Abstract
Rheumatoid arthritis (RA) is closely associated with HLA-DRB1 alleles that code a five-amino acid sequence motif in positions 70–74 of the HLA-DRβ chain, called the shared epitope (SE). The mechanistic basis of SE-RA association is unknown. We have recently found that the SE functions as an allele-specific signal transducing ligand that activates a nitric oxide (NO)-mediated pathway in other cells. To better understand the role of the SE in the immune system, here we have examined its effect on T cell polarization in mice. In CD11c+CD8+ dendritic cells (DCs) the SE inhibited the enzymatic activity of indoleamine 2,3 dioxygenase (IDO), a key enzyme in immune tolerance and T cell regulation, while in CD11c+CD8− DCs the ligand activated robust production of IL-6. When SE-activated DCs were co-cultured with CD4+ T cells, the differentiation of Foxp3+ T regulatory (Treg) cells was suppressed, while Th17 cells were expanded. The polarizing effects could be seen with SE-positive synthetic peptides, but even more so, when the SE was in its natural tri-dimensional conformation as part of HLA-DR tetrameric proteins. In vivo administration of the SE ligand resulted in higher abundance of Th17 cells in the draining lymph nodes and increased IL-17 production by splenocytes. Thus, we conclude that the SE acts as a potent immune-stimulatory ligand that can polarize T cell differentiation toward Th17 cells, a T cell subset that has been recently implicated in the pathogenesis of autoimmune diseases, including RA.
INTRODUCTION
Rheumatoid arthritis (RA) is a chronic inflammatory disease that leads to joint destruction and early death (1, 2). The pathogenesis of the disease is not fully understood, but previous studies have shown that RA is closely associated with HLA-DRB1 alleles that code a five amino acid sequence motif in residues 70–74 of the DRβ chain (3, 4) – commonly referred to as the “shared epitope” (SE). The disease in SE-positive patients begins earlier and is more erosive than in SE-negative individuals (5). The mechanism underlying the effect of SE in RA is unclear. Based on the known role of MHC molecules in antigen presentation, the prevailing hypotheses postulate that either presentation of arthritogenic self-peptides (6), molecular mimicry with foreign antigens (7), or T cell repertoire selection (8) are involved.
Although these hypotheses are plausible, evidence to support them is inconclusive. We have recently discovered a novel functional role of the SE: acting as a signal transduction ligand that activates innate immune signaling in other cells. Our data have shown that whether expressed in its native conformation on the cell surface, or as a cell-free HLA-DR tetrameric molecule, or engineered into large recombinant proteins, or as a short synthetic peptide, the SE activated in all cases nitric oxide (NO)-mediated signaling in trans in a strictly allele-specific manner (9–11).
SE-triggered signaling is transduced via cell surface calreticulin (CRT) (12), a known innate immunity receptor (13), which is expressed on the surface of many cells (14, 15). CRT serves as the signal-transducing receptor for members of the collectin family and other innate immune system ligands (16). Importantly, CRT plays a pivotal role in the junction between tolerance and autoimmunity due to its critical role in elimination of apoptotic cells (17). Aberrant activation of CRT-mediated pathway can lead to autoimmunity as exemplifies by conditions that involve defective CRT-mediated clearance of apoptotic cells (18).
CRT is expressed on dendritic cells (DCs), which are believed to play a role in the pathogenesis of RA (19). DCs are strategically positioned in the interface between the innate and adaptive immune systems. In addition to their antigen presentation role, DCs also induce tolerance through cross-talk with regulatory T (Treg) cells (20). A growing body of evidence indicates that the tolerogenic effect of DCs is mediated to a large extent by indoleamine 2,3 dioxygenase (IDO), an enzyme that catabolizes tryptophan (21). IDO is inducible by IFNγ (22) and by CTLA-4 (23), while NO (24, 25) and IL-6 (26) potently inhibit its activity. Relevant to RA, activation of IDO in DCs by Treg-expressed CTLA-4 has been shown to inhibit Th17 cells (27), a T cell subset that is believed to play a key role in RA pathogenesis (28).
To gain insights into the role of the SE in immune regulation, in this study we have undertaken to examine its functional effects on DCs. We show here that the SE inhibits IDO activity in the CD11c+CD8+ subset of murine DCs and increases IL-6 production by CD11c+CD8− DCs. This leads to enhanced differentiation and expansion of Th17 cells with a reciprocal effect on Treg cells.
MATERIALS AND METHODS
Mice and reagents
All mice were from Jackson Laboratory. Experiments were carried out in 5–10 week-old male DBA/1, Balb/c, C57BL/6, or a DBA/1 mouse line carrying transgenic (Tg) collagen type II (CII)-specific TCR [D1Lac.Cg-Tg(TCRa,TCRb)24Efro/J]. For brevity, the latter mouse line is designated here as “CII-TCR Tg mice”. The animals were housed in the University of Michigan Unit for Laboratory Animal Medicine facility. All experiments were performed in accordance with protocols approved by University of Michigan Committee on Use and Care of Animals.
Monoclonal antibodies against mouse CD3 (clone 2C11), IL-4 (clone 11B11), IFNγ (clone R46A2), and IL-2 (clone S4B6) were purified from the supernatants of hybridomas obtained from the University of Michigan Hybridoma Core Facility. Purified anti-mouse CD28 (clone 37.51) and murine rIL-23 were purchased from e-Bioscience (San Diego, CA). Human rTGFβ and rIFNγ, as well as murine rIL-4, rIFNγ, rGM-CSF and rIL-6 were purchased from Peprotech (Rocky Hill, NJ).
Peptides were synthesized and HPLC-purified to > 90% by the University of Michigan Protein Structure Facility as previously described (9, 12). SE-expressing 15mer peptides, designated as 65–79*0401 (aa sequence 65-KDLLEQKRAAVDTYC-79), or 65–79*0404 (aa sequence 65-KDLLEQRRAAVDTYC-79), corresponded to the third allelic hypervariable region (HVR3) of the DRβ chain encoded by of SE-positive HLA-DRB1*0401 or HLA-DRB1*0404 alleles, respectively. Control 15mer peptides 65–79*0402 (65-KDILEDERAAVDTYC-79) and 65–79*0403 (65-KDLLEQRRAEVDTYC-79) corresponded to the HVR3 of the DRβ chain encoded by of SE-negative HLA-DRB1*0402 or HLA-DRB1*0403 alleles, respectively. The CII259–273 peptide, which corresponds to residues 259–273 of chicken CII, was kindly provided by Dr. Steven Lundy.
Chimeric hepatitis B core (HBc) particles engineered to express the HVR3 of the HLA-DRβ chain were prepared as previously described (9) at the Latvian Biomedical Research and Study Center, (Riga, Latvia). HBc particles expressing a SE-positive HVR3, encoded by HLA-DRB1*0401 (designated here as HBc*0401) or a SE-negative HVR3, encoded by HLA-DRB1*0402 (designated here as HBc*0402) were used in this study as previously described (9). SE-positive HLA-DR tetramers DRB1*0401/DRA1*0101 (designated here as T-DRB1*0401), SE-negative DRB1*1501/DRA1*0101 (T-DRB1*1501), and SE-negative DRB1*0301/DRA1*0101 (T-DRB1*0301), all containing identical class II-associated invariant chain peptide (CLIP) in the peptide-binding groove, were generated by the National Institutes of Health Tetramer Core Facility as previously described (26). Unless stated otherwise, all chemicals were from Sigma-Aldrich (St. Louis, MO).
Isolation and culture of cells
Murine L cell transfectants expressing human HLA-DRα/β heterodimers (29) and human fibroblast line M1 (9) were maintained as we previously described. For generation of CD11c+DCs, mouse bone barrow cells were plated in culture flasks (2 × 106 cells/ml per T150, Costar, Corning, NY) in RPMI 1640 medium containing 2 mM L-glutamine, 10% FBS, 1% Penicillin-Streptomycin, 10 mM HEPES buffer solution, 10 mM Sodium Pyruvate, 50 mM 2-mercaptoethanol, GM-CSF (10 ng/ml) and IL-4 (10 ng/ml). On day 3, half of the medium was removed and fresh medium containing GM-CSF (10 ng/ml) and IL-4 (10 ng/ml) were added. After 5–7 days, DCs were purified using positive selection columns with CD11c microbeads (Miltenyi Biotec Inc, CA, USA) as previously described (30). For preparation of CD11c+CD8+ and CD11c+CD8− DCs, freshly isolated splenic DCs were subjected to positive selection with CD11c and CD8α microbeads. Purified DC subsets were then cultured in RPMI 1640 medium containing 2 mM L-glutamine, 10% FBS, 1% Penicillin-Streptomycin, 10 mM HEPES buffer solution, 10 mM Sodium Pyruvate and 50 mM 2-mercaptoethanol. CD4+ T cells were isolated from the spleen, using a negative selection immunomagnetic isolation kit (EasySep®, Stem Cell technology, Vancouver, Canada) according to the manufacture’s instructions. To purify CD4+CD25−CD62L+CD44− naïve T cells, CD4+T cells were incubated with FITC anti-mouse CD4 and a mixture of PE-labeled anti-CD25, APC-labeled anti-CD62L and Pe-Cy7-labeled anti-CD44 antibodies (all from Biolegend, San Diego, CA). CD4+CD25−CD62L+CD44− naïve T cells were sorted using a FACSDiva™ instrument (Becton Dickinson, Franklin Lakes, NJ) with a purity > 98%.
Measurement of NO production, IDO activity and cytokine secretion
To determine the rate of NO production, cells were loaded with 20 µM of the fluorescent NO probe 4,5-diaminofluorescein diacetate (DAF-2DA) and the fluorescence level was recorded every 5 minutes over a period of 500 minutes using a Fusion αHT system (PerkinElmer Life Sciences) at an excitation wavelength of 488 nm and emission wavelength of 515 nm. To determine IDO enzymatic activity, the generation of its product, kynurenine was measured as previously reported (31). Cytokine concentrations were measured in cell culture supernatants using a Luminex platform (Millipore Corporation, Danvers, MA). In some experiments, cytokines were determined using ELISA (Quantikine®, R&D Systems, Minneapolis, MN) following the manufacture’s instruction.
Determination of surface CRT expression on DCs
Splenic cells from DBA/1 mice were isolated followed by purification of DC subtypes using positive selection columns with CD11c and CD8α microbeads. Purified cells were stained for flow cytometry analysis using PE anti-mouse CD8 (clone 53-6.7, BD Pharmigen, San Jose, CA), FITC anti-rabbit CRT (ABR - Affinity Bioreagents, Rockford, IL) and isotype controls (Biolegend, San Diego, CA).
Treg differentiation
DBA/1 bone marrow-derived CD11c+DCs were placed in 24-well plates (BD Biosciences, San Jose, CA) at a density of 2.5 × 105 cells per well and cultured overnight with or without 50 µg/ml of peptidic (65–79*0401 or 65–79*0402), or 2 µg/ml of tetrameric (T-DR1*0401, T-DR1*0301, or T-DR1*1501) ligands at 37°C. On the following day, 5.0 × 105 CD4+ T cells or CD4+CD25−CD62L+CD44− naïve T cells, isolated as described above, were added to each well in addition to anti-CD3 antibodies (5 µg/ml) and rhTGFβ (2.5 ng/ml).
After 5 days in culture, cells were harvested and stained for flow cytometric analysis using FITC anti-mouse CD4 (clone XMG1.2), PE anti-mouse CD25 (clone PC61) and isotype controls (Biolegend, San Diego, CA). Next, cells were permeabilized and fixed using a Cytofix/Cytoperm kit (BD Biosciences, San Jose, CA) as recommended by the manufacturer. After permeabilization, cells were stained using an APC-conjugated anti-mouse Foxp3 antibody (clone FLK-16S from e-Bioscience, San Diego, CA) and analyzed by FACScalibur flow cytometer using the CELLQuest™ software (Becton Dickinson, Franklin Lakes, NJ).
Th17 differentiation
Bone marrow-derived CD11c+DCs (2.5×105 cells per well) were cultured overnight in 24-well plates with or without SE ligands or controls as above. Then, 5 × 105 CD4+ T cells or CD4+CD25−CD62L+CD44− naïve T cells were added at the ratio of 2:1 in the presence of Th17-polarazing cytokine/antibodies cocktail containing: anti-IL4 (2 µg/ml), anti-IFNγ (2 µg/ml), anti-IL2 (3 µg/ml), rhTGFβ (5 ng/ml), rmIL-6 (20 ng/ml), rmIL-23 (10 ng/ml), anti-CD3 (5 µg/ml) and anti-CD28 (1 µg/ml) as previously described (32).
After 6 days, cells were stimulated with PMA (5 ng/ml) and ionomycin (500 ng/ml) for the last 6 hrs of culture. Brefeldin A (10 µg/ml) was added to the culture for the last 5 hrs. Cells were then harvested and stained for surface marker using PercP anti-mouse CD4 or isotype control (Biolegend, San Diego, CA) followed by fixation and permeabilization using a Cytofix/Cytoperm™ kit. Intracellular staining was performed using PE-conjugated anti-mouse IL-17A mAb (clone TC11-18H 10.1 from Biolegend, San Diego, CA). Mean florescence intensity and percentages of stained cells were determined by flow cytometry.
Proliferation assays
Cells were labeled with 1 µM of CFSE (Molecular Probes™, Invitrogen Corporation, Carlsbad, CA), stained with CD4-PercP, CD25-Pe, and Foxp3-APC or IL-17A-APC antibodies (Biolegend, San Diego, CA) and proliferation was determined by measuring the percentages of CFSE-labeled cycling CD4+ T, CD4+CD25+Foxp3+ Treg or CD4+IL17A+ Th17 cells, using a FACS analysis.
Determination of the SE polarizing effect In vivo
Mice were injected subcutaneously in the footpad with 100 µg of chicken collagen type II (CII) (Chondrex, Inc, Redmond, WA) emulsified in CFA (4 mg/ml). The inoculums contained 10 µg of either SE-positive 65–79*0401 or SE-negative 65–79*0402 ligands in PBS, or an equal volum of PBS alone. Animals were sacrificed 7 days after immunization. For Th17 quantification studies, inguinal and popliteal lymph nodes were collected and single cell suspensions were prepared. Unfractionated lymph node cells were cultured with PMA, Ionomycin and Brefeldin A for 6 hours as above. Cells were stained with FITC anti-mouse CD4 or isotype controls, followed by fixation and permeabilization using a Cytofix-Cytoperm™ kit. After permeabilization, intracellular staining was performed using PE-conjugated anti-mouse IL17A and APC-conjugated anti-mouse IFN-γ and cells were analyzed by flow cytometry as above. To measure IL-17 production, splenocytes from mice immunized as above were stimulated in vitro with 5 µg of CII259–273 peptide. At different time points thereafter, supernatants were collected and assayed for IL-17 by ELISA as above.
Data analysis
A 2-tailed Student’s t-test was used. Unless stated otherwise, an asterisk indicates p < 0.05.
RESULTS
The SE inhibits IDO activity
In previous studies we have demonstrated that the SE ligand indiscriminately activates NO signaling in different cell lineages from several species (9–12). As a prelude to investigating the immune regulatory effect of the SE, we first confirmed that it activates NO signaling in DCs. As expected, the SE activated robust NO production in CD11c+DCs from several mouse strains in a strictly allele-specific manner (Supplemental Figure 1). Thus, similar to its effect in many other cell lineages, the SE activates NO signaling in mouse DCs as well.
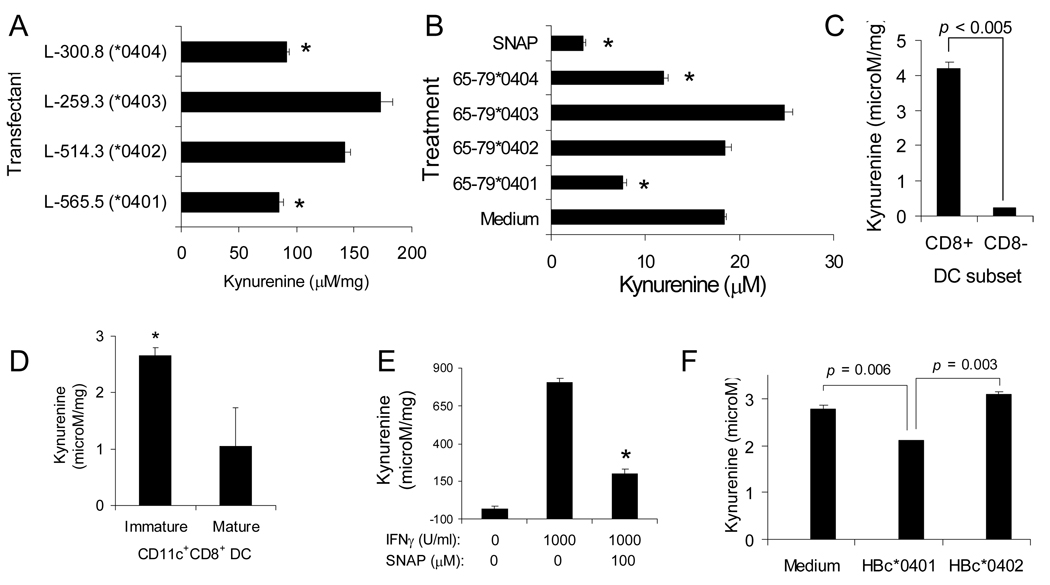
Given the known inhibitory effect of NO on IDO activity (24), we next examined whether the SE could affect IDO enzymatic activity. In addition to a small subset of DCs (33), IDO is expressed in several other cell lineages, including fibroblasts. Given the much greater abundance of fibroblasts over IDO-producing DCs, we first determined the effect of the SE on IDO activity in murine fibroblast L-cells transfectants expressing functionally and structurally intact HLA-DRα/β heterodimeric molecules on their surface through cDNA transfection (29). As can be seen in Fig. 1A, transfectants expressing SE-positive HLA-DR molecules on their surface (lines L-565.5 and L-300.8, expressing the SE-positive DRβ 0401 or DRβ 0404 molecules, respectively) produced significantly less kynurenine in response to IFNγ, compared to transfectants expressing SE-negative HLA-DR molecules (lines L-514.3 and L-259.3 expressing SE-negative DRβ 0402 or DRβ 0403 molecules, respectively). An identical pattern was seen when M1 fibroblasts were stimulated with SE peptidic ligands 65–79*0401 or 65–79*0404. As can be seen in Fig. 1B, the SE ligands strongly inhibited IFNγ-induced IDO activity. SE-negative controls 65–79*0402 and 65–79*0403 did not inhibit IDO activity. Consistent with previous studies (24, 34) the NO-donor S-nitroso-N-acetylpenicillamine (SNAP) inhibited IDO activity too. Thus, these results demonstrate that the SE, whether physiologically expressed on the cell surface, or added as a cell-free ligand effectively and specifically inhibits the activity of the tolerogenic enzyme IDO in human and murine cells.
FIGURE 1. Inhibition of IDO activity by the SE ligand.
(A) Murine L cells expressing either SE-positive (L-565.5, or L-300.8) or SE-negative (L-514.3, or L-259.3) functional HLA-DR molecules on their surface through cDNA transfection were incubated for 48 hrs with rhIFNγ (1000 U/ml) and cellular IDO activity was determined. *, p < 0.001. (B) M1 fibroblasts were incubated overnight with medium, NO donor SNAP, or 100 µg/ml of SE ligands (65–79*0401, 65–79*0404), or with SE-negative controls (65–79*0402, or 65–79*0403). Cells were cultured for 48 hrs with rhIFNγ and cellular IDO activity was determined. *, p < 0.005. (C) The CD11c+CD8+ and CD11c+ CD8− DCs subsets were purified from DBA/1 spleens and their IDO activity in response to IFNγ was determined. (D) CD11c+CD8+ DCs were purified from DBA/1 spleens, maturated or not with LPS, and their IDO activity in response to IFNγ was determined. (E) DBA/1 splenic CD11c+CD8+ DCs were activated with or without IFNγ, in the presence of absence of the NO donor SNAP. IDO activity was determined at 48h as above. (F) DBA/1 splenic CD11c+CD8+ DCs were pre-incubated for 1hr with or without HBc particles engineered to express the 65–79 region of DRβ chains, encoded by either SE-positive (HBc*0401) or SE-negative (HBc*0402) HLA-DRB1 alleles. DCs were subsequently stimulated with IFNγ and IDO activity was determined as above.
In order to proceed with the studies reported here that are focused on DBA/1 mice, we first confirmed (Fig. 1C) that IFNγ-induced IDO activity in this strain is found in CD11c+CD8+ DCs, but not in CD11c+CD8− DCs, similar to published reports in other strains (33). To examine the effect of maturation on IDO activity in DBA/1 mice, CD11c+CD8+ DCs were incubated for 24 hrs with or without LPS (1 µg/ml) and IFNγ-induced IDO activity was determined. As shown in Fig. 1D, activation of IDO in immature DCs was significantly more potent than in mature cells. We also confirmed that, similar to other mouse strains, IDO activation in DBA/1 mice is inhibited by NO by demonstrating that IFNγ-induced IDO activity in DBA/1 immature CD11c+CD8+ DCs is potently inhibited by the NO-donor SNAP (Fig. 1E). Finally, to determine the effect of the SE on IDO activity in DCs, DBA/1 immature CD11c+CD8+ DCs were pre-incubated with HBc particles engineered to express the HVR3 (residues 65–79) encoded by the SE-positive allele DRB1*0401 (designated HBc*0401), or the HVR3 encoded by the SE-negative allele DRB1*0402 (HBc*0402). Cells were then stimulated with IFNγ and IDO activity was determined as above. As can be seen in Fig. 1F, SE-positive HBc*0401 but not SE-negative HBc*0402 particles significantly inhibited IDO activity in DCs. Thus, our data indicate that the SE ligand inhibits IDO activity in CD11c+CD8+ DCs.
Cytokine production by SE-stimulated DCs
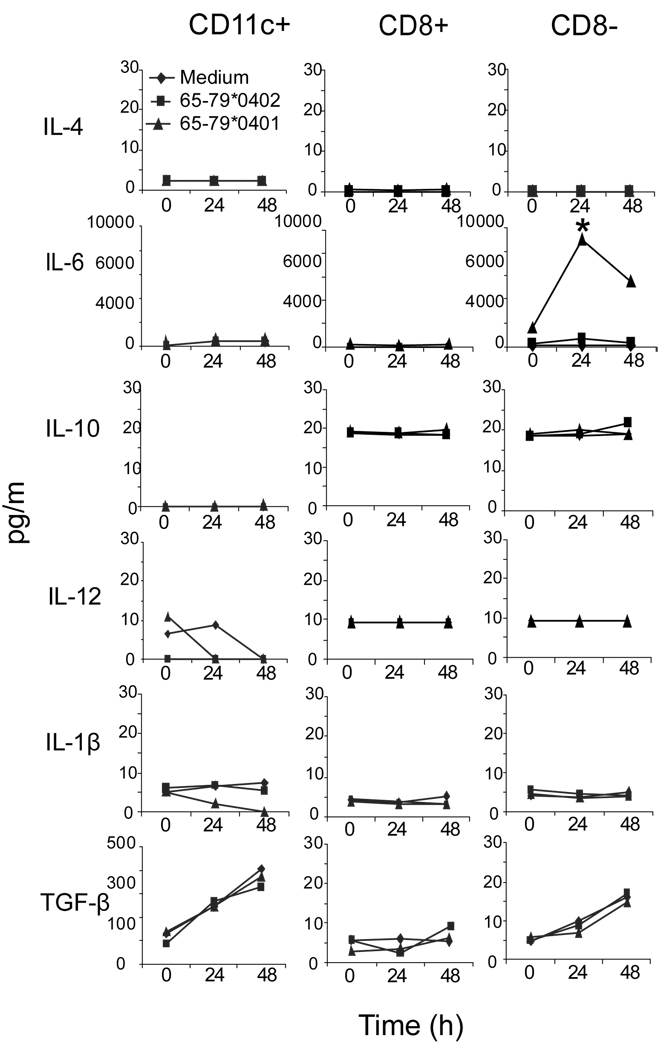
In addition to IDO-mediated T cell regulation, DCs can affect immune responses by producing cytokines capable of activating or expanding particular subsets of T cells, thereby polarizing the immune response. For example, in mice, the combination of IL-6 and TGFβ facilitates differentiation of Th17 cells, while IL-23 is involved in the expansion of this subset (35). In order to determine whether SE-activated signaling in DCs induces cytokine production, we studied supernatants of SE-stimulated DCs. As can be seen in Fig. 2, the SE ligand 65–79*0401 activated a robust production of IL-6 in CD11c+CD8− DCs, but not in the CD11c+CD8+ subset. IL-6 levels peaked at a relatively early time point (24h) and later declined. This pattern is likely a result of short half life of the peptidic ligand due to rapid degradation in tissue culture conditions. The SE-negative control 65–79*0402 did not trigger any cytokine production. Other cytokines (IL-4, IL-10, IL-12 IL-1β, TGFβ) did not show any increased production, attesting to the specificity of the SE effect (Fig. 2). Thus, while in CD8+ DCs the SE inhibited IDO activity (Fig. 1) its IL-6 production effect was restricted to the CD8− DCs subset (Fig. 2). The reason for this dichotomy is presently unknown, but may be attributed to differential abundance of the receptor CRT on the surface of these two DC subsets (Supplemental Figure 2).
FIGURE 2. The SE Activates IL-6 production in CD8−DCs.
DBA/1 splenic unfractionated CD11c+ DCs (CD11c+), or their purified CD11c+CD8+ DCs (CD8+) or CD11c+CD8− DCs (CD8−) subsets were cultured with the SE ligand 65–79*0401 or SE-negative control 65–79*0402 or medium. Supernatants were collected at different time points and assayed for cytokine content using a Luminex platform. Data are from a representative experiment, one of three replications.
It is worth pointing out that SE-activated IL-6 production could be seen in CD11c+CD8− DCs only when they were separated from the CD11c+CD8+ subset, but not when unfractionated CD11c+ DCs were assayed (Fig. 2). This could be explained by the fact that although CD11c+CD8+ DCs are a small subset (~5–15% of splenic CD11c+ cells), once activated by the SE, they could exert potent inhibitory effect on the activation of CD11c+CD8− DCs, consistent with previously reported DCs suppressive effects (36). Recent studies have indeed shown that IDO produced by a small subset of DCs can dominantly suppress production of IL-6 in other DCs (37, 38). We should point out however that in preliminary studies using pharmacologic IDO inhibitors we could not reverse the apparent suppressive effect of IDO-producing DCs (data not shown). More experimental effort is required to conclusively address this question.
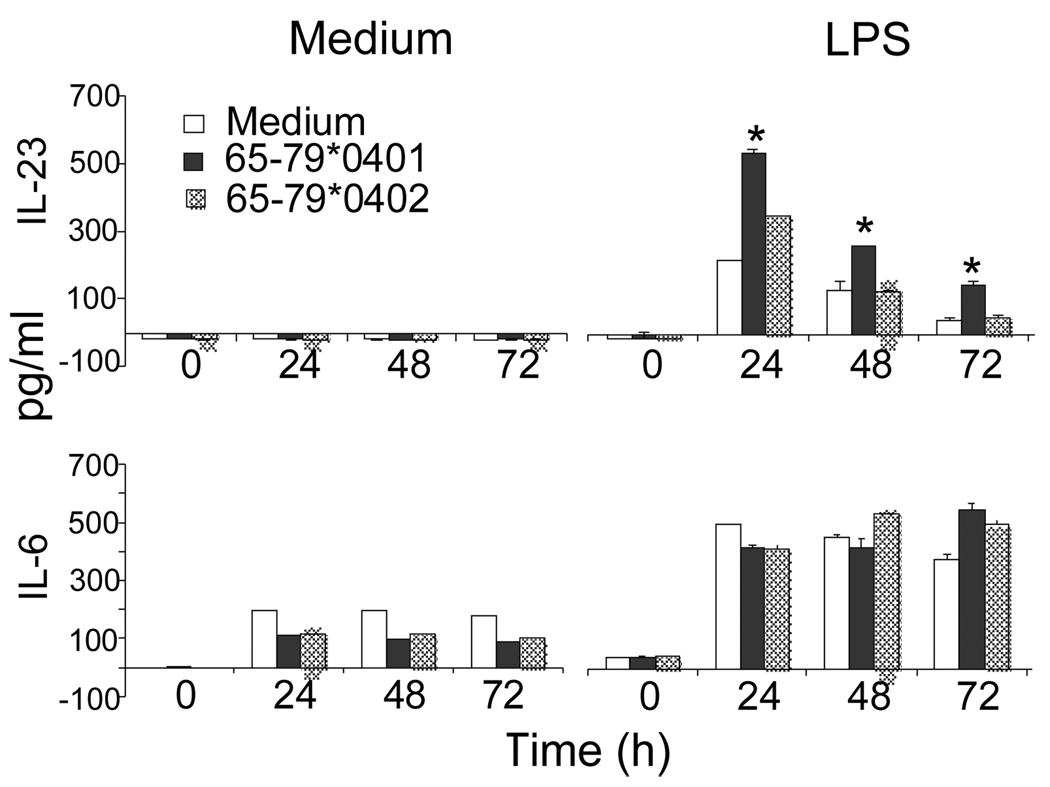
IL-23 levels in DCs did not increase following stimulation with the SE ligand (Fig. 3). However, in the presence of LPS (100 ng/ml), the SE had a prolonged synergistic effect in CD11c+DCs. As can be seen, the SE had no effect when applied alone, but in the presence of LPS it had a synergistic effect, which lasted for up to 72 hours after stimulation, long after LPS effect had subsided (Fig.3). The effect was specific for IL-23, since no synergism was found in the production of another LPS-inducible cytokine, IL-6 (Fig. 3, bottom).
FIGURE 3. The SE augments IL-23 production in LPS-stimulated CD11c+DCs.
DBA/1 bone marrow-derived CD11c+DCs were cultured with or without 100 ng/ml LPS in the presence or absence of SE-positive or SE-negative 15mer peptides (50 µg/ml). Supernatants were collected at different time points and assayed for IL-23 and IL-6 content by ELISA. Data shown are representative of two duplicate experiments.
Inhibition of Treg differentiation by the SE
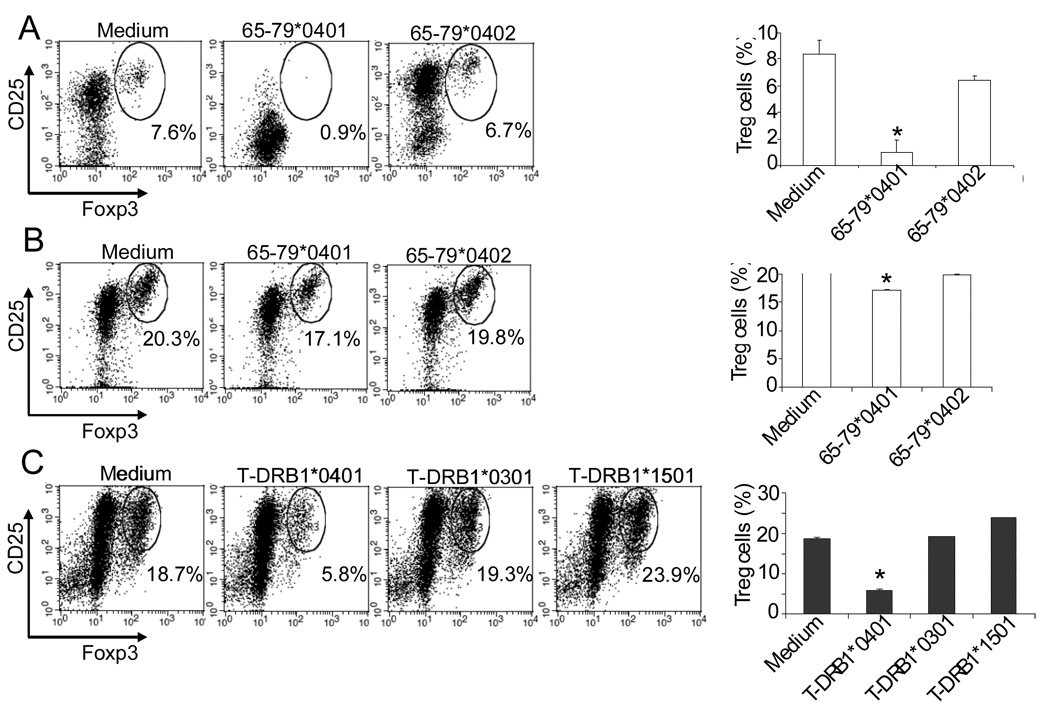
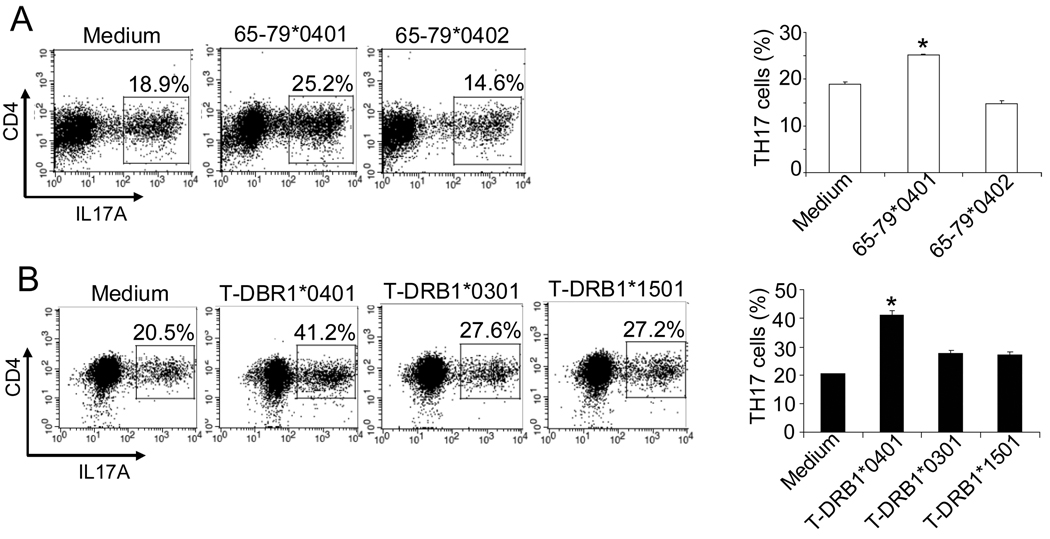
It has been previously demonstrated that IDO inhibition (39) or increased IL-6 levels (40) inhibit Treg cells. As shown above, the SE inhibited IDO activity in CD11c+CD8+ DCs and increased IL-6 production in CD11c+CD8− DCs. Therefore, our next step was to determine whether the SE interferes with Treg differentiation or expansion. Accordingly, DBA/1 CD11c+DCs were first incubated overnight with the SE ligand 65–79*0401, or SE-negative control 65–79*0402, or with medium. DCs were then co-cultured with purified syngeneic CD4+ T cells (Fig. 4A) or CD4+CD25−CD62L+CD44− naïve T cells (Fig. 4B) in the presence of TGF-β (2.5 ng/ml) and anti-CD3 antibodies (5.0 µg/ml). After 5 days, CD4+CD25+Foxp3+ Treg abundance was determined by flow cytometry. As shown in Figs. 4A and 4B, the SE ligand 65–79*0401 significantly inhibited Treg expansion and differentiation, respectively. The inhibitory effect of 65–79*040 on Treg differentiation was statistically significant, yet modest. We have previously observed that peptidic SE ligands exert weaker signaling effects due to their flexible conformation in solution (9, 10, 12). To address this possibility, similar Treg differentiation experiments were performed using SE-positive HLA-DR tetramer (designated T-DRB1*0401), or control, SE-negative HLA-DR tetramers (T-DRB1*1501 or T-DRB1*0301) instead of soluble peptides. The HLA-DR molecule in tetramers is folded in its natural tri-dimensional conformation and therefore better preserves the physiologic function of the protein. Fig. 4C shows that the SE-positive tetramer T-DRB1*0401 indeed had a specific and much more potent inhibitory effect on Treg differentiation.
FIGURE 4. The SE inhibits Treg generation.
(A) DBA/1 bone marrow-derived CD11c+DCs were cultured overnight with 50 µg/ml SE ligand 65–79*0401 or SE-negative control 65–79*0402, or medium. Syngeneic splenic CD4+ T cells were then added to the culture, and incubated with anti-CD3 and TGF-β for 5 days. On the left, flow cytometry dot plots showing percentages of CD25+Foxp3+ cells obtained from gated CD4+ T cells in each treatment. Each plot is representative of three experiments. On the right, bar graphs present results as mean percentage ± SD of replicate samples. *, p < 0.01, compared to both Medium and 65–79*0402. (B) Cultures were performed as in (A), with the exception that CD4+CD25−CD62L+CD44− naïve T cells, instead of CD4+ T cells were added to the CD11c+DCs. *, p < 0.05, compared to both Medium and 65–79*0402. (C) DBA/1 bone marrow-derived CD11c+DCs were incubated overnight with 2 µg/ml tetramers (SE-positive T-DRB1*0401, versus SE-negative T-DRB1*0301, or T-DRB1*1501). Syngeneic CD4+CD25−CD62L+CD44− naïve T cells, anti-CD3 and TGF-β were then added to the culture and incubated for 5 days and analyzed as above. *, p < 0.0005, compared to Medium, T-DRB1*0301 and T-DRB1*1501.
SE-activated DCs facilitate Th17 differentiation
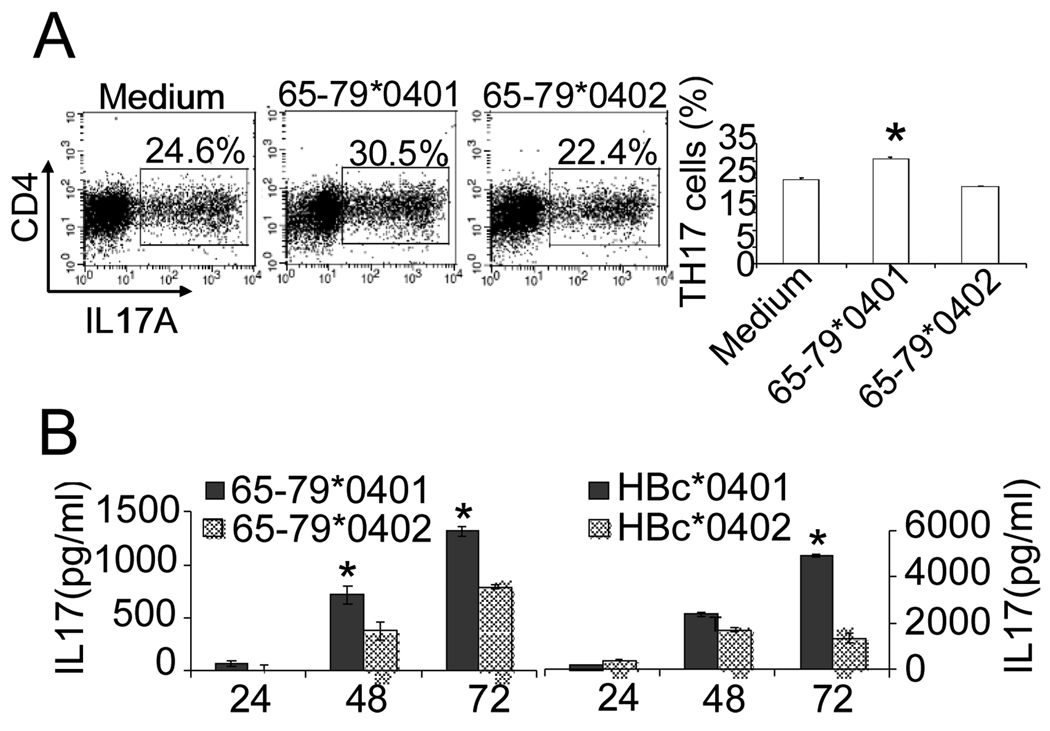
As shown above, in the CD11c+CD8− DC subset, the SE ligand 65–79*0401 triggered a robust production of IL-6, an obligatory cytokine for Th17 differentiation. IL-23 production by LPS-treated DCs was also augmented by 65–79*0401. We therefore next determined whether the SE ligand can facilitate Th17 differentiation or activation. To determine whether the SE affects Th17 differentiation, CD11c+DCs were stimulated overnight with either peptidic or tetrameric ligands as above. Next, CD4+CD25−CD62L+CD44− naïve T cells were added and cultured in the presence of a Th17-polarizing cocktail of cytokines and antibodies as discussed in the Methods section. After 6 days, cells were collected and analyzed by flow cytometry. As can be seen in Fig. 5A, the SE ligand 65–79*0401 induced a significant increase in the differentiation of CD4+IL17A+ T cells. A more robust SE-induced Th17 polarization effect could be seen when DCs were stimulated with SE-positive HLA-DR tetramers (Fig. 5B).
FIGURE 5. The SE facilitates Th17 differentiation.
DBA/1 bone marrow-derived CD11c+DCs were cultured overnight in the presence or absence of (A) SE ligand 65–79*0401 or SE-negative control 65–79*0402, or (B) SE-positive T-DRB1*0401 tetramers, versus SE-negative T-DRB1*0301, or T-DRB1*1501 tetramers. Syngeneic splenic CD4+CD25−CD62L+CD44− naïve T cells plus a Th17-differentiation cytokine/antibody cocktail were then added to the culture and incubated for 6 days. Intracellular IL17A was determined by flow cytometry. On the left: a representative experiment, one of three repetitions, showing percentages of CD4+IL17A+ cells as dot plots. On the right: bar graphs show results presented as mean percentage ± SD of replicate samples. *, p < 0.001.
To determine whether SE-activated DCs can increase IL-17 production in CD4+ T cells, CD11c+DCs were incubated overnight with SE ligands or control reagents. Cells were then co-cultured with total CD4+ T cells. As shown in the Fig. 6, SE-activated DCs induced higher intracellular (Fig. 6A) and extracellular (Fig. 6B) IL-17 expression in co-cultured CD4+ T cells, compared to cells cultured with DCs pre-incubated with a control reagents or medium. The effect was seen both with a soluble SE peptidic ligand, as well as with conformationally preserved ligand in the form of an HBc particle (Fig. 6B).
FIGURE 6. Activation of IL-17 production in CD4+ T cells by the SE.
(A) DBA/1 bone marrow-derived CD11c+ DCs were cultured overnight with different peptidic ligands as above. Syngeneic splenic CD4+ T cells plus a Th17-polarizing cocktail were added to the culture and incubated for 6 days. IL-17A-positive cells were quantified by flow cytometry. On the left: dot plots showing percentage of CD4+IL17A+ T cells. On the right: bar graphs (mean ± SD) of replicate samples. *, p < 0.005. (B) To measure IL-17 secretion, DBA/1 bone marrow-derived CD11c+DCs were treated overnight with peptidic or particular ligands or medium and then co-cultured with CD4+ T cells as above. Supernatants were assayed for IL-17 content by ELISA. Data is representative of two experiments. *, p < 0.05.
We have also found that the SE affected the proliferation of Th17 and Tregs in a reciprocal manner. Enhanced expansion through increased proliferative activity of Th17 cells and decreased proliferative activity of Tregs could be seen in both TCR-independent (Supplemental Figure 3) and TCR-mediated (Supplemental Figure 4) T cell activation conditions.
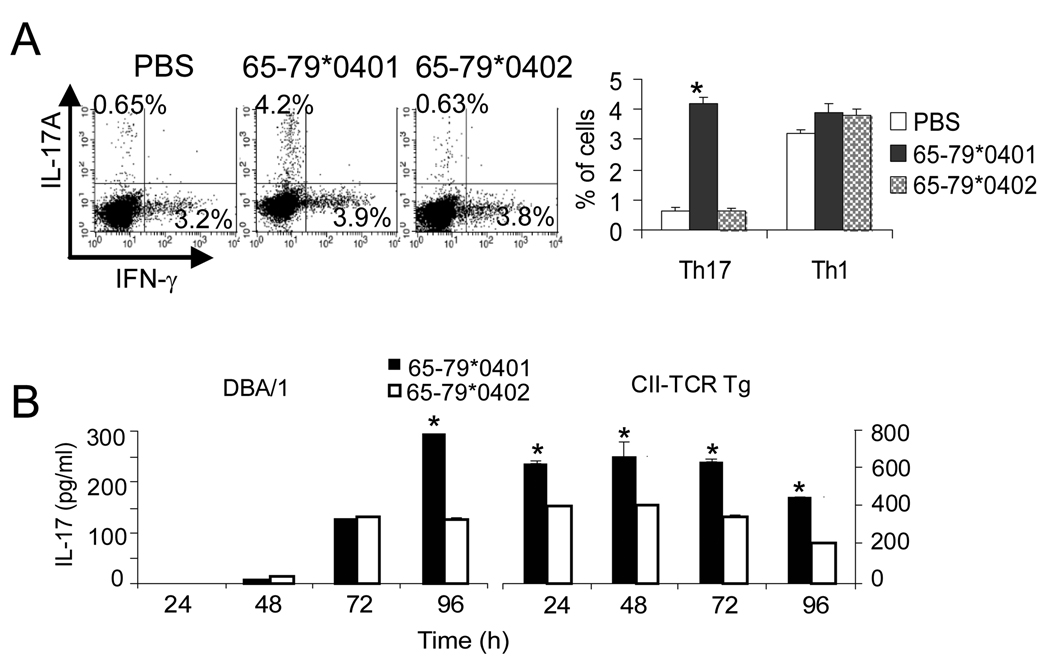
Finally, to determine the biologic significance of the in vitro data shown above, we have undertaken to characterize the SE polarizing effect In vivo (Fig. 7). Draining lymph nodes of control DBA/1 mice immunized with CII had a 0.65% abundance of Th17 cells, consistent with published data showing frequencies of less than 1% of Th17 in draining lymph nodes (41, 42). Co-administration of the SE-negative 65–79*0402 had no effect on Th17 abundance. However co-administration of the SE ligand 65–79*0401 dramatically increased the frequency of these cells (Fig. 7A). As can be seen, the SE-induced expansion was specific for Th17 cells since there was no change in the frequency of Th1 (IFNγ-positive) cells (Fig 7A). Additionally, splenocytes from DBA/1 mice immunized with CII in the presence of the SE ligand 65–79*0401 showed significantly more robust CII-stimulated IL-17 production, compared to splenocytes obtained from mice co-immunized with the SE-negative control 65–79*0402 (Fig 7B). Thus, when taken collectively, our data indicate that the SE facilitates Th17 polarization both in vitro and In vivo.
FIGURE 7. The SE facilitates Th17 polarization In vivo.
(A) DBA/1 mice were immunized with CII in CFA in the presence of PBS, 65–79*0401 or 65–79*0402. On day 7, cells from draining lymph nodes were isolated, cultured for 6 hours with PMA, Ionomycin and Brefeldin A and then stained with anti-mouse CD4, IL17A and IFN-γ as above. On the left: Representative dot plots showing percentages of IL17A+ and IFN-γ+ cells obtained from gated CD4+ cells. On the right, bar graphs showing results as mean ± SD of duplicate experiments. *, p < 0.001 compared to Medium and 65–79*0402. (B) DBA/1 and CII-TCR Tg mice were immunized as in (A). On day 7, splenic cells were isolated and cultured with the synthetic peptide CII260–267 (5 µg/ml). Supernatants were collected every 24 hours and assayed for IL-17 levels by ELISA. Bar graphs show data as mean ± SD of duplicate samples. *, p < 0.005.
DISCUSSION
In prior studies we have demonstrated that the SE activates innate immune signaling in many cell lineages (12). Here we show that when murine DCs were exposed to the SE ligand, they enhanced Th17 differentiation and inhibited the generation of Treg cells. The SE effect in DCs was found to be allele-specific and particularly potent when the ligand was presented in its physiologic tri-dimensional conformation.
The functional dichotomy between DC subsets relative to their response to the SE ligand is intriguing. As shown here, in CD11c+CD8− DCs, the SE triggered robust production of the pro-inflammatory and immunostimulatory cytokine, IL-6, while in CD11c+CD8+DCs it inhibited induction of IDO. These data demonstrate that identical ligands can produce markedly different functional effects in different cellular contexts. The variability of the functional response may conceivably be attributed to differential abundance of the receptor, as suggested in Supplemental Figure 2, or modulatory effects by other signaling pathways. Such factors could vary markedly among different lineages in different tissue sites. Specifically relevant to immune regulation, the data shown here reiterate the known functional heterogeneity of different DC subsets and add new insights into the distinct effects of the SE in those subsets. It is worth mentioning that several reports have shown that IDO is selectively expressed by a small subset of murine plasmacytoid DCs that express the B cell marker CD19 (43, 44). The relevance of that small DC subset to the findings shown here is presently unknown. More research is needed in order to dissect the fine phenotypic and functional characteristics of different DC subpopulations relative to their role in SE-triggered IDO inhibition.
Although the SE ligand alone did not activate IL-23 production in DCs in the in vitro experiments described here, it produced a potent synergistic effect in the presence of low concentrations of LPS (Fig. 4). The effect was specific for the SE ligand, because a SE-negative control reagent failed to produce it. The synergistic effect with LPS shown here is reminiscent to LPS effect on high molecular group binding protein-mediated activation (45). Different from that synergism, however, the SE-LPS effect reported here selectively involved IL-23 production, but not that of IL-6. Since IL-23 has been previously shown to induce Th17 cell expansion, it is tempting to speculate that under certain co-stimulatory In vivo conditions, the SE may accelerate pathogen-triggered expansion of Th17 cells.
To simulate stringent structural conditions, the T cell polarization studies were repeated with physiologically-folded SE ligands in the form of tetrameric molecules (Fig. 4C, 5B). It should be noted that tetramers are made of four identical units of the HLA-DR heterodimeric molecule, each folded in its native tri-dimensional conformation. To assure consistency among different tetramers, they were all loaded with identical groove peptide, CLIP (26). Likewise, SE-expressing HBc particles have been engineered to allow the SE to preserve its native α helical structure outside of the HLA-DR molecule context and without the presence of groove peptide (10). Therefore, the significance of the experiments using these reagents is two fold: First, they demonstrate high consistency among data generated with the SE in several different formulations and, most importantly, given their intact tri-dimensional conformation, the results attest to the physiologic relevance of the findings.
The data presented here provide the first direct indication that the SE may be acting as an immune stimulatory ligand. The study demonstrates that the SE, independent of groove peptides, activates pathogenically-relevant immune system events. In addition to its generic antigen-independent Th17 polarization effects, the SE particularly facilitates antigen-specific immune responses. One scenario to consider is that the SE could allow promiscuous augmentation of various antigen-specific immune responses and increase their autoimmune potential. Depending on genetic and environmental co-factors, the nature of the specific antigens and their tissue distribution, the SE could conceivably catapult the onset of distinct autoimmune diseases. This scenario could provide a plausible explanation for the counterintuitive, promiscuous association of the SE - a highly specific HVR3 sequence - with many antigenically-unrelated human diseases and animal models (discussed in 10).
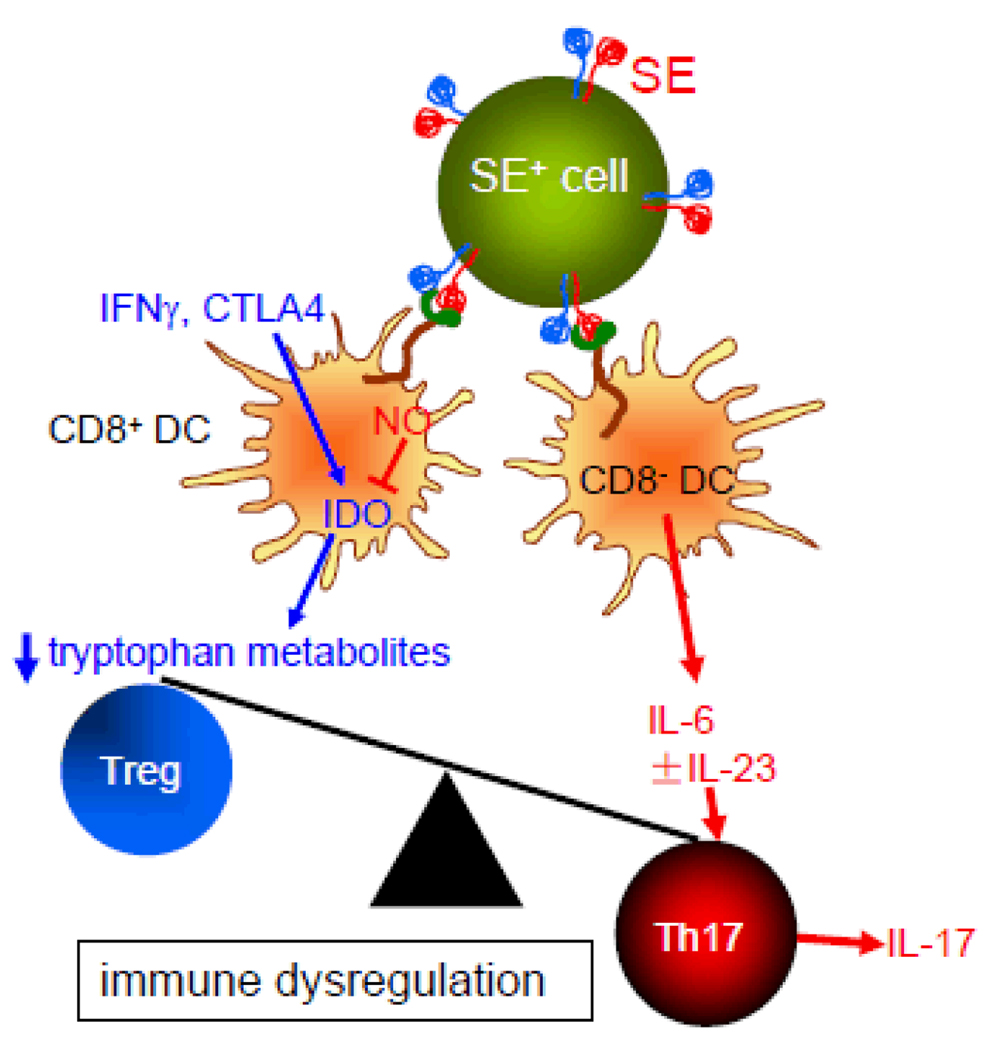
Based on the findings of this study, in Figure 8 we propose a model describing the SE immune polarizing effects. Given the superior potency of physiologically-folded tetramers shown here, we propose that the tri-dimensional conformation of the SE plays a significant role in its immune effect. Accordingly, our model predicts that SE-expressing HLA-DR molecules are most effective In vivo when they are expressed on the cell surface and interact in trans with CRT expressed on other cells. It should be clarified, however, that we cannot rule out a scenario in which SE-positive HLA-DR molecules that are shed to the circulation (or SE-expressing peptidic fragments of such molecules) may exert similar effects. Additionally, SE-CRT interaction on the same cell surface (in cis) cannot be ruled out as well.
FIGURE 8. A proposed model of the role of the SE ligand in autoimmunity.
The effect of the SE ligand is the end result of its interaction with two DC subsets. The SE ligand, expressed on MHC class II-positive cells, interacts with cell surface CRT (green) with resultant NO production. In CD8+ DCs, increased NO levels inhibit the enzymatic activity of IDO, which leads to decrease tryptophan metabolites and reduced generation of Tregs. In CD8− DCs, the SE ligand activates production of IL-6 and, in the presence of LPS, could increase production of IL-23 as well. The end result is immune dysregulation that involves increased numbers of Th17 cells and increased production of the pro-arthritogenic and autoimmunogenic cytokine IL-17.
Our model predicts that as a result of CRT ligation by the SE, two events take place: 1. In CD11c+CD8+ DCs, the SE ligand inhibits activation of IDO. Consequently, tryptophan catabolism diminishes with resultant inhibition of Treg differentiation; 2. In CD11c+CD8− DCs SE-actiavted signaling leads to over production of Th17-promoting cytokines IL-6 and - in the presence of LPS - IL-23. The end result of the SE dual effect on DCs is immune imbalance and Th17 polarization.
The balance between Treg and Th17 has been previously shown to play a role in the pathogenesis of inflammatory arthritis. For example, IL-17 has been found to enhance angiogenesis, matrix metalloproteinase activation, osteoclastogenesis, leukocyte recruitment and inflammation (46). Additionally, both IL-17-producing cells and IL-17 are abundantly expressed in the RA joint (47, 48) and IL-17 neutralization prevents experimental arthritis development. Treg cells, on the other hand, have been shown to modulate disease severity in the collagen-induced arthritis model (49, 50). Thus, based on the well documented impact of Treg/Th17 balance on autoimmune arthritis, we propose that the mechanism described here may contribute to SE-associated RA severity.
In summary, we demonstrate here a previously uncharacterized immune polarization by the SE ligand both in vitro and In vivo. The effect is highly allele-specific and particularly potent with physiologically-folded ligands. Studies are presently under way to determine whether the polarizing effect of the SE ligand can facilitate autoimmunity in mice.
Supplementary Material
ACKNOWLEDGMENTS
We thank the NIH Tetramer Core Facility for providing tetrameric HLA-DR reagents. Special thanks to Laura Tesmer for technical advice and Dr. Steven Lundy for providing CII-TCR Tg mice and a CII synthetic peptide.
Abbreviations used in this paper
- CRT
calreticulin
- DC
dendritic cell
- FU
fluorescence units
- HBc
hepatitis B core
- HVR3
third allelic hypervariable region
- RA
rheumatoid arthritis
- SE
shared epitope
- SNAP
S-nitroso-N-acetylpenicillamine
- Treg
regulatory T cell
Footnotes
Supported with grants awarded to JH by the US National Institutes of Health (AR55170, UL1RR02498, AI47331, GM088560, AR20557, AR48310), the University of Michigan Global Reach Collaborative Research Fund, by a Basic Research Grant from the Arthritis Foundation and an Innovative Basic Science Award from the American College of Rheumatology Research and Education Foundation.
REFERENCES
- 1.Firestein GS, Zvaifler NJ. How important are T cells in chronic rheumatoid synovitis? Arthritis Rheum. 1990;33:768–773. doi: 10.1002/art.1780330602. [DOI] [PubMed] [Google Scholar]
- 2.Panayi GS, Lanchbury JS, Kingsley GH. The importance of the T cell in initiating and maintaining the chronic synovitis of rheumatoid arthritis. Arthritis Rheum. 1992;35:729–735. doi: 10.1002/art.1780350702. [DOI] [PubMed] [Google Scholar]
- 3.Gregersen PK, Silver J, Winchester RJ. The shared epitope hypothesis. An approach to understanding the molecular genetics of susceptibility to rheumatoid arthritis. Arthritis Rheum. 1987;30:1205–1213. doi: 10.1002/art.1780301102. [DOI] [PubMed] [Google Scholar]
- 4.Turesson C, Schaid DJ, Weyand CM, Jacobsson LT, Goronzy JJ, Petersson IF, Sturfelt G, Nyhall-Wahlin BM, Truedsson L, Dechant SA, Matteson EL. The impact of HLA-DRB1 genes on extra-articular disease manifestations in rheumatoid arthritis. Arthritis Res Ther. 2005;7:R1386–R1393. doi: 10.1186/ar1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moreno I, Valenzuela A, Garcia A, Yelamos J, Sanchez B, Hernanz W. Association of the shared epitope with radiological severity of rheumatoid arthritis. J Rheumatol. 1996;23:6–9. [PubMed] [Google Scholar]
- 6.Wucherpfennig KW, Strominger JL. Selective binding of self peptides to disease-associated major histocompatibility complex (MHC) molecules: a mechanism for MHC-linked susceptibility to human autoimmune diseases. J Exp Med. 1995;181:1597–1601. doi: 10.1084/jem.181.5.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.La Cava A, Nelson JL, Ollier WE, MacGregor A, Keystone EC, Thorne JC, Scavulli JF, Berry CC, Carson DA, Albani S. Genetic bias in immune responses to a cassette shared by different microorganisms in patients with rheumatoid arthritis. J Clin Invest. 1997;100:658–663. doi: 10.1172/JCI119577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bhayani HR, Hedrick SM. The role of polymorphic amino acids of the MHC molecule in the selection of the T cell repertoire. J Immunol. 1991;146:1093–1098. [PubMed] [Google Scholar]
- 9.Holoshitz J, Ling S. Nitric oxide signaling triggered by the rheumatoid arthritis shared epitope: a new paradigm for MHC disease association? Ann N Y Acad Sci. 2007;1110:73–83. doi: 10.1196/annals.1423.009. [DOI] [PubMed] [Google Scholar]
- 10.Ling S, Lai A, Borschukova O, Pumpens P, Holoshitz J. Activation of nitric oxide signaling by the rheumatoid arthritis shared epitope. Arthritis Rheum. 2006;54:3423–3432. doi: 10.1002/art.22178. [DOI] [PubMed] [Google Scholar]
- 11.Ling S, Li Z, Borschukova O, Xiao L, Pumpens P, Holoshitz J. The rheumatoid arthritis shared epitope increases cellular susceptibility to oxidative stress by antagonizing an adenosine-mediated anti-oxidative pathway. Arthritis Res Ther. 2007;9:R5. doi: 10.1186/ar2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ling S, Pi X, Holoshitz J. The rheumatoid arthritis shared epitope triggers innate immune signaling via cell surface calreticulin. J Immunol. 2007;179:6359–6367. doi: 10.4049/jimmunol.179.9.6359. [DOI] [PubMed] [Google Scholar]
- 13.Okuyama Y, Cho JH, Nakajima Y, Homma K, Sekimizu K, Natori S. Binding between azurocidin and calreticulin: its involvement in the activation of peripheral monocytes. J Biochem. 2004;135:171–177. doi: 10.1093/jb/mvh020. [DOI] [PubMed] [Google Scholar]
- 14.Henson PM, Bratton DL, Fadok VA. The phosphatidylserine receptor: a crucial molecular switch? Nat Rev Mol Cell Biol. 2001;2:627–633. doi: 10.1038/35085094. [DOI] [PubMed] [Google Scholar]
- 15.Zeng G, Aldridge ME, Tian X, Seiler D, Zhang X, Jin Y, Rao J, Li W, Chen D, Langford MP, Duggan C, Belldegrun AS, Dubinett SM. Dendritic cell surface calreticulin is a receptor for NY-ESO-1: direct interactions between tumor-associated antigen and the innate immune system. J Immunol. 2006;177:3582–3589. doi: 10.4049/jimmunol.177.6.3582. [DOI] [PubMed] [Google Scholar]
- 16.Gardai SJ, Bratton DL, Ogden CA, Henson PM. Recognition ligands on apoptotic cells: a perspective. J Leukoc Biol. 2006;79:896–903. doi: 10.1189/jlb.1005550. [DOI] [PubMed] [Google Scholar]
- 17.Gardai SJ, Xiao YQ, Dickinson M, Nick JA, Voelker DR, Greene KE, Henson PM. By binding SIRPalpha or calreticulin/CD91, lung collectins act as dual function surveillance molecules to suppress or enhance inflammation. Cell. 2003;115:13–23. doi: 10.1016/s0092-8674(03)00758-x. [DOI] [PubMed] [Google Scholar]
- 18.Donnelly S, Roake W, Brown S, Young P, Naik H, Wordsworth P, Isenberg DA, Reid KB, Eggleton P. Impaired recognition of apoptotic neutrophils by the C1q/calreticulin and CD91 pathway in systemic lupus erythematosus. Arthritis Rheum. 2006;54:1543–1556. doi: 10.1002/art.21783. [DOI] [PubMed] [Google Scholar]
- 19.Lakey RL, Morgan TG, Rowan AD, Isaacs JD, Cawston TE, Hilkens CM. A novel paradigm for dendritic cells as effectors of cartilage destruction. Rheumatology (Oxford) 2009;48:502–507. doi: 10.1093/rheumatology/kep040. [DOI] [PubMed] [Google Scholar]
- 20.Paust S, Cantor H. Regulatory T cells and autoimmune disease. Immunol Rev. 2005;204:195–207. doi: 10.1111/j.0105-2896.2005.00247.x. [DOI] [PubMed] [Google Scholar]
- 21.Hwu P, Du MX, Lapointe R, Do M, Taylor MW, Young HA. Indoleamine 2,3-dioxygenase production by human dendritic cells results in the inhibition of T cell proliferation. J Immunol. 2000;164:3596–3599. doi: 10.4049/jimmunol.164.7.3596. [DOI] [PubMed] [Google Scholar]
- 22.Terness P, Chuang JJ, Opelz G. The immunoregulatory role of IDO-producing human dendritic cells revisited. Trends Immunol. 2006;27:68–73. doi: 10.1016/j.it.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 23.Fallarino F, Grohmann U, Hwang KW, Orabona C, Vacca C, Bianchi R, Belladonna ML, Fioretti MC, Alegre ML, Puccetti P. Modulation of tryptophan catabolism by regulatory T cells. Nat Immunol. 2003;4:1206–1212. doi: 10.1038/ni1003. [DOI] [PubMed] [Google Scholar]
- 24.Alberati-Giani D, Malherbe P, Ricciardi-Castagnoli P, Kohler C, Denis-Donini S, Cesura AM. Differential regulation of indoleamine 2,3-dioxygenase expression by nitric oxide and inflammatory mediators in IFN-gamma-activated murine macrophages and microglial cells. J Immunol. 1997;159:419–426. [PubMed] [Google Scholar]
- 25.Hucke C, MacKenzie CR, Adjogble KD, Takikawa O, Daubener W. Nitric oxide-mediated regulation of gamma interferon-induced bacteriostasis: inhibition and degradation of human indoleamine 2,3-dioxygenase. Infect Immun. 2004;72:2723–2730. doi: 10.1128/IAI.72.5.2723-2730.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Day CL, Seth NP, Lucas M, Appel H, Gauthier L, Lauer GM, Robbins GK, Szczepiorkowski ZM, Casson DR, Chung RT, Chung RT, Bell S, Harcourt G, Walker BD, Klenerman P, Wucherpfennig KW. Ex vivo analysis of human memory CD4 T cells specific for hepatitis C virus using MHC class II tetramers. J Clin Invest. 2003;112:831–842. doi: 10.1172/JCI18509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.De Luca A, Montagnoli C, Zelante T, Bonifazi P, Bozza S, Moretti S, D'Angelo C, Vacca C, Boon L, Bistoni F, Puccetti P, Fallarino F, Romani L. Functional yet balanced reactivity to Candida albicans requires TRIF, MyD88, and IDO-dependent inhibition of Rorc. J Immunol. 2007;179:5999–6008. doi: 10.4049/jimmunol.179.9.5999. [DOI] [PubMed] [Google Scholar]
- 28.Lundy SK, Sarkar S, Tesmer LA, Fox DA. Cells of the synovium in rheumatoid arthritis. T lymphocytes. Arthritis Res Ther. 2007;9:202. doi: 10.1186/ar2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Olson RR, Reuter JJ, McNicholl J, Alber C, Klohe E, Callahan K, Siliciano RF, Karr RW. Acidic residues in the DR beta chain third hypervariable region are required for stimulation of a DR(alpha, beta 1*0402)-restricted T-cell clone. Hum Immunol. 1994;41:193–200. doi: 10.1016/0198-8859(94)90036-1. [DOI] [PubMed] [Google Scholar]
- 30.Grohmann U, Belladonna ML, Bianchi R, Orabona C, Ayroldi E, Fioretti MC, Puccetti P. IL-12 acts directly on DC to promote nuclear localization of NF-kappaB and primes DC for IL-12 production. Immunity. 1998;9:315–323. doi: 10.1016/s1074-7613(00)80614-7. [DOI] [PubMed] [Google Scholar]
- 31.Takikawa O, Kuroiwa T, Yamazaki F, Kido R. Mechanism of interferon-gamma action. Characterization of indoleamine 2,3-dioxygenase in cultured human cells induced by interferon-gamma and evaluation of the enzyme-mediated tryptophan degradation in its anticellular activity. J Biol Chem. 1988;263:2041–2048. [PubMed] [Google Scholar]
- 32.Zhou L, Ivanov II, Spolski R, Min R, Shenderov K, Egawa T, Levy DE, Leonard WJ, Littman DR. IL-6 programs T(H)-17 cell differentiation by promoting sequential engagement of the IL-21 and IL-23 pathways. Nat Immunol. 2007;8:967–974. doi: 10.1038/ni1488. [DOI] [PubMed] [Google Scholar]
- 33.Fallarino F, Vacca C, Orabona C, Belladonna ML, Bianchi R, Marshall B, Keskin DB, Mellor AL, Fioretti MC, Grohmann U, Puccetti P. Functional expression of indoleamine 2,3-dioxygenase by murine CD8 alpha(+) dendritic cells. Int Immunol. 2002;14:65–68. doi: 10.1093/intimm/14.1.65. [DOI] [PubMed] [Google Scholar]
- 34.Thomas SR, Mohr D, Stocker R. Nitric oxide inhibits indoleamine 2,3-dioxygenase activity in interferon-gamma primed mononuclear phagocytes. J Biol Chem. 1994;269:14457–14464. [PubMed] [Google Scholar]
- 35.Ivanov II, Zhou L, Littman DR. Transcriptional regulation of Th17 cell differentiation. Semin Immunol. 2007;19:409–417. doi: 10.1016/j.smim.2007.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ardavin C, Amigorena S, Reis e Sousa C. Dendritic cells: immunobiology and cancer immunotherapy. Immunity. 2004;20:17–23. doi: 10.1016/s1074-7613(03)00352-2. [DOI] [PubMed] [Google Scholar]
- 37.Sharma MD, Hou DY, Liu Y, Koni PA, Metz R, Chandler P, Mellor AL, He Y, Munn DH. Indoleamine 2,3-dioxygenase controls conversion of Foxp3+ Tregs to TH17− like cells in tumor-draining lymph nodes. Blood. 2009;113:6102–6111. doi: 10.1182/blood-2008-12-195354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baban B, Chandler PR, Sharma MD, Pihkala J, Koni PA, Munn DH, Mellor AL. IDO activates regulatory T cells and blocks their conversion into Th17-like T cells. J Immunol. 2009;183:2475–2483. doi: 10.4049/jimmunol.0900986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sharma MD, Baban B, Chandler P, Hou DY, Singh N, Yagita H, Azuma M, Blazar BR, Mellor AL, Munn DH. Plasmacytoid dendritic cells from mouse tumor-draining lymph nodes directly activate mature Tregs via indoleamine 2,3-dioxygenase. J Clin Invest. 2007;117:2570–2582. doi: 10.1172/JCI31911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Korn T, Bettelli E, Gao W, Awasthi A, Jager A, Strom TB, Oukka M, Kuchroo VK. IL-21 initiates an alternative pathway to induce proinflammatory T(H)17 cells. Nature. 2007;448:484–487. doi: 10.1038/nature05970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Iwanami K, Matsumoto I, Yoshiga Y, Inoue A, Kondo Y, Yamamoto K, Tanaka Y, Minami R, Hayashi T, Goto D, Ito S, Nishimura Y, Sumida T. Altered peptide ligands inhibit arthritis induced by glucose-6-phosphate isomerase peptide. Arthritis Res Ther. 2009;11:R167. doi: 10.1186/ar2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Notley CA, Inglis JJ, Alzabin S, McCann FE, McNamee KE, Williams RO. Blockade of tumor necrosis factor in collagen-induced arthritis reveals a novel immunoregulatory pathway for Th1 and Th17 cells. J Exp Med. 2008;205:2491–2497. doi: 10.1084/jem.20072707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mellor AL, Baban B, Chandler PR, Manlapat A, Kahler DJ, Munn DH. Cutting edge: CpG oligonucleotides induce splenic CD19+ dendritic cells to acquire potent indoleamine 2,3-dioxygenase-dependent T cell regulatory functions via IFN Type 1 signaling. J Immunol. 2005;175:5601–5605. doi: 10.4049/jimmunol.175.9.5601. [DOI] [PubMed] [Google Scholar]
- 44.Munn DH, Sharma MD, Hou D, Baban B, Lee JR, Antonia SJ, Messina JL, Chandler P, Koni PA, Mellor AL. Expression of indoleamine 2,3-dioxygenase by plasmacytoid dendritic cells in tumor-draining lymph nodes. J Clin Invest. 2004;114:280–290. doi: 10.1172/JCI21583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hreggvidsdottir HS, Ostberg T, Wahamaa H, Schierbeck H, Aveberger AC, Klevenvall L, Palmblad K, Ottosson L, Andersson U, Harris HE. The alarmin HMGB1 acts in synergy with endogenous and exogenous danger signals to promote inflammation. J Leukoc Biol. 2009;86:655–662. doi: 10.1189/jlb.0908548. [DOI] [PubMed] [Google Scholar]
- 46.Tesmer LA, Lundy SK, Sarkar S, Fox DA. Th17 cells in human disease. Immunol Rev. 2008;223:87–113. doi: 10.1111/j.1600-065X.2008.00628.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chabaud M, Durand JM, Buchs N, Fossiez F, Page G, Frappart L, Miossec P. Human interleukin-17: A T cell-derived proinflammatory cytokine produced by the rheumatoid synovium. Arthritis Rheum. 1999;42:963–970. doi: 10.1002/1529-0131(199905)42:5<963::AID-ANR15>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 48.Ziolkowska M, Koc A, Luszczykiewicz G, Ksiezopolska-Pietrzak K, Klimczak E, Chwalinska-Sadowska H, Maslinski W. High levels of IL-17 in rheumatoid arthritis patients: IL-15 triggers in vitro IL-17 production via cyclosporin A-sensitive mechanism. J Immunol. 2000;164:2832–2838. doi: 10.4049/jimmunol.164.5.2832. [DOI] [PubMed] [Google Scholar]
- 49.Morgan ME, Sutmuller RP, Witteveen HJ, van Duivenvoorde LM, Zanelli E, Melief CJ, Snijders A, Offringa R, de Vries RR, Toes RE. CD25+ cell depletion hastens the onset of severe disease in collagen-induced arthritis. Arthritis Rheum. 2003;48:1452–1460. doi: 10.1002/art.11063. [DOI] [PubMed] [Google Scholar]
- 50.Nakae S, Nambu A, Sudo K, Iwakura Y. Suppression of immune induction of collagen-induced arthritis in IL-17-deficient mice. J Immunol. 2003;171:6173–6177. doi: 10.4049/jimmunol.171.11.6173. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.