SUMMARY
We previously proposed that POT1 prevents ATR signaling at telomeres by excluding RPA from the single-stranded TTAGGG repeats. Here we use a Shld1-stabilized degron-POT1a fusion (DD-POT1a) to study the telomeric ATR kinase response. In the absence of Shld1, DD-POT1a degradation resulted in rapid and reversible activation of the ATR pathway in G1 and S/G2. ATR signaling was abrogated by shRNAs to ATR and TopBP1, but shRNAs to the ATM kinase or DNA-PKcs did not affect the telomere damage response. Importantly, ATR signaling in G1 and S/G2 was reduced by shRNAs to RPA. In S/G2, RPA was readily detectable at dysfunctional telomeres and both POT1a and POT1b were required to exclude RPA and prevent ATR activation. In G1, the accumulation of RPA at dysfunctional telomeres was strikingly less and POT1a was sufficient to repress ATR signaling. These results support an RPA exclusion model for the repression of ATR signaling at telomeres.
INTRODUCTION
Mammalian telomeres evade DNA damage checkpoints through the agency of shelterin, a six-subunit protein complex that binds to telomeres (reviewed in (Palm and de Lange, 2008; de Lange, 2009)). One of several pathways repressed by shelterin is the ATR kinase signaling cascade, which responds to single-stranded DNA. ATR signaling is activated by recessed 5’ ends, which occur at stalled replication forks and at DNA double-stranded breaks (DSBs) that have been processed (reviewed in (Cimprich and Cortez, 2008)). Telomeres are at risk of inappropriately activating the ATR kinase because they have a substantial segment of single-stranded TTAGGG repeats, either at their 3’ end or in the form of the displacement loop (D loop) at the base of the t-loop (McElligott and Wellinger, 1997; Makarov et al., 1997; Chai et al., 2006; Griffith et al., 1999). The single-stranded region is estimated to be 50–400 nt in length which is sufficient for the activation of the ATR kinase pathway in vitro (MacDougall et al., 2007).
The activation of ATR signaling requires RPA, an abundant heterotrimeric single-stranded DNA binding protein that functions in DNA replication, homology-directed DSB repair, and DNA damage signaling. The RPA-coated single-stranded DNA interacts with the ATRIP component of the ATR kinase complex thereby recruiting the ATR kinase to DNA lesions (Zou and Elledge, 2003). Activation of the RPA-bound ATR kinase requires TopBP1, a multiple BRCT-domain protein that interacts with Rad9 in the complex that is formed on the 5’ end flanking the single-stranded DNA by the Rad17 clamploader and the Rad9-Hus1-Rad1 clamp (Furuya et al., 2004; Kumagai et al., 2006; Delacroix et al., 2007; Lee et al., 2007; Mordes et al., 2008). The dual interaction of TopBP1 with Rad17/9-1-1 and ATR/ATRIP allows the ATR activating domain (AAD) of TopBP1 to activate the kinase (Kumagai et al., 2006; Mordes et al., 2008). The ATR kinase phosphorylates S/TQ sites on a large number of target proteins (Matsuoka et al., 2007), including factors that mediate cell cycle arrest, DNA repair, replisome stability, and replication restart. Constitutive activation of these pathways at the natural ends of mammalian chromosomes would not be compatible with cell viability and proliferation.
The repression of ATR signaling at vertebrate telomeres requires POT1, the component of shelterin that specifically binds single-stranded TTAGGG repeats (Hockemeyer et al., 2006; Churikov and Price, 2008; Lazzerini Denchi and de Lange, 2007; Guo et al., 2007). POT1 is bound to telomeres through its interaction with TPP1, which in turn associates with the double-stranded telomeric DNA binding proteins (TRF1 and TRF2) via TIN2 (reviewed in (Palm and de Lange, 2008)). Although POT1 proteins rely on TPP1, TRF1, TRF2, and TIN2 for their accumulation at telomeres, the converse is not true. Deletion of POT1 proteins from mouse cells does not perturb the other shelterin subunits, allowing assignment of specific functions to POT1 based on gene deletion experiments in mouse cells (Hockemeyer et al., 2006).
Unlike most vertebrates, including humans, mice and other rodents have two POT1 genes, coding for distinct telomeric proteins, POT1a and POT1b (Hockemeyer et al., 2006). POT1a and POT1b are similar in abundance, have the same affinity for telomeric DNA, and both require TPP1 for their recruitment to telomeres. Despite these similarities, POT1a and POT1b have largely non-overlapping functions that are specified by the N-terminal OB-fold DNA binding domains (Hockemeyer et al., 2006; Palm et al., 2009; Hockemeyer et al., 2008). POT1a is required for the repression of the ATR kinase whereas the primary function of POT1b is to protect telomeres from excessive shortening of the 5’ ended C-rich telomeric strand and accompanying extention of the 3’ overhangs. Deletion of POT1a induces phosphorylation of Chk1 and the formation of telomere dysfunction induced foci (TIFs) at telomeres in a subset of the cells (Hockemeyer et al., 2006; Lazzerini Denchi and de Lange, 2007). Cells lacking POT1b do not show these indices of telomere deprotection but have a 2–4 fold increase in the amount of single-stranded TTAGGG repeats at telomeres (Hockemeyer et al., 2006; Hockemeyer et al., 2008). However, deletion of both POT1a and POT1b exacerbates the DNA damage response, extending the TIF response to most cells (Hockemeyer et al., 2006). Such POT1a/b DKO cells also show an increased rate of telomere fusions in G2, undergo telomere recombination when Ku70/80 is absent, and show a high rate of endoreduplication (Palm et al., 2009; Hockemeyer et al., 2006; Wu et al., 2006). To study the repression of ATR signaling at telomeres, we have developed a switchable allele of POT1a that allows rapid and reversible deprotection of chromosome ends. Previous inducible inactivation of shelterin components relied on doxycyclin-inducible expression, Cre-mediated gene deletion, shRNA-mediated knockdown, and a temperature sensitive mutant. The Shld1 system has the advantage that the effect is rapid, reversible, and can be applied to most proteins by constructing a simple fusion protein (Banaszynski et al., 2006).
RESULTS AND DISCUSSION
Reversible telomere deprotection with a Shld1-controlled POT1a
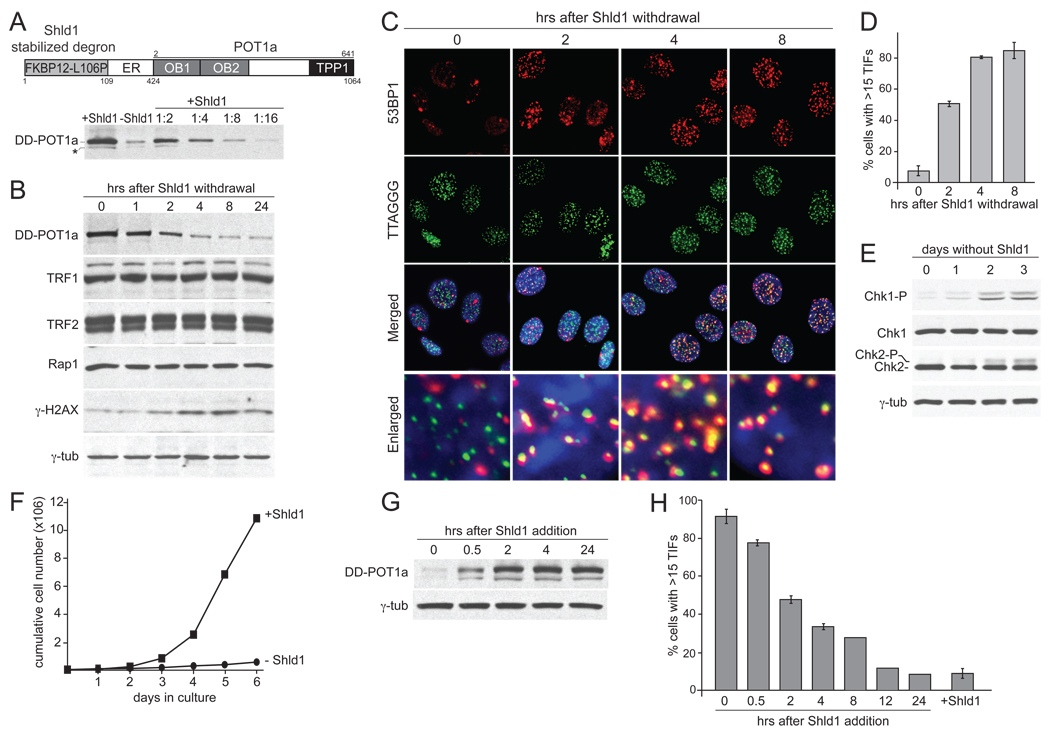
A switchable POT1a allele was generated by fusion to the destabilizing domain (DD) of the inherently unstable FKBP12-L106P mutant (Banaszynski et al., 2006) (Fig. 1A). The unfolding and concomitant degradation of this FKBP12 degron can be blocked by addition of the stabilizer Shield-1 (Shld1) to cell culture media (Banaszynski et al., 2006). The DD-POT1a fusion was expressed in POT1a/b DKO cells from which the endogenous POT1 proteins were subsequently deleted with Cre recombinase. A clonal derivative lacking the endogenous POT1a/b (c223) was used for the studies below (Suppl. Fig. 1A). The DD-POT1a fusion protein was expressed at a slightly higher level than the endogenous POT1a (see Figure 6 below). As anticipated, DD-POT1a levels gradually decreased over 6–8 hrs to ~10% of the original expression level when Shld1 was absent (Fig. 1A, B). At later time points, DD-POT1a reached a new, lower, steady-state level reflecting its faster degradation in the absence of Shld1 (Fig. 1B). When stabilized by Shld1, DD-POT1a was functional as evidenced by several indices of telomere protection and cells proliferated at a normal rate (Fig. 1B–F and data not shown). Telomeres became rapidly deprotected upon withdrawal of Shld1, resulting in the formation of TIFs and phosphorylation of H2AX and MCM2 within 6 hours (Fig. 1B–D; Suppl. Fig. 1B). The extent of telomere dysfunction reached a maximal level at 4–8 hours after removal of Shld1 and persisted for at least 6 days (Fig. 1D and Suppl. Fig.1B). At later time points, the cells showed phosphorylation of Chk1 and Chk2 and stopped proliferating (Fig. 1E, F). Upon re-addition of Shld1, DD-POT1a was re-expressed and reached maximal levels after 2 hours (Fig. 1G). This rise in POT1a was accompanied by the restoration of telomere function in most cells although full telomere protection was not achieved until ~12 hours after addition of Shld1 (Fig. 1H).
Figure 1. Reversible telomere deprotection with a switchable POT1a.
(A) Schematic of DD-POT1a and quantitative immunoblotting for DD-POT1a after Shld1 withdrawal. The complete POT1a ORF (starting at aa 2) was fused to the estrogen receptor domain (ER; not used in this study) and the Shld1-dependent mutated FKBP12 domain. The immunoblot shows DD-POT1a after Shld1 was removed from POT1a/b DKO cells expressing DD-POT1a (clone 223, isolated after Cre treatment) for 6 hrs. The last 4 lanes show serial dilutions of the Shld1-stabilized cell lysate. DD-POT1a was detected with POT1a Ab1221. The asterisk marks a 28 aa shorter form of DD-POT1a due to initiation at a second ATG. (B) Time course of DD-POT1a, other shelterin components, and γ-H2AX after Shld1 withdrawal for the indicated times. γ-tubulin serves as a loading control. (C) TIF response after Shld1 withdrawal. IF-FISH for 53BP1 (red) and telomeric TTAGGG DNA (green). The merged images include DAPI staining of DNA (blue). (D) Quantification of the TIF response in C. Average TIF response values and SDs were derived from three independent experiments. (E) Phosphorylation of Chk1 and Chk2 after several days without Shld1. Cells were removed of Shld1 for the indicated times and processed for immunoblotting. (F) Proliferation of the DD-POT1a c223 clone in the presence or absence of Shld1. (G) Re-expression of DD-POT1a after re-addition of Shld1. Shld1 was removed for 16 hrs and then added back for the indicated times. (H) Dissipation of the DNA damage response upon re-expression of DD-POT1a. The TIF response after Shld1 addition was determined as in panel C and quantified as in panel D. See Suppl. Fig. 1 for related data.
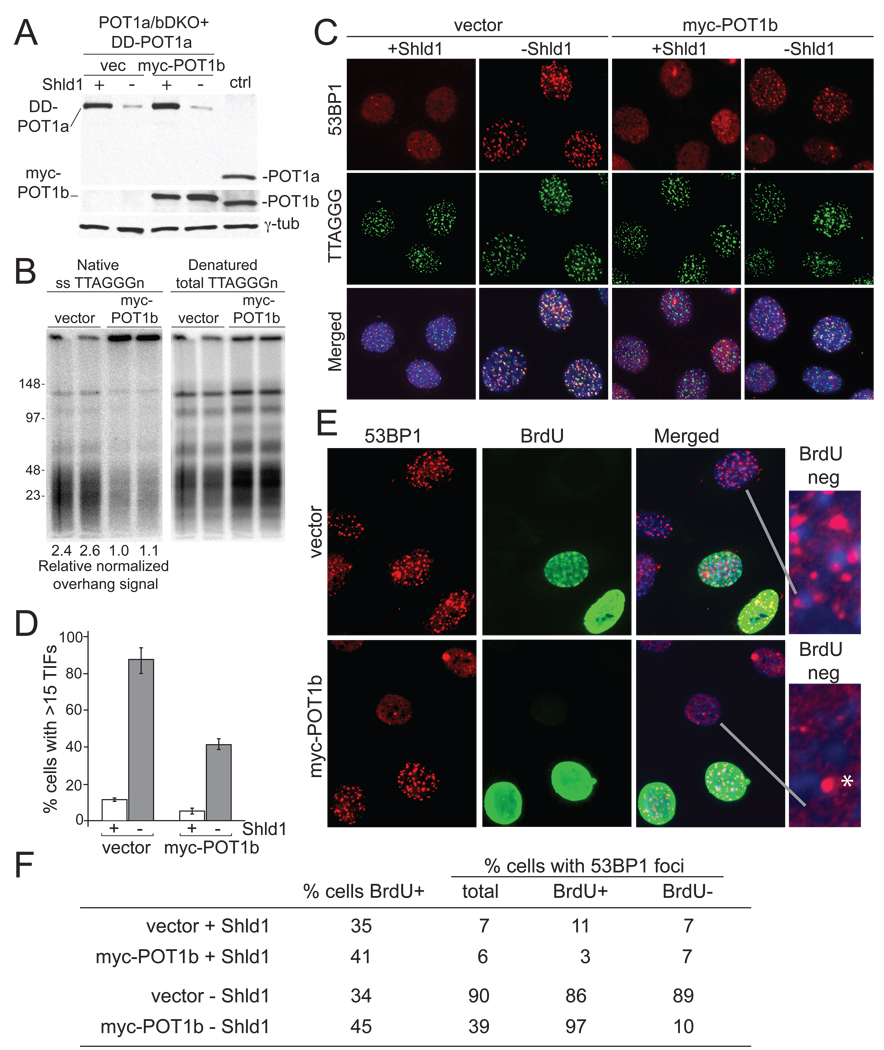
Figure 6. Different requirements for POT1-mediated telomere protection in G1 and S/G2.
(A) Immunoblotting comparing the expression of DD-POT1a and myc-POT1b in clone c223 to the endogenous POT1a and POT1b in the parental POT1a/bDKO cells (lane marked ctrl). Myc-POT1b (or empty vector) transduced c223 cells were incubated in the presence or absence of Shld1 for 16 hours and processed for immunoblotting. POT1a was detected with Ab 1221p; POT1b was detected with Ab 1223. (B) Rapid restoration of telomeric overhangs by myc-POT1b. Cells were collected at 48 hrs after infection with myc-POT1b (or the empty vector) and processed for telomeric DNA analysis in duplicate. Overhang signals were quantified with ImageQuant software and normalized to the denatured TTAGGG signal in the same lane. The numbers below the lanes show relative values of the normalized overhang signals with the value for lane 3 set to 1.0. (C) Effect of myc-POT1b expression on the TIF response after DD-POT1a depletion. Cells were collected as described in panel A and processed for IF-FISH as in Figure 1. (D) Quantification of the TIF response in C. Bars show average values and standard deviations derived from three experiments (>100 nuclei/experiment). (E) 53BP1 and BrdU co-staining in c223 cells with and without myc-POT1b. Asynchronous cultures were incubated without Shld1 for 4 hrs in the presence of 10 µM BrdU and then processed for IF for 53BP1 (red) and BrdU (green). Enlarged images show examples of 53BP1 pattern in nuclei lacking BrdU. Asterisk indicates the type of single 53BP1 foci often observed in untreated G1 cells. The foci are not indicative of a telomeric DNA damage response. (F) Quantification of the 53BP1 foci in BrdU positive and negative c223 cells with or without myc-POT1b. Cells were processed as in panel E and examined for 53BP1 foci. Cells with 15 or more 53BP1 foci were scored positive and evaluated for BrdU staining. Values are based on 150–250 cells. Similar data was obtained in a second independent experiment.
Specific activation of the ATR pathway
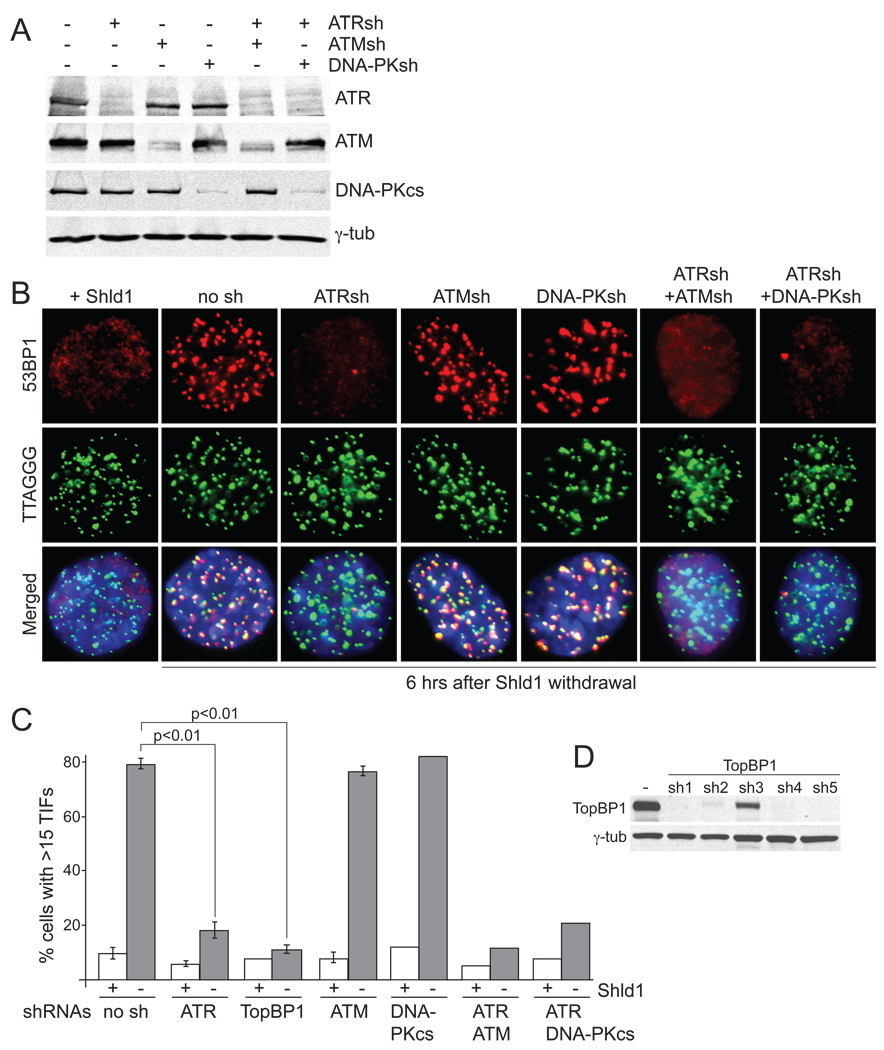
Previous studies showed that ATR signaling is activated within 3–4 days after deletion of POT1a or POT1a and -b. Inhibition of the ATR kinase with an shRNA in this context diminished the response but did not fully abrogate it, indicating either incomplete ATR inhibition or signaling through another pathway (Lazzerini Denchi and de Lange, 2007). To determine whether ATR is in fact the only transducer of telomere damage response in this setting, DD-POT1a cells were infected with shRNAs against ATM, ATR, or DNA-PKcs individually or in combination (Fig. 2A) and the damage response was measured by TIF analysis at 6 hours after Shld1 withdrawal (Fig. 2B,C). Whereas ATM and DNA-PKcs knockdown did not diminish the TIF response, knockdown of ATR severely decreased this outcome of telomere deprotection. The effect of ATR was not further exacerbated by simultaneous knockdown of ATM or DNA-PKcs, indicating that neither of these kinases played a substantial role in the response to the telomere damage generated by POT1a loss. The involvement of ATR in this telomere damage response was confirmed by inhibiting TopBP1 with several shRNAs, which resulted in a dramatic reduction in the TIF response to a level barely above background (Fig. 2C,D and data not shown). Thus, it is likely that the ATR kinase is the only transducer of the telomeric DNA damage signal after POT1 deletion. This result is consistent with the view that loss of POT1a and -b exposes the single-stranded telomeric DNA while other aspects of telomere protection, including repression of ATM signaling, remain intact.
Figure 2. DNA damage signaling after DD-POT1a depletion depends on ATR.
(A) Immunoblots to examine the effects of shRNAs against ATR, ATM, and DNA-PKcs. Cells were collected at 84 hrs after the first shRNA infection. (B) Effects of shRNAs against ATR, ATM, and DNA-PKcs on the TIF response. Cells were collected 84 hrs after the first shRNA infection, for the last 6 hrs, cells were incubated either in the presence or absence of Shld1 and processed for IF-FISH as in Fig. 1. (C) Quantification of the TIF response measured as in panel B. Average TIF response values and SDs for shRNAs against ATR, ATM, and TopBP1 (sh2, see (D)) were derived from three independent experiments. A similar reduction in TIF response was observed with TobBP1 sh4 and sh5. (D) Immunoblot showing reduction in TopBP1 level in shRNA treated cells. See Suppl. Fig. 2 for related data.
Activation of ATR signaling in G1 and S/G2
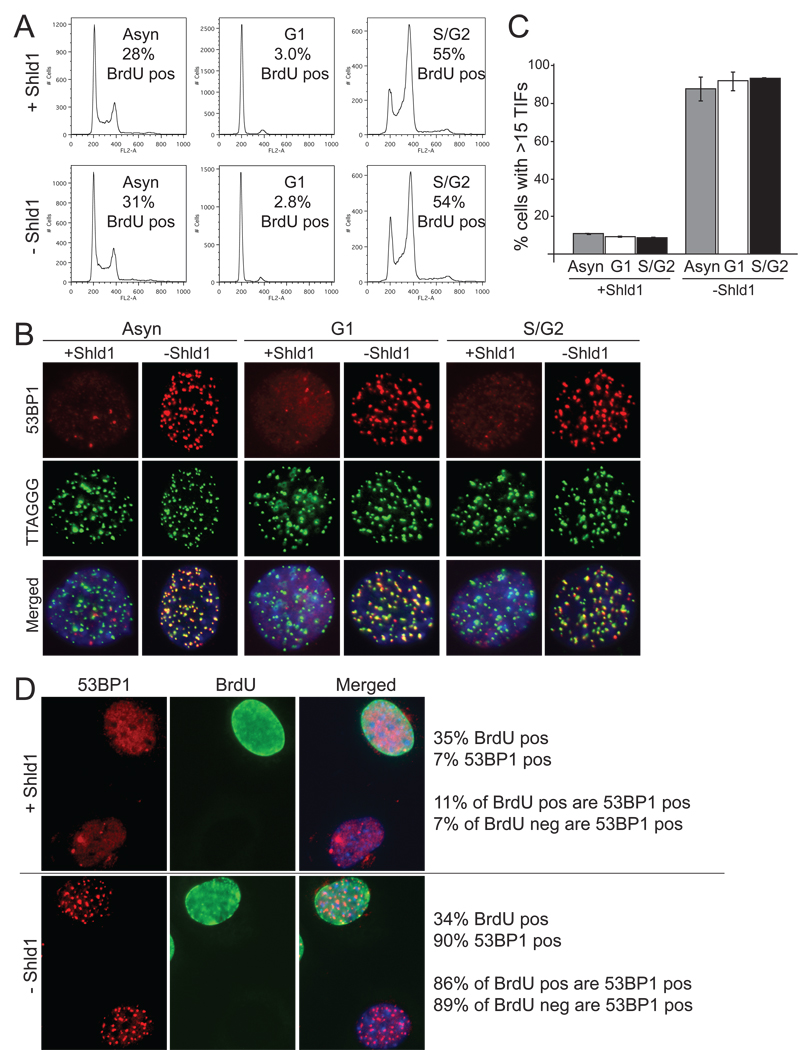
The rapid degradation of DD-POT1a allowed dissection of the cell cycle aspects of ATR kinase signaling. Using mitotic shake-off, M phase cells were harvested and allowed to progress through the cell cycle for 4 hours, resulting in a population of cells in G1 as evidenced by their DNA content and minimal incorporation of BrdU (Fig. 3A). A mixture of S and G2 cells was obtained 4 hours after release from a double thymidine block (Fig. 3A). Upon Shld1 withdrawal, the TIF response was not significantly different in G1 versus S/G2 and indistinguishable from the TIF response in asynchronous cells (p>0.05; Fig. 3B, C).
Figure 3. Activation of ATR signaling in G1 and S/G2.
(A) FACS profiles and BrdU incorporation of asynchronous, G1, and S/G2 cells with and without Shld1. For the asynchronous cells, Shld1 was removed for 4 hrs with BrdU added during the last hour. G1 cells were collected after mitotic shake-off and plated in the presence or absence of Shld1 for 4hrs with BrdU added during the last hour. S/G2 cells were synchronized with a double thymidine block. After the 2nd thymidine release, cells were incubated in the presence or absence of Shld1 for 4 hrs with BrdU added during the last hour. % of cells showing BrdU incorporation was determined based on BrdU IF. (B) TIF response in asynchronous, G1, and S/G2 cells with and without Shld1. Cells were collected as described in A and processed for IF-FISH as in Figure 1. (C) Quantification of the TIF response in B. Averages and SDs were derived from three experiments. The values in absence of Shld1 are not significantly different (p >0.05). (D) 53BP1 and BrdU co-staining after Shld1 withdrawal. Cells were incubated in the presence or absence of Shld1 together with 10 µM BrdU for 4 hrs and processed for IF for 53BP1 (red) and BrdU (green). The values to the right of the images were derived from analysis of ~200 cells that were evaluated for BrdU staining (BrdU pos and neg) and the presence of >15 53BP1 foci (referred to as 53BP1 pos).
The activation of ATR signaling in G1 was further confirmed in an asynchronous population of cells that was labeled with BrdU for 4 hours in the absence of Shld1. Because of the duration of the labeling period, both S phase cells and cells that were in G2 at the time of harvest are expected to be BrdU positive. Withdrawal of Shld1 did not affect the fraction of cells that incorporated BrdU (~35%) (Fig. 3D). Importantly, DNA damage foci occurred in close to 90% of the cells, regardless of whether they were BrdU positive (S/G2) or BrdU negative (G1) (Fig. 3D). Thus, POT1a inactivation in absence of POT1b resulted in the induction of ATR signaling in G1 cells as well as in S/G2.
RPA required for ATR activation in G1 and in S/G2
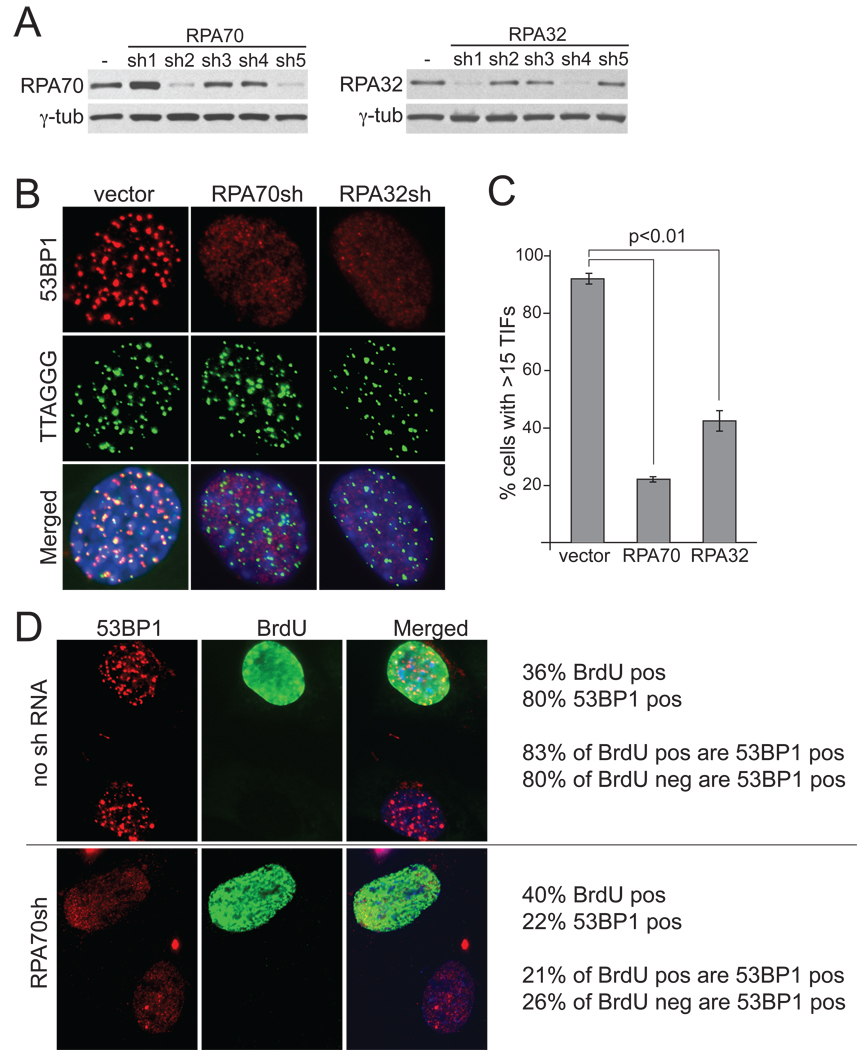
ShRNA-mediated knockdown was employed to query the role of RPA in the activation of the ATR pathway. Lowering the expression levels of RPA32 or RPA70 significantly diminished the formation of TIFs at telomeres lacking POT1a and POT1b (Fig. 4A–C). The ATR signaling was not fully abrogated by the RPA shRNAs, presumably because of the essential nature of RPA which precludes more complete knockdown.
Figure 4. Telomeric ATR signaling depends on RPA in G1 and S/G2.
(A) Immunoblotting to monitor the effects OF RPA70 and RPA32 shRNAs. Clone c223 cells were collected 84 hrs after the first infection with shRNA expressing retroviruses. (B) Effects of RPA70 and RPA32 shRNAs on the TIF response after DD-POT1a depletion. Cells were treated as in A (sh5 for RPA70 and sh4 for RPA32) and IF analysis was executed as in Figure 1 at 6 hours after removal of Shld1. (C) Quantification of the TIF response in B. Average TIF response values and SDs were derived from three independent experiments (>100 nuclei/experiment). (D) RPA70 shRNA lowers the incidence of cells with 53BP1 foci in G1 and S/G2. At 84 hrs after infection with RPA70 shRNA expressing retrovirus (or vector), cells were incubated for 4 hours in media containing 10 µM BrdU but no Shld1 and processed for IF for 53BP1 (red) and BrdU (green). The values to the right of the images were derived from analysis of ~200 cells that were evaluated for BrdU staining (BrdU pos and neg) and the presence of >15 53BP1 foci (referred to as 53BP1 pos). See Suppl. Fig. 3 for related data.
We next asked whether RPA was required for ATR signaling in both G1 and S/G2 cells. To address this question, we again used a 4-hour BrdU pulse to identify cells that had not replicated their DNA in the 4 hours prior to analysis and therefore were most likely in G1 at the time of harvesting. If RPA was required for ATR activation in S/G2 but not in G1, we would expect to observe a difference in the effect of RPA shRNA on the occurrence of 53BP1 foci in BrdU positive and negative cells. The overall incorporation rate of BrdU was not affected by the RPA shRNA. The telomeric DNA damage response was reduced to a similar extent in BrdU positive and negative cells, implying that RPA70 is required to activate ATR both during and outside S phase (Fig. 4D). A similar result was obtained with the shRNA to RPA32 (data not shown). These results demonstrate that RPA directly contributes to ATR activation at dysfunctional telomeres generated by POT1a/b loss both in G1 and in S/G2.
Different levels of RPA accumulation at telomeres in G1 and S/G2
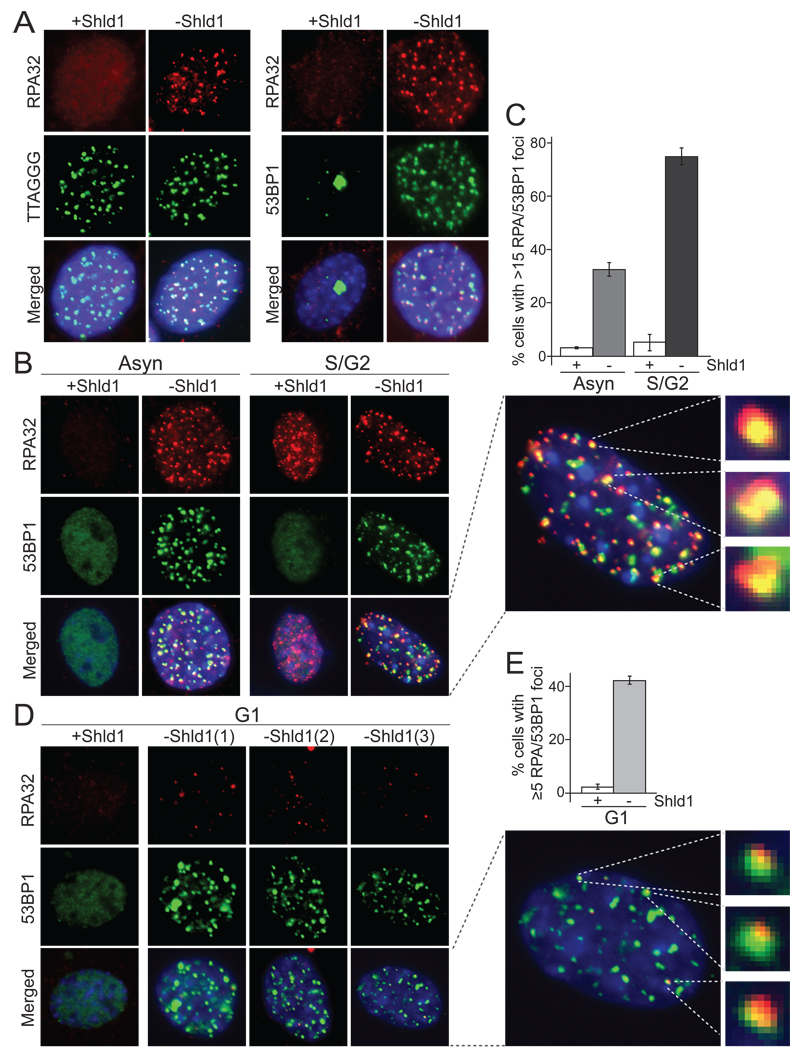
A previous report noted RPA co-localizing with telomeres upon inhibition of human POT1 but the data were difficult to interpret due to the high level of pan-nuclear RPA signals remaining after the fixation conditions used (Barrientos et al., 2008). We therefore examined the localization of RPA at telomeres using the DD-POT1a system and a well-established protocol for detection of RPA foci that involves pre-extraction of nucleoplasmic proteins (Dimitrova et al., 1999; Mirzoeva and Petrini, 2001). When DDPOT1a was stabilized with Shld1, RPA foci were absent from most cells (Fig. 5A). As expected, ~30% of the cells exhibited the numerous small RPA32 foci indicative ongoing DNA replication, but these foci did not co-localize with telomeric DNA (data not shown) and did not contain 53BP1 (Fig. 5B). When DD-POT1a was removed, RPA32 formed readily detectable foci that co-localized with 53BP1 and telomeric DNA (Fig. 5A). After Shld1 removal, ~35% of the cells in the asynchronous population had more than 15 RPA foci that co-localized with 53BP1 and therefore were presumed to represent the accumulation of RPA at dysfunctional telomeres (Fig. 5C). In the S/G2 cell population, 70–80% of the cells showed RPA foci coinciding with 53BP1 (Fig. 5B, C), indicating that most of the S/G2 cells with 53BP1 at their telomeres (~90%, see Fig. 3C) also showed telomeric accumulation of RPA.
Figure 5. RPA at dysfunctional telomeres upon DD-POT1a depletion.
(A) Formation of RPA foci at 6 hrs after Shld1 withdrawal from c223 cells. Left panels, RPA32 IF (red) and FISH for telomeric TTAGGG repeats (green). Right panels, IF for RPA32 (red) and 53BP1 (green). The merged images include DAPI staining of DNA (blue). (B) IF for RPA (red) and 53BP1 (green) in asynchronous and S/G2 cells after Shld1 withdrawal. An enlarged nucleus is shown on the right together with three randomly selected 53BP1/RPA foci. (C) Quantification of the % of cells containing RPA/53BP1 foci as shown in B. (D) IF for RPA32 (red) and 53BP1 (green) in G1 cells after Shld1 withdrawal. Cells were collected as described in Figure 3. Three independent images of G1 cells after Shld1 withdrawal are shown. An enlarged nucleus is shown on the right together with three 53BP1 foci that also contain RPA. The enlarged images in B and D were processed identically so that the relative intensities of 53BP1 and RPA in the foci can be compared. (E) Quantification the fraction of G1 cells with 53BP1 foci that contain RPA as detected as in D.Cells with 5 or more foci were scored positive. The bargraphs represents the average from three independent experiment and the standard deviations. See Suppl. Fig. 4 for related data.
A different result was obtained with G1 cells. Although 90% of G1 cells had 53BP1 at their telomeres after Shld1 removal (Fig. 3C), the RPA foci in G1 were weak and only about 40% cells showed more than 5 RPA foci co-localizing with 53BP1 (Fig. 5D, E). The intensity of RPA staining at the telomeres of the remainder of the TIF positive G1 cells appeared too low to be detected under these conditions. Together, these experiments establish that RPA localizes to most telomeres that have lost protection by POT1a/b, regardless of the cell cycle stage.
Interestingly, RPA is more readily detectable at deprotected telomeres in S/G2, presumably indicating that the amount of RPA per telomere is greater. Why RPA loading on the dysfunctional telomeres is increased in S/G2 is not clear. The ~2-fold increase in the single-stranded DNA at telomeres in S phase (Suppl. Fig. 4) seems insufficient to explain the much more intense RPA staining. It is not known whether the ability of RPA to load onto ss DNA is increased in S phase. Trimeric RPA is expressed equally throughout the cell cycle (Loo and Melendy, 2000) and while RPA32 is phosphorylated in S phase and in response to DNA damage, the effect of this modification on its ability to associate with ssDNA is not known (reviewed in (Binz et al., 2004)).
Different requirements for POT1-mediated protection in G1 and S/G2
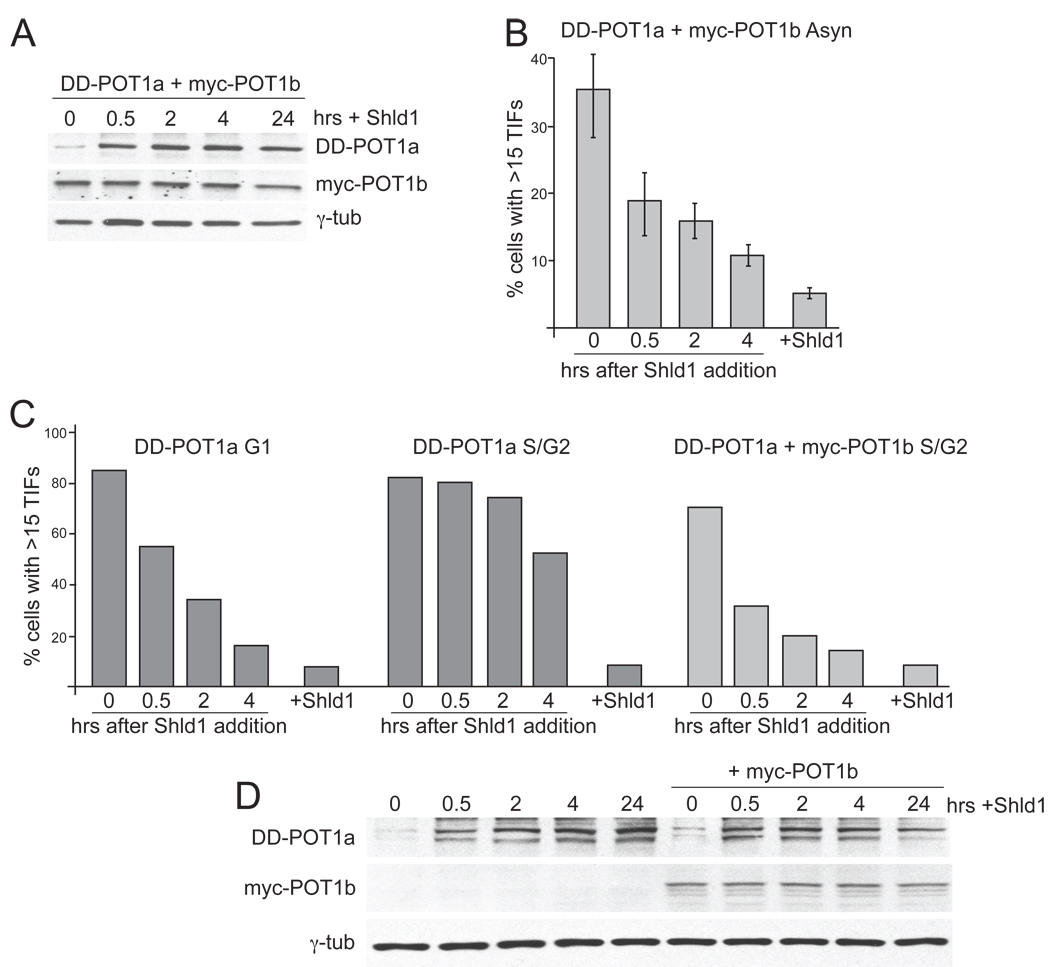
Given the prominent difference in RPA staining at dysfunctional telomeres in G1 and S/G2, we determined the effect of cell cycle stage on the ability of POT1a and -b to protect telomeres. Previous data showed that the two POT1 proteins differ in their ability to repress the DNA damage response at the telomeres. Whereas deletion of both genes leads to telomere deprotection in all cells, only 30–40% of POT1a deficient cells show TIFs. To determine whether these cells represent a specific cell cycle stage, Myc-tagged POT1b was expressed in the c223 DD-POT1a clone lacking both of the endogenous POT1 proteins (Fig. 6A). Immunoblotting showed that DD-POT1a and myc-POT1b are expressed at a level slightly higher than the endogenous proteins (Fig. 6A). As expected, the expression of myc-POT1b rapidly reduced the excessive overhang signal associated with the absence of POT1b (Fig. 6B) and limited the TIF response to ~40% of the cells (Fig. 6C and D).
To determine whether the residual TIF response was limited to a specific cell cycle phase, cells were examined after a 4-hour BrdU pulse to differentiate G1 cells from those in S and G2. The expression of POT1b did not affect the BrdU uptake of cells (Fig. 6E,F). When POT1b was absent, nearly 90% of the cells contained 53BP1 foci, regardless of whether they were in G1 or S/G2. However, when POT1b was present, the 53BP1 foci occurred primarily in cells containing BrdU, indicating that the TIF response was limited to S/G2 cells (Fig. 6E,F). Thus, when POT1b is present, POT1a is required to repress ATR signaling in S/G2 but not in G1.
Different requirements for restoration of telomere protection in G1 versus S/G2
We next tested the effect of cell cycle stage on the restoration of telomere protection in cells re-expressing POT1a with or without POT1b (Fig. 1G, H and Fig. 7A,B). Using cells synchronized in G1 and S/G2, the dissipation of the telomere damage response was monitored over 4 hours after addition of Shld1. The expression of myc-POT1b did not affect the cell cycle profile of the cells (Suppl. Fig. 5). The re-establishment of telomere protection by POT1a alone was nearly complete in G1 cells but substantially delayed in S/G2 cells (Fig. 7C and Suppl. Fig. 3). After 4 hours, more than 50% of the S/G2 population remained TIF positive and full restoration of telomere protection required a time-period that would include progression through mitosis. In contrast, when POT1b was also present, telomere protection was re-established efficiently in S/G2 cells (Fig. 7C). Restoration of the telomeres appeared nearly complete in 4 hours (Fig. 7C) even though the kinetics of POT1a re-expression in the S/G2 cells was not altered (Fig. 7D). Thus, POT1a alone is sufficient to rapidly re-establish telomere protection in G1. In contrast, rapid restoration of telomere function in S/G2 requires re-expression of POT1a in a POT1b proficient setting. It is likely that the presence of POT1b facilitates the restoration of telomere protection in S/G2 by both limiting the extent of the single-stranded DNA at telomere ends and by binding to the TTAGGG repeats.
Figure 7. Restoration of telomere protection in G1 and S/G2.
(A) Immunoblot for DD-POT1a and myc-POT1b expression after Shld1 addition. Cells were generated as in Figure 6 and deprived of Shld1 for 16 hrs. Shld1 was added back for the indicated times and the cells were processed for immunoblotting (POT1a, Ab1221; POT1b, Ab1223). (B) Quantification of the TIF response after Shld1 addition in POT1b proficient cells. Cells were collected as described in A and processed for IF-FISH to score TIFs as in Figure 1. (C) Quantification of the TIF response after Shld1 addition in synchronized cells deficient or proficient for POT1b as indicated. G1 cells were collected after mitotic shake-off and plated in the absence of Shld1 for 4 hrs after which Shld1 was added back for the indicated times before analysis of the TIF response. S/G2 cells were obtained with a double thymidine block. After the release from the 2nd thymidine block, cells were incubated in the absence of Shld1 for 4 hrs. Shld1 was then added back for the indicated times before processing for TIF analysis. (D) Time course of DD-POT1a and myc-POT1b levels of the S/G2 cells described in C. See Suppl. Fig. 5 for related data.
In absence of POT1b, the restoration of telomere protection by POT1a is much faster in G1 than in S/G2 (Fig. 7C). This difference may be due to a temporary increase in the single-stranded overhangs in S phase (see Suppl. Fig. 2), titration of POT1a to single-stranded DNA at the replication fork, or a change in POT1a (e.g. modification) altering its ability to bind to telomeres. A fourth possibility, which we favor, is that the slower restoration of telomeres in S/G2 is related to the apparently greater amount of RPA loaded on telomeres (Fig. 5) which would require higher levels of POT1a in order to effectively remove RPA from the single-stranded DNA.
Conclusions
A priori, there are a number of ways in which POT1 proteins might block ATR signaling. As a protein positioned close to (or at) the 5’ recessed end of the telomere, POT1 could prevent the loading of the 9-1-1 clamp or interfere with the interaction between 9-1-1 and TopBP1. POT1 could also act further downstream in the signaling cascade, as has been proposed for its protection of telomeres in fission yeast (M. Godinho Ferreira, pers. comm.). The current data support a simpler model, previously proposed by us and others (Lazzerini Denchi and de Lange, 2007; Barrientos et al., 2008; Guo et al., 2007; Churikov and Price, 2008), in which POT1 prevents activation of the ATR pathway by blocking the binding of RPA to the single-stranded TTAGGG repeats. In agreement with this RPA exclusion model, RPA appears at telomeres and is required for activation of the ATR kinase once POT1a/b are removed. Furthermore, consistent with POT1a competing with RPA, telomere protection by POT1a can be re-established in a few hours and does not require progression through S phase or mitosis. Finally, RPA is more prominent at dysfunctional telomeres in S/G2 versus G1 and this difference correlates with the increased requirement for POT1 proteins at telomeres in S/G2. The future challenge will be to establish the mechanism by which POT1 proteins, which are not abundant (Takai et al., 2010), exclude the abundant RPA from the telomeric DNA.
EXPERIMENTAL PROCEDURES
Plasmid constructs
DD-POT1a was expressed from pBabe-DD-ER-POT1a encoding full-length human FKBP12, estrogen-receptor (ER) ligand-binding domain, and full-length mouse POT1a was generated by PCR and ligated into a pBabe-puro retroviral vector. The F36V and L106P mutations were introduced into FKBP12 using QuickChange Site-Directed Mutagenesis Kit (Stratagene) according to the manufacturer’s instructions. Although the fusion construct contained the ER domain, this feature was not employed in the experiments since preliminary data indicated poor regulation of POT1a with tamoxifen. pLPC-N-myc-POT1b encoding full-length mouse POT1b preceded by a myc tag was generated from the pLPC-N-myc retroviral vector.
Generation of DD-POT1a expressing cells
To generate a clonal DD-POT1a expression cell line, pBabe-DD-ER-POT1a was introduced into SV-LT POT1aSTOP/FLOXPOT1bSTOP/FLOX MEFs (Hockemeyer et al., 2006) using 4 retroviral infections at 12-hr intervals using supernatant from transfected ecotropic Phoenix cells. The endogenous POT1a and -b genes were deleted with Hit&Run Cre recombinase and clones were then isolated with cloning cylinders in the presence of 1 µM Shld1 (BD Biosciences). One clone, c223, was selected for further study based on the absence of the endogenous POT1a/b and its rapid response after Shld1 withdrawal. To express POT1b in this clone, pLPC-N-myc-POT1b was introduced by 4 retroviral infections at 12-hr intervals using supernatant from transfected ecotropic Phoenix cells.
To confirm the absence of the endogenous POT1a and POT1b, genotyping PCRs were preformed using standard DNA isolation techniques and Takara Taq polymerase (Madison, WI, USA). POT1a FLOX PCR: 6-FRTfw-2 TGAGCCCAGAAAGCGAAGGAG; 6-FRTbw1 ACAAACCCACCCCGTCAGAGTAAG. POT1a Δex3 PCR: 6-allfw-2 CTTCCCTGTTTGCCCTCCTTTACT; 6-allbw-2 TTCCCCCTTTCATTTTCTTTTCTC. POT1b FLOX PCR: 17wtfw-1 CGCTGGGGAGGGTATCGTAG; 6-FRTfw-2 TGAGCCCAGAAAGCGAAGGAG. POT1b Δex3 PCR: 17-allfw-1 GTTGCCCCTATCATCCTACACG; 17-FRTbw-2 TGTGTTGGGAGAGGAAGTGAAAGA. PCRs were performed for 32 cycles (94°Cfor 45 s, 60°C for 45 s, 72°C for 60 s).
shRNAs
Previously published shRNAs against ATRsh3-1 (GGAGATGCAACTCGTTTAA, (Lazzerini Denchi and de Lange, 2007)), ATMsh3 (GGAAGTCAAGGAACAACAACTA), and DNA-PKcssh3 (CGGATCCCTGGTAGAACAGT) were introduced by 4 infections at 12 hr intervals using pSuperior-hygro (ATM sh3 and ATR sh3-1) or pSuper-puro (DNA-PKcs sh3) retroviral vectors. For combined knockdown of ATR and ATM, or ATR and DNA-PKcs, shRNAs against ATM or DNA-PKcs were introduced first by 4 infections at 12 hr intervals using supernatant from transfected ecotropic Phoenix cells and 72 hrs later, shRNAs against ATR was introduced by another 4 infections at 12 hr intervals. Cells were collected 84 hrs (or 156 hrs for combined shRNA treatment) after first infection for analysis.shRNAs (Open Biosystems) against RPA70 (sh1, CCCTGTTTGAAGATAGCAGAA; sh2, GCCCTGAAGATCGCTAACAAA; sh3, CGCATGATCTTATCGGCAAAT; sh4, CGTTGGATTAAAGATTGGGAA; sh5, CGCGAACATCAGGAAGAACAT); RPA32 (sh1, GCACTTTCCTTCCCTTGTCTT; sh2, CCCAGCATATTGTGCCCTGTA; sh3, GAATAACTTCATGCCAGCAAA; sh4, GAATTGGAGATGTCGAGATTT; sh5, GATCACTTTAAGTCTACAGAT); TopBP1 (sh1, CCTGAATTTGAATCACTGGTT; sh2, GCTCTTAGAAACTGCGAGAAT; sh3, GCTTTATATCTGTGACCGTTT; sh4, CGCTTTATATCTGTGACCGTT; sh5, GCCAGAAGAGTTTCCTTGTTT) were introduced by 4 lentiviral infections at 12 hr intervals using supernatant from transfected 293T cells. Infections with the empty vector pLK0.1 were used as a negative control.
Immunoblotting
Cells were lysed in 2xLaemmli buffer (100 mM Tris-HCl, pH 6.8, 200 mM dithiothreitol, 3% sodium dodecyl sulfate, 20% glycerol, 0.05% bromophenol blue), denatured at 100°C for 5 min, sheared with an insulin needle, and subjected to SDS-PAGE. Equivalent of 2 × 105 cells per lane was loaded. After immunoblotting, membranes were blocked in PBS or TBS with 5% milk and 0.1% Tween20, and incubated with the following primary antibodies in 5% milk and 0.1% Tween20: affinity-purified rabbit antibody raised against mTRF1, #1449; mTRF2, #1254; mRAP1, #1252; γ-tubulin (clone GTU 488, Sigma); γH2AX (mouse monoclonal, Upstate Biotechnology); phosphorylated Chk1 S-345 (rabbit monoclonal, Cell Signaling Technology); Chk2 (mouse monoclonal, BD Biosciences); RPA32 (rabbit polyclonal, Bethyl Laboratories); RPA70 (rabbit polyclonal, Bethyl Laboratories); TopBP1 (rabbit polyclonal, Abcam); ATM (mouse monoclonal, Sigma); ATR (goat polyclonal, Santa cruz Biotechnology); DNA-PKcs (mouse monoclonal, Neomarkers); MCM2-serine108 (rabbit polyclonal, Bethyl Laboratories). Immunoblots for POT1a and POT1b were performed using the renaturation protocol described previously (Loayza and de Lange, 2003) with affinity-purified antibody raised against POT1a (#1221) and POT1b (#1223). Immunoblot for detection of MCM2 at serine108 was performed using the NETN buffer (400 mM NaCl, 20 mM Tris-HCl, pH 8.0, 1 mM EDTA, 0.5% NP40), and 5 × 103 cells were loaded per lane. Secondary antibodies were horseradish-peroxidase-conjugated, donkey anti-mouse or anti-rabbit IgGs. Blots were developed ECL (Amersham).
IF and IF-FISH
One million cells were grown on 12 mm coverslips and fixed for 10 min in 2% paraformaldehyde at room temperature. Cells were rinsed with PBS, permeabilized with 0.5% NP-40 in PBS for 10 min, blocked with PBG (1% BSA, 0.2% cold fish gelatin in PBS) for 30 min, and incubated with antibody against 53BP1 (rabbit polyclonal; 100-304A, Novus Biologicals) diluted in PBG for 1 h at room temperature. After PBS washes, cells were incubated with Alexa Fluor 555 goat anti rabbit secondary antibody (Invitrogen) for 1 hr. For IF-FISH, cells were fixed again with 2% paraformaldehyde for 8 min, then dehydrated consecutively in 70%, 95%, and 100% ethanol for 5 min each, and allowed to dry completely. Hybridizing solution (70% formamide, 1 mg ml−1 blocking reagent (Roche), 10 mM Tris-HCl, pH 7.2, containing PNA probe FITC-OO-(AATCCC)3 (Applied Biosystems)) was added to each coverslip and the cells were denatured by heating for 10 min at 80°C on a heat block. After 2 h incubation at room temperature in the dark, cells were washed twice with washing solution (70% formamide, 10 mM Tris- HCl, pH 7.2) and twice in PBS. For BrdU co-staining, cells were fixed again with 2% paraformaldehyde for 8 min, followed by incubation of denaturing solution 4 N HCl for 10 min. After extensive wash in PBS, cells were incubated with FITC-conjugated anti-BrdU (BD Biosciences) for 1 hr. For RPA and 53BP1 co-staining, the in situ cell fractionation protocol was used as described (Mirzoeva and Petrini, 2001). RPA32 was detected with a mouse monoclonal antibody (Abcam). DNA was counterstained with DAPI and slides were mounted in anti-fade reagent (ProLong Gold, Invitrogen). Digital images were captured with a Zeiss Axioplan II microscope with a Hamamatsu C4742-95 camera using Improvision OpenLab software.
Cell synchronization and FACS analysis
For mitotic shake-off, 2–3×106 cells were plated in T150 flasks and incubated for 16–24 hrs. Mitotic cells were harvested by tapping the flasks manually and the supernatant was transferred to a 250 ml centrifuge bottles and kept on ice. The selecting process was repeated twice to collect sufficient numbers of mitotic cells over a 1 hr period. Cells were then centrifuged at 1000 rpm for 5 min, and replated for experiments and FACS analysis.
For double-thymidine block, 5×105 cells were plated on 10 cm dishes and grown for 16–24 hrs. Thymidine was added to a final concentration of 2 mM and cells were incubated for 14–16 hrs. Cells were washed three times with PBS and grown in fresh medium for 10–12 hrs. A second thymidine block was performed the same way and cells were incubated for another 14–16 hrs. Finally, cells were washed three times with PBS and released into fresh medium.
For FACS analysis, 1×106 cells were collected, washed in PBS, and fixed in ice cold 70% ethanol for at least 24 hr. Cells were recovered by centrifugation and resuspended in 0.5 ml of 0.5% BSA in PBS containing 5 µg propidium-iodide and 100 µg RNase A per ml. The samples were analyzed on a FACScalibur flow cytometer (Becton Dickinson). Data analysis was performed using FlowJo software.
Analysis of telomeric DNA
Mouse telomeric DNA was analyzed on CHEF gels as described previously (Celli and de Lange, 2005). Cells were resuspended in PBS and mixed 1:1 (v/v) with 2% agarose (SeaKem agarose) to obtain 5 × 105 cells per agarose plug. Plugs were digested overnight with 1 mg ml−1 Proteinase K (in buffer containing 100 mM EDTA, 0.2% sodium deoxycolate, 1% sodium lauryl sarcosine), washed extensively with TE buffer (10 mM Tris-HCl, pH 8.0, 1 mM EDTA), equilibrated with MboI digestion buffer for 1 hr, and incubated overnight at 37°C with 60 U MboI in 0.5 ml per plug. The following day, the plugs were washed once in TE and once in water, and were equilibrated in 0.5 × TBE. Plugs were loaded on a 1% agarose/0.5 × TBE gel and run for 24 h using CHEF-DRII PFGE apparatus (BioRad) in 0.5 × TBE running buffer. The settings were as follows: initial pulse, 5 min; final pulse, 5 min; 6 V cm−1; 14°C. In-gel hybridization of the native gel with a 32P-ATP end-labelled (CCCTAA)4 oligonucleotide and subsequent denaturation and hybridization steps were performed as described. Gels were exposed onto a PhosphoImager screen overnight, and the single-stranded G-overhang signal was quantified with ImageQuant software and normalized to the total telomeric DNA quantified after denaturation.
HIGHLIGHTS.
A Shld1-dependent POT1a allows rapid and reversible deprotection of telomeres
Loss of POT1 protection activates RPA- and TopBP1-dependent ATR kinase signaling
RPA accumulates at deprotected telomeres in S/G2 but less so in G1
Distinct requirements for POT1a in S/G2 and G1 support an RPA exclusion model
Supplementary Material
ACKNOWLEDGEMENTS
We thank Thomas Wandless for initial supplies of Shld1 and members of the de Lange lab for comments on this work. This work was supported by grants from the NIH to TdL (OD000379 and AG016642) and by an Anderson Cancer Center Postdoctoral fellowship award to YG.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Banaszynski LA, Chen LC, Maynard-Smith LA, Ooi AG, Wandless TJ. A rapid, reversible, and tunable method to regulate protein function in living cells using synthetic small molecules. Cell. 2006;126:995–1004. doi: 10.1016/j.cell.2006.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrientos KS, Kendellen MF, Freibaum BD, Armbruster BN, Etheridge KT, Counter CM. Distinct functions of POT1 at telomeres. Mol Cell Biol. 2008;28:5251–5264. doi: 10.1128/MCB.00048-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binz SK, Sheehan AM, Wold MS. Replication protein A phosphorylation and the cellular response to DNA damage. DNA Repair (Amst) 2004;3:1015–1024. doi: 10.1016/j.dnarep.2004.03.028. [DOI] [PubMed] [Google Scholar]
- Celli G, de Lange T. DNA processing not required for ATM-mediated telomere damage response after TRF2 deletion. Nat Cell Biol. 2005;7:712–718. doi: 10.1038/ncb1275. [DOI] [PubMed] [Google Scholar]
- Chai W, Du Q, Shay JW, Wright WE. Human telomeres have different overhang sizes at leading versus lagging strands. Mol Cell. 2006;21:427–435. doi: 10.1016/j.molcel.2005.12.004. [DOI] [PubMed] [Google Scholar]
- Churikov D, Price CM. Pot1 and cell cycle progression cooperate in telomere length regulation. Nat Struct Mol Biol. 2008;15:79–84. doi: 10.1038/nsmb1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimprich KA, Cortez D. ATR: an essential regulator of genome integrity. Nat Rev Mol Cell Biol. 2008;9:616–627. doi: 10.1038/nrm2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lange T. How telomeres solve the end-protection problem. Science. 2009;326:948–952. doi: 10.1126/science.1170633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delacroix S, Wagner JM, Kobayashi M, Yamamoto K, Karnitz LM. The Rad9-Hus1-Rad1 (9-1-1) clamp activates checkpoint signaling via TopBP1. Genes Dev. 2007;21:1472–1477. doi: 10.1101/gad.1547007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrova DS, Todorov IT, Melendy T, Gilbert DM. Mcm2, but not RPA, is a component of the mammalian early G1-phase prereplication complex. J Cell Biol. 1999;146:709–722. doi: 10.1083/jcb.146.4.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuya K, Poitelea M, Guo L, Caspari T, Carr AM. Chk1 activation requires Rad9 S/TQ-site phosphorylation to promote association with C-terminal BRCT domains of Rad4TOPBP1. Genes Dev. 2004;18:1154–1164. doi: 10.1101/gad.291104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith JD, Comeau L, Rosenfield S, Stansel RM, Bianchi A, Moss H, de Lange T. Mammalian telomeres end in a large duplex loop. Cell. 1999;97:503–514. doi: 10.1016/s0092-8674(00)80760-6. [DOI] [PubMed] [Google Scholar]
- Guo X, Deng Y, Lin Y, Cosme-Blanco W, Chan S, He H, Yuan G, Brown EJ, Chang S. Dysfunctional telomeres activate an ATM-ATR-dependent DNA damage response to suppress tumorigenesis. EMBO J. 2007;26:4709–4719. doi: 10.1038/sj.emboj.7601893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockemeyer D, Daniels JP, Takai H, de Lange T. Recent expansion of the telomeric complex in rodents: Two distinct POT1 proteins protect mouse telomeres. Cell. 2006;126:63–77. doi: 10.1016/j.cell.2006.04.044. [DOI] [PubMed] [Google Scholar]
- Hockemeyer D, Palm W, Wang RC, Couto SS, de Lange T. Engineered telomere degradation models dyskeratosis congenita. Genes Dev. 2008;22:1773–1785. doi: 10.1101/gad.1679208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai A, Lee J, Yoo HY, Dunphy WG. TopBP1 activates the ATR-ATRIP complex. Cell. 2006;124:943–955. doi: 10.1016/j.cell.2005.12.041. [DOI] [PubMed] [Google Scholar]
- Lazzerini Denchi E, de Lange T. Protection of telomeres through independent control of ATM and ATR by TRF2 and POT1. Nature. 2007;448:1068–1071. doi: 10.1038/nature06065. [DOI] [PubMed] [Google Scholar]
- Lee J, Kumagai A, Dunphy WG. The Rad9-Hus1-Rad1 checkpoint clamp regulates interaction of TopBP1 with ATR. J Biol Chem. 2007;282:28036–28044. doi: 10.1074/jbc.M704635200. [DOI] [PubMed] [Google Scholar]
- Loayza D, de Lange T. POT1 as a terminal transducer of TRF1 telomere length control. Nature. 2003;424:1013–1018. doi: 10.1038/nature01688. [DOI] [PubMed] [Google Scholar]
- Loo YM, Melendy T. The majority of human replication protein A remains complexed throughout the cell cycle. Nucleic Acids Res. 2000;28:3354–3360. doi: 10.1093/nar/28.17.3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDougall CA, Byun TS, Van C, Yee MC, Cimprich KA. The structural determinants of checkpoint activation. Genes Dev. 2007;21:898–903. doi: 10.1101/gad.1522607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarov VL, Hirose Y, Langmore JP. Long G tails at both ends of human chromosomes suggest a C strand degradation mechanism for telomere shortening. Cell. 1997;88:657–666. doi: 10.1016/s0092-8674(00)81908-x. [DOI] [PubMed] [Google Scholar]
- Matsuoka S, Ballif BA, Smogorzewska A, McDonald ERr, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, Shiloh Y, Gygi SP, Elledge SJ. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- McElligott R, Wellinger RJ. The terminal DNA structure of mammalian chromosomes. EMBO J. 1997;16:3705–3714. doi: 10.1093/emboj/16.12.3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirzoeva OK, Petrini JH. DNA damage-dependent nuclear dynamics of the Mre11 complex. Mol Cell Biol. 2001;21:281–288. doi: 10.1128/MCB.21.1.281-288.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mordes DA, Glick GG, Zhao R, Cortez D. TopBP1 activates ATR through ATRIP and a PIKK regulatory domain. Genes Dev. 2008;22:1478–1489. doi: 10.1101/gad.1666208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm W, Hockemeyer D, Kibe T, de Lange T. Functional dissection of human and mouse POT1 proteins. Mol Cell Biol. 2009;29:471–482. doi: 10.1128/MCB.01352-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm W, de Lange T. How shelterin protects mammalian telomeres. Ann Rev Genetics. 2008;42:301–334. doi: 10.1146/annurev.genet.41.110306.130350. [DOI] [PubMed] [Google Scholar]
- Takai KK, Hooper S, Blackwood S, Gandhi R, de Lange T. In vivo stoichiometry of shelterin components. J Biol Chem. 2010;285:1457–1467. doi: 10.1074/jbc.M109.038026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Multani AS, He H, Cosme-Blanco W, Deng Y, Deng JM, Bachilo O, Pathak S, Tahara H, Bailey SM, Deng Y, Behringer RR, Chang S. Pot1 deficiency initiates DNA damage checkpoint activation and aberrant homologous recombination at telomeres. Cell. 2006;126:49–62. doi: 10.1016/j.cell.2006.05.037. [DOI] [PubMed] [Google Scholar]
- Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.