slightly favors a C3′-endo conformation or becomes less dynamic at low pH. Further indication of pH-induced backbone rearrangements at the C2556 residue comes from HP-COSY spectra, in which the 31P chemical shift of G2553 changes as a function of pH (data not shown). pH-dependent changes occur in both modified and unmodified A-loop RNA molecules. The U2552 modification, although influencing the C2556-U2552 pair, does not greatly alter its conformational freedom.
Figure 4.
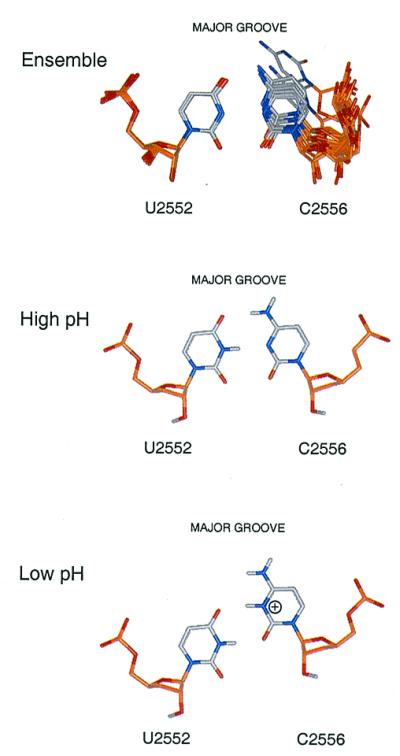
Upper illustrates superposition at U2552 of unmodified A-loop RNA structures showing the degree of variability in C2556-U2552 base-pairing geometry. Lower indicates our model of C-U pair restructuring that may occur on protonation at the N3 position of C2556.
U2555 Proton Resonances Are Influenced by U2552 2′-O-Methylation.
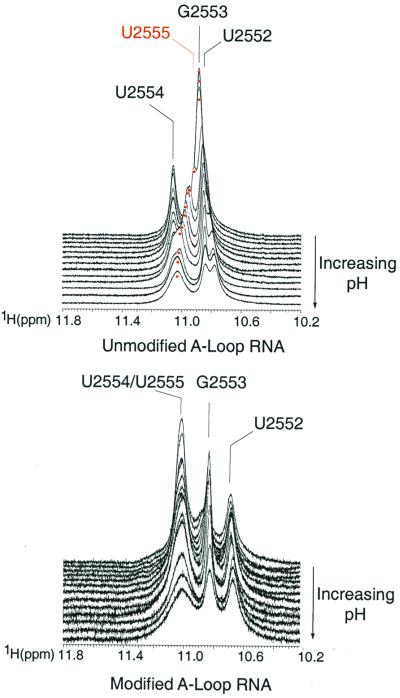
The imino proton spectrum of the unmodified A-loop RNA is more affected by pH changes than that of the modified RNA. The imino resonances of the modified A-loop RNA broaden with increasing pH but do not change chemical shift. By contrast, the imino proton chemical shift of U2555 and U2552 changes significantly in the unmodified A-loop RNA (≈0.3 ppm downfield shift for U2555 and ≈0.2 ppm upfield shift for U2552) with increasing pH (Fig. 5). These data are consistent with greater conformational changes occurring throughout the A loop in the absence of 2′-O-methylation, as discussed above.
Figure 5.
Imino proton spectra taken as a function of pH for both unmodified (Upper) and modified (Lower) A-loop RNA molecules. Resonances are labeled as indicated. The imino proton of U2555 shifts 0.3 ppm in the unmodified A-loop RNA from pH 7.5 (front) to pH 5.0 (back).
Discussion
The A-loop structure is consistent with both phylogenetic conservation of loop nucleotides, and general principles of RNA-loop structure. A conserved noncanonical base pair closes the hairpin loop, causing distortions that lead to reversal of the RNA backbone. Similar themes of noncanonical pairs capping helical sequence and local backbone reversals have been observed in many other RNA-loop structures (22, 23). Narrow base–base interactions such as U-U pairing or sheared G-A pairing are not observed in place of the C2556-U2552 pair of the A loop, suggesting that additional features of the C-U pair are required for ribosome function. We observed a shifted pKa for C2556 (≈6.4), whose protonation is coupled to changes in base-pairing geometry with U2552 and subtle changes in loop geometry. Protonation of C2556 seems to affect directly the U2555 base and may therefore be coupled with U2555 rearrangement.
The solution structure of the A loop presents the base-pairing faces of G2553 and U2555 toward the major groove. These functionally critical residues are conformationally dynamic within the loop structure, a feature that may be required for substrate binding and subsequent accommodation into the A site. Ribose methylation at U2552 has little effect on the overall loop fold but affects local conformational features of loop residues, including the configuration of C2556 and U2555 within the loop fold. Transversion of U2554 or U2555 to cytidine would have little consequence on the A-loop fold, whereas transition to a purine at either position would likely lead to distortion of backbone geometries to accommodate the larger base moiety.
Solution and Crystal Structures of the A Loop.
The solution structure of the A loop differs greatly from that observed in the crystal structure (rms deviation for positions 2584–2594 is 4.86 Å). The primary reason for this dramatic difference is that G2553, U2555, and C2556 are found on the minor-groove side of the A loop in the crystal structure. Reorientation of these bases in the crystal structure with respect to the helical axis is principally brought about by the polarity of the U2555 ribose. The backbone polarity at U2555 is not reversed in the crystal structure, whereas in solution, U2555's polarity reversal forces the loop backbone toward the minor groove. The observed conformational flexibility of U2555, C2556, and G2553 may be an important feature contributing to this conformational change. In the ensemble of structures, G2553 samples both major and minor groove. In addition, C2556 appears to slide freely from the major to minor groove side of the helical axis. Sliding of C2556 toward the minor groove is accompanied by a U2555 rearrangement that yields a solution structure with a U2555 and C2556 conformation similar to that of the crystal structure. These structures are eliminated from the ensemble because they possess higher total and restraint violation energies by the one SD cutoff.
In the crystal structure, all five residues of the A loop are modeled as having a C3′-endo sugar-pucker conformation. In solution, U2554 and U2555 possess strongly C2′-endo sugar-pucker conformations; G2553 is highly dynamic but adopts a predominantly C2′-endo sugar pucker. Trans-ɛ torsion angles observed in solution for U2554, U2555, and C2556 are not modeled in the crystallographically derived data. Moreover, unusual α, β, and γ torsion angles modeled in the crystal structure do not agree with the solution data. These differences can be attributed to large conformational changes of the A-loop structure on docking into the peptidyltransferase center. In particular, interaction of the sheared face of G2553 and the base-pairing face of C2556 with residues in helices 90 and 71, respectively, secure these residues in a conformation that is distinct from that found in solution. Additional packing interactions of the A loop with helices 61, 71, and 90 prevent G2553 and U2555 from occupying the major groove side of the A-loop helix.
Possible Function of 2′-O-Methylation at U2552 of the A Loop.
The U2552 2′-O-methyl modification has little effect on the global A-loop fold. However, U2552 ribose methylation influences conformational properties of critical loop residues—U2552, U2555, and C2556—that mediate tertiary interactions of the A loop within ribosome structure. C2556 in the modified A-loop RNA adopts a predominantly C2′-endo ribose conformation compared with a predominantly C3′-endo sugar conformation in unmodified A-loop RNA. This change of ribose conformation probably shifts the position of the C2556 base within the loop structure. Coupled to the change of C2556 sugar pucker is a change in the position of U2555. The chemical shift of the U2555 imino proton, although unchanged as a function of pH in the modified A-loop RNA, changes dramatically with pH in the unmodified RNA. U2555's change is accompanied by a similar change in the imino proton chemical shift of U2552 (Fig. 5). Absence of these chemical-shift changes in the modified A-loop RNA may reflect reduced conformational dynamics.
The conserved U2552 methylation is functionally important for the ribosome, as mutation of the U2552 methylase in E. coli leads to severe growth defects (24). The 2′-O-methylation of U2552 may be required for rRNA tertiary structure, the A-loop fold, or in the interaction of the A loop with A-site tRNA. The crystal structure of the 50S subunit suggests that the ribose modification does not directly mediate tertiary packing interaction of the A loop with the larger ribosome structure. Thus, the principle function of the U2552 modification must be on the A-loop fold itself, which indirectly influences tertiary packing or tRNA binding. U2554 packs on the minor groove face of the A loop in both solution and crystal structures. Stacking of U2554 on the ribose methyl group of U2552 would stabilize this configuration.
Shifted pKa Values and the Mechanism of Peptidyl Transfer.
The unusually shifted pKa found for the N3 of C2556 may be of importance to reversible conformational changes that occur at the A loop during one or more steps of translation. Such rearrangements may be required for substrate binding or docking of the A loop into the peptidyltransferase center or the peptidyltransferase reaction itself. Preliminary results from our lab suggest that a C2556U mutation is viable and that the pKa shift is not important to the catalytic activity of the 50S subunit (unpublished data). Although the observed pKa shift may be peculiar to the A-loop RNA oligonucleotide, the conformational plasticity of C2556 may be of functional importance to the mechanism of translation.
A-Loop and tRNA Accommodation at the A Site.
The solution structure of the A loop provides important insights into the mechanism of translation. The conformational flexibility evidenced in this region of the ribosome may reflect conformational changes undergone by the 50S subunit during tRNA selection, accommodation, and translocation. In particular, reversible undocking of the A loop may be required to bind tRNA and deliver it properly to the peptidyltransferase center. Indeed, such a rearrangement would account for the slow kinetics of tRNA accommodation into the A site. Evidence for translation cycle-dependent rearrangement of L12 in this region of the ribosome is known from electron microscopic studies (25). L12 remodeling on the ribosome is an elongation factor-dependent process. Likewise, undocking of the A loop from the peptidyltransferase center may be elongation factor dependent.
Supplementary Material
Acknowledgments
We thank R. Green and E. Viani Puglisi for many useful discussions on all aspects of the manuscript, as well as Peter Lukavsky and Stephen Lynch for contributing through generous technical support and discussion regarding NMR spectroscopic methods. This work was supported by National Institutes of Health Grant GM51266 and by a grant from the Lucille and David Packard Foundation. The Stanford Magnetic Resonance Facility is supported by funds from the Stanford University School of Medicine.
Abbreviations
- A-
aminoacyl
- P-
peptidyl
- COSY
correlated spectroscopy
- NOE
nuclear Overhauser effect
- TOCSY
total correlation spectroscopy
Footnotes
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID codes 1I3X and 1I3Y).
References
- 1.Ban N, Nissen P, Hansen J, Moore P B, Steitz T A. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- 2.Green R R, Switzer C, Noller H F. Science. 1998;280:286–289. doi: 10.1126/science.280.5361.286. [DOI] [PubMed] [Google Scholar]
- 3.Kim D F, Green R. Mol Cell. 1999;4:859–864. doi: 10.1016/s1097-2765(00)80395-0. [DOI] [PubMed] [Google Scholar]
- 4.Samaha R R, Green R, Noller H F. Nature (London) 1995;377:309–314. doi: 10.1038/377309a0. [DOI] [PubMed] [Google Scholar]
- 5.Gutell R R. Curr Opin Struct Biol. 1993;3:313–322. doi: 10.1016/s0959-440x(02)00339-1. [DOI] [PubMed] [Google Scholar]
- 6.Pape T, Wintermeyer W, Rodnina M V. EMBO J. 1998;17:7490–7497. doi: 10.1093/emboj/17.24.7490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pape T, Wintermeyer W, Rodina M W. EMBO J. 1999;18:3800–3807. doi: 10.1093/emboj/18.13.3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O'Connor M, Willis N M, Bossi L, Gesteland R F, Atkins J F. EMBO J. 1993;12:2559–2566. doi: 10.1002/j.1460-2075.1993.tb05911.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Porse B T, Garrett R A. J Mol Biol. 1995;249:1–10. doi: 10.1006/jmbi.1995.0276. [DOI] [PubMed] [Google Scholar]
- 10.Liu M, Horowitz J. Proc Natl Acad Sci USA. 1994;91:10389–10393. doi: 10.1073/pnas.91.22.10389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Green R, Noller H F. Annu Rev Biochem. 1997;66:679–716. doi: 10.1146/annurev.biochem.66.1.679. [DOI] [PubMed] [Google Scholar]
- 12.O'Connor M O, Dahlberg A E. J Mol Biol. 1995;254:838–847. doi: 10.1006/jmbi.1995.0659. [DOI] [PubMed] [Google Scholar]
- 13.Sirum-Connolly K, Mason T L. Science. 1993;262:1886–1889. doi: 10.1126/science.8266080. [DOI] [PubMed] [Google Scholar]
- 14.Sirum-Connolly K, Peltier J M, Crain P F, McCloskey J A, Mason T L. Biochimie. 1995;77:30–39. doi: 10.1016/0300-9084(96)88101-6. [DOI] [PubMed] [Google Scholar]
- 15.Wyatt J R, Chastain M, Tinoco I J. BioTechniques. 1991;11:764–769. [PubMed] [Google Scholar]
- 16.Batey R T, Inada M, Kujawinski E, Puglisi J D, Williamson J R. Nucleic Acids Res. 1992;20:4515–4523. doi: 10.1093/nar/20.17.4515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marino J P, Deiner J L, Moore P B, Griesinger C. J Am Chem Soc. 1997;119:7361–7366. [Google Scholar]
- 18.Dingley A J, Grzesiek S. J Am Chem Soc. 1998;120:8293–8297. [Google Scholar]
- 19.Puglisi E V, Puglisi J D. In: mRNA Metabolism and Post-Transcriptional Gene Regulation. Harford J B, Morris D R, editors. New York: Wiley; 1996. [Google Scholar]
- 20.Saenger W. Principles of Nucleic Acid Structure. New York: Springer; 1984. [Google Scholar]
- 21.Varani G, Tinoco I, J. Q Rev Biophys. 1991;24:479–532. doi: 10.1017/s0033583500003875. [DOI] [PubMed] [Google Scholar]
- 22.Puglisi E, Green R, Noller H F, Puglisi J D. Nat Struct Biol. 1997;10:775–778. doi: 10.1038/nsb1097-775. [DOI] [PubMed] [Google Scholar]
- 23.Battiste J L, Mao H, Rao N S, Tan R, Muhandiram D R, Kay L E, Frankel A D, Williamson J R. Science. 1996;273:1547–1551. doi: 10.1126/science.273.5281.1547. [DOI] [PubMed] [Google Scholar]
- 24.Bügl H, Fauman E B, Staker B L, Zheng F, Kushner S R, Saper M A, Bardwell J C A, Jakob U. Mol Cell. 2000;6:349–360. doi: 10.1016/s1097-2765(00)00035-6. [DOI] [PubMed] [Google Scholar]
- 25.Frank J, Agrawal R K. Nature (London) 2000;406:318–321. doi: 10.1038/35018597. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.