SUMMARY
Replication timing profiles are cell type-specific and reflect genome organization changes upon differentiation. In this protocol we describe how to analyze replication timing genome-wide in mammalian cells. Asynchronously cycling cells are pulse labeled with the nucleotide analog 5-bromo-2-deoxyuridine (BrdU) and sorted into S-phase fractions based on DNA content using flow cytometry. BrdU-labeled DNA from each fraction is immunoprecipitated, amplified, differentially labeled, and co-hybridized to a whole-genome CGH microarray, which is currently more cost effective than high-throughput sequencing and equally capable of resolving features at the biologically relevant level of tens to hundreds of kilobases. We also present a guide to analyzing the resulting datasets, based on methods we use routinely. Subjects include normalization, scaling, and data quality measures, loess (local polynomial) smoothing of replication timing values, segmentation of data into domains, and assignment of timing values to gene promoters. Finally, we cover clustering methods and means to relate changes in the replication program to gene expression and other genetic and epigenetic datasets. Some experience with R or similar programming languages is assumed. Altogether, the protocol takes approximately 3 weeks to complete.
Keywords: RT, genome-wide, FACS
INTRODUCTION
Although the mechanisms that specify the timing and placement of origin firing in higher eukaryotes remain a mystery, all eukaryotes have a defined replication timing program that is largely conserved between closely related species1, including human and mouse2,32,3. Analyses of replication timing in various cell types have yielded insights into genome organization and repackaging events during development, suggesting an important role for the timing program itself or 3D genome organization in regulating developmental gene expression1,3,4. In this protocol, we describe approaches for measuring replication timing genome-wide. As data processing and analysis are often a bottleneck in these studies, the protocol also covers methods used routinely in our lab for downstream analysis3,5,6. Although this protocol emphasizes mammalian cells as applied to analyze replication timing changes in various mouse and human cell types3,5,6, it can be adapted to any proliferating cell type, and such variations have been used to analyze replication timing in Drosophila7–9, Arabidopsis10, and budding yeast11.
Overview of the Procedure: Generating experimental data (steps 1–61)
This first portion of the protocol describes how to derive raw data for genome-wide replication timing analysis. Given that the protocol measures the timing of events during the cell cycle, some form of synchronization is required. Synchronization can be achieved either prospectively, prior to cell collection, or retroactively, after the cells have been collected. In yeasts, prospective synchrony methods are well established, and in many cases the same method can be used to compare different strains12,13. However, most synchronization schemes for multi-cellular organisms are cumbersome and optimized for specific cell lines14–16, and most require the use of metabolic inhibitors that can interfere with normal regulation of replication17,18. By contrast, retroactive synchronization using a fluorescence activated cell sorter (FACS) to select cells based upon the increase in DNA content during S-phase can be applied to any proliferating cell population without the need for any prior manipulation beyond dissociation of cells into a single-cell suspension19. Moreover, most prospective synchronization regimes for studying replication timing verify the quality of synchronization by FACS analysis of DNA content; since DNA content defines S phase interval, selection of cells for DNA content is the most direct means to the desired end. The resolution of S phase intervals is determined by the fineness of DNA content windows selected. The only situations in which the above synchronization alternatives may need to be considered are for cells that are very difficult to dissociate or those that are severely aneuploid, such that DNA content does not reflect the time during S phase.
In the original method20,21. cells were labeled with BrdU for a fraction of S-phase and sorted into several different time points during S-phase. BrdU-substituted DNA could then be isolated either based on its increased density or using anti-BrdU-antibodies and specific loci could be examined by hybridization or PCR20–22. With microarray analysis, replication of the entire genome can be queried in a single array hybridization by limiting the analysis to two differentially labeled samples, allowing all probes to be assigned one internally normalized relative replication timing value and rapid comparison of many samples3,5,6,23,24. One limitation of assigning one replication timing value per map position is that it cannot distinguish cases where homologous loci replicate asynchronously, a situation that is estimated to occur for a few percent of the genome19. However, the protocol can be readily adapted for analysis of these genomic segments by dividing and sorting S phase into finer fractions19
The two most popular variations of retroactive synchronization by FACS are described in the Procedure below. In the first method, BrdU-labeled cells are divided into early and late S-phase fractions, and BrdU-labeled DNA synthesized either early or late can then be labeled and hybridized to a microarray. This method produces a high signal to noise ratio since immunoprecipitation (BrdU-IP) substantially enriches for DNA synthesized in each half of S-phase. However, BrdU-IP efficacy can fluctuate and must be closely monitored. In the second method, unlabeled cells are sorted into total S-phase vs. G1-phase populations and DNA from these stages is differentially labeled and used as the target. This obviates BrdU-IP, but the dynamic range is limited to the 2-fold copy number increase during S-phase. Both methods give similar results, evidenced by a direct comparison in the same cell line in one study6. In both methods, DNA from each fraction is differentially labeled with Cy3 and Cy5 dyes and then cohybridized to a whole-genome oligonucleotide microarray. The ratio of the abundance of each probe in each fraction is then used to generate a replication-timing profile.
Overview of the Procedure: Normalization and computational analysis of replication timing datasets (steps 62–88)
In this section of the protocol we focus on methods specifically useful for replication timing analysis using whole-genome comparative genome hybridization (CGH) microarrays25., which we have used to investigate the type, degree, and mechanism of replication timing changes in mouse and human cell lines3,5,6,23,24. General methods for normalizing and analyzing microarray experiments for chromatin modifications or transcription at gene promoters have been described in detail in other works26–29. Similar to two-color microarray designs comparing an experimental sample to a reference, our replication timing experiments employ a two-channel design comparing early versus late fraction enrichment for each target. Typically, we include two dye-swap replicates per sample to address bias due to dye-specific effects, such as more rapid photobleaching of Cy5 dye than Cy3. Our philosophy is to minimize the number of transformations applied to the data and apply only minimally invasive global methods for removing bias and scaling datasets to allow comparisons between them.
All of the analysis described here uses the R framework for statistical computing30–32. Through user-submitted packages that facilitate a wide variety of methods, R has become an indispensible tool for many common computational tasks. Although R has an initially steep learning curve due to its command line interface, help is available in many locations and forms, including books33–35, online manuals (http://cran.r-project.org/), and mailing lists aggregated in the R Mailing Lists Archive (http://tolstoy.newcastle.edu.au/R/). Help can also be found within R itself; str() is often helpful for viewing the structure of variables and datasets, and the ? operator (e.g., ?data.frame() ) provides a help page for the corresponding function. We use the R package LIMMA (linear models for microarray data), also available with a user interface through the limmaGUI package, for normalization and scaling27,36. The steps for this process are straightforward, and illustrated using two biological replicate datasets of mouse L1210 lymphoblast cells, which are available in raw form in Supplementary Data and after normalization and smoothing at www.ReplicationDomain.org.
We provide this section as a verified route for extracting information from the microarray experiments described in the Procedure; however, users with sufficient experience with R or having different requirements for their data are free to modify the analysis as needed, and a wide array of alternative and additional methods are available through Bioconductor31. While our methods for downstream analyses were tested primarily with NimbleGen CGH microarrays, most are applicable to any data format containing chromosome, genomic position, and replication timing information for each probe.
EXPERIMENTAL DESIGN
BrdU Incorporation
The nucleotide analog 5-bromo-2′-deoxyuridine (BrdU) can be used to pulse-label newly synthesized DNA during S-phase. For mammalian cell types that have 8–12 hour S phases, incubation with BrdU for two hours has been empirically determined to provide sufficient incorporation to ensure successful BrdU-IP in subsequent steps yet be short enough to identify even subtle differences in replication timing, such as between female cells with one vs. two early replicating X chromosomes5. Success has also been achieved with BrdU labeling times as short as one hour, but subsequent BrdU-IP can be problematic as there is very little substituted DNA relative to background of unsubstituted DNA that will contribute to noise in the BrdU-IP6. The BrdU-labeling times for cells with S-phase lengths significantly different to mammalian cells, such as amphibian20 or fly8 cells, should be adjusted appropriately.
FACS sorting fractions of S-phase
For first time users, it is recommended that at least 5 × 106 cells be used; however, with experience and a sufficient fraction of S-phase cells, as few as 0.5–1 × 106 starting cells can be successfully profiled. The important parameter is to obtain 20,000 – 30,000 cells in each of the early and late S-phase fractions. As described in Procedure step 1A, ethanol-fixed cells can be stained with propidium iodide (PI) and sorted based on DNA content. Alternative fluorochromes that do not require RNase digestion, such as chromomycin A3, can also be used with ethanol fixed cells20,21. Some cell types tend to clump or produce a lot of cellular debris when fixed in ethanol. For these cell types, the fixation step can be skipped and DNA can be stained with DAPI in permeabilized cells, as described in Procedure step 1B. The advantage of the method described in step 1A is that cells fixed in ethanol can be stored at −20°C (empirically determined to be the optimal temperature) or shipped to collaborators. Shipping should be done on dry ice, with a partition between the tube and the dry ice to prevent cell freezing. All steps, particularly storage, should be performed in the dark since BrdU-substituted DNA is light sensitive.
During FACS analysis forward and side scatter analyses should be used to select an appropriate population of cells free of doublets or cell debris, both of which can hinder accurate sorting of desired populations. Lasers used in this protocol include 488 Blue to detect PI or 407 Violet to detect DAPI in cells that have been stained for DNA content. Two separate fractions of S phase, early and late, are chosen to be collected, but more can be collected if desired20,21.
Immunoprecipitation of BrdU labeled DNA
DNA from BrdU-labeled cells should be sonicated into fragments ranging from 250bp to 2kb and then immunoprecipitated using an anti-BrdU antibody Sonication into fragments of this size helps eliminate immunoprecipitation of DNA that has not been BrdU labeled. If samples have been stored at −20°C prior to beginning the immunoprecipitation, thaw samples in a 56°C water bath to completely dissolve SDS and add 200μl of SDS-PK Buffer pre-warmed to 56°C with 0.05mg/mL glycogen to each sample prior to performing the phenol-chloroform extraction in Procedure step 13.
Quality control check of S-phase DNA
Due to the sensitivity and large number of steps involved, BrdU-IP is one of the trickiest parts of the protocol. To ensure quality, screen BrdU-IPs by PCR amplification using primers specific to DNA markers of known relative replication time (i.e. early or late). Although real-time PCR can be performed, we find gel electrophoresis to be sufficient to evaluate enrichment of DNA in each IP sample. Importantly, as PCR results can vary between aliquots of the same sample, and replication timing can vary between cell types3,5, consistency across multiple samples from the same cell type is the best way to verify quality. Use the primer sets listed in Table 1 for mouse or human cell types, or substitute suitable alternatives to screen several IPs from both early and late S phase fractions.
TABLE 1.
Primers used for human and mouse BrdU IP screen
| Name | Sequence | Base pairs |
|---|---|---|
| Human test regions | ||
| Mitochondrial DNA | Forward 5′-CCTAGGAATCACCTCCCATTCC-3′ Reverse 5′-GTGTTTAAGGGGTTGGCTAGGG-3′ |
168 bp |
| α-globin | Forward 5′-GACCCTCTTCTCTGCACAGCTC-3′ Reverse 5′-GCTACCGAGGCTCCAGCTTAAC-3′ |
257 bp |
| β-globin | Forward 5′-CCTGAGGAGAAGTCTGCCGTTA-3′ Reverse 5′-GAACCTCTGGGTCCAAGGGTAG-3′ |
241 bp |
| MMP15 | Forward 5′-CAGGCCTCTGGTCTCTGTCATT-3′ Reverse 5′-AGAGCTGAGAAACCACCACCAG-3′ |
249 bp |
| BMP1 | Forward 5′-GATGAAGCCTCGACCCCTAGAT-3′ Reverse 5′-ACCCGTCAGAGACGAACTTGAG-3′ |
177 bp |
| hPTGS2 | Forward 5′-GTTCTAGGCTGGTGTCCCATTG-3′ Reverse 5′-CTTTCTGTACTGCGGGTGGAAC-3′ |
230 bp |
| hNETO1 | Forward 5′-GGAGGTGGAATGCTAGGGACTT-3′ Reverse 5′-GCTGAGTGTGGCCTTAAGAGGA-3′ |
286 bp |
| hSLITRK6 | Forward 5′-GGAGAACATGCCTCCACAGTCT-3′ Reverse 5′-GTCCTGGAAGTTGAGTGGATGG-3′ |
281 bp |
| hZFP42 | Forward 5′-CTTGTGGGGACACCCAGATAAG-3′ Reverse 5′-AACCACCTCCAGGCAGTAGTGA-3′ |
233 bp |
| hDPPA2 | Forward 5′-AGGTGGACAGCGAAGACAGAAC-3′ Reverse 5′-GGCCATCAGCAGTGTCCTAAAC-3′ |
168bp |
| Mouse test regions | ||
| Mitochondrial DNA | Forward 5′-GACATCTGGTTCTTACTTCA-3′ Reverse 5′-GTTTTTGGGGTTTGGCATTA-3′ |
346 bp |
| α-globin | Forward 5′-AAGGGGAGCAGAGGCATCA-3′ Reverse 5′-AGGGCTTGGGAGGGACTG-3′ |
439 bp |
| β-globin | Forward 5′-CAGTAAGCCACAGATCCTATTG-3′ Reverse 5′-CCCATAGTGACTATTGACTGTG-3′ |
369 bp |
| Pou5f1 | Forward 5′-CCCTCCCTAAGTGCCAGTTTCT-3′ Reverse 5′-GTAATCGCCCTCAGCAGTGTCT-3′ |
194 bp |
| Mmp15 | Forward 5′-AACAGAAGGCCTGCCTTGAC-3′ Reverse 5′-TGCATAGCACGACAGCATTG-3′ |
360 bp |
| Zfp42 | Forward 5′-TGAGATTAGCCCCGAGACTGAG-3′ Reverse 5′-CGTCCCCTTTGTCATGTACTCC-3′ |
211 bp |
| Dppa2 | Forward 5′-CCACAGGAAGACAGGAAGCAGT-3′ Reverse 5′-AGCCAGACAGGAGCCCTAGAGT-3′ |
199 bp |
| Ptn | Forward 5′-CTGGAATGAGTTACTGACGGGG-3′ Reverse 5′-CTGGCCCCACTGTGTAATAAGC-3′ |
230 bp |
| Mash1 | Forward 5′-GAAGATGAGCAAGGTGGAGACG-3′ Reverse 5′-AGTAGGACGAGACCGGAGAACC-3′ |
182 bp |
| Akt3 | Forward 5′-GAAGTGTGGGTTGAACCTCTGG-3′ Reverse 5′-GCACCCTCTCCACTGTTCTGAT-3′ |
173 bp |
Amplification methods for immunoprecipitated single-stranded DNA
Once purified by immunoprecipitation and screened for sample quality, BrdU incorporated DNA must be amplified to obtain sufficient amounts for array hybridization. If multiple samples pass PCR screening, pool DNA from parallel immunoprecipitations to use as the starting material for whole genome amplification; otherwise, use a single screen immunoprecipitation. Perform whole-genome amplification as desired (we use the GenomePlex Complete Whole Genome Amplification and Reamplification Kits from Sigma) then load amplified samples onto a gel to determine size range and screen once more by PCR to ensure that no bias was introduced during amplification.
Labeling and hybridization of amplified samples
The specific steps required in this section will largely depend on the chosen array platform. While we focus on NimbleGen products in order to avoid the ambiguity inherent to generalized methods, the products can be applied successfully to additional platforms8,9, including deep sequencing of BrdU-IP DNA37. Currently, mammalian replication timing data generated from microarray hybridization and deep sequencing is of equal quality3,6, while the microarray method remains more cost-effective and the bioinformatics are considerably less demanding for the typical laboratory. Future advances reducing BrdU-labeling times and sequencing limitations may make this method more cost-effective and accessible38.
Once a platform is chosen, the labeling and hybridization steps are fairly straightforward. Briefly, 1 μg of early or late replicating DNA may be labeled with either Cy5 or Cy3 random 9-mer dyes by Klenow reaction, precipitated with isopropanol, and resuspended and quantified in nuclease-free water. Finally, equal quantities of labeled early and late fraction DNA should be combined (specific quantity will depend upon array design).
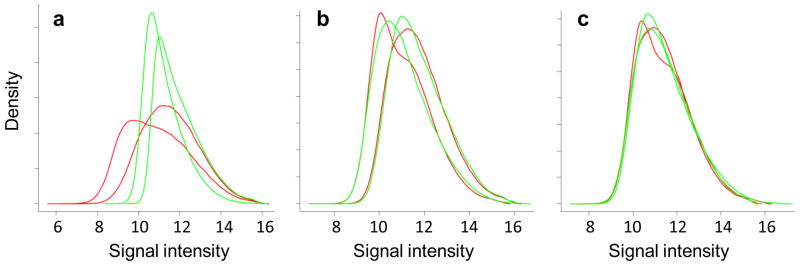
Array design
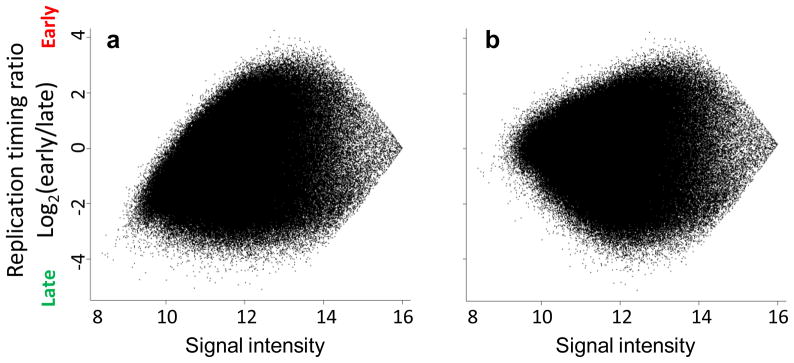
Array design is also an important consideration, and the nature of your study should be a guide in selecting between the variety of available standard and custom designs. For our genome-wide studies in both mouse and human, 385K and 3X720K-feature Comparative Genomic Hybridization (CGH) tiling arrays have sufficient probe densities, showing no disadvantage compared to high density 2.1M CGH tiling arrays5,6, but have considerable cost and convenience advantages. Tiling designs with roughly evenly spaced probes also facilitate the interpretation and analysis of genetic features.
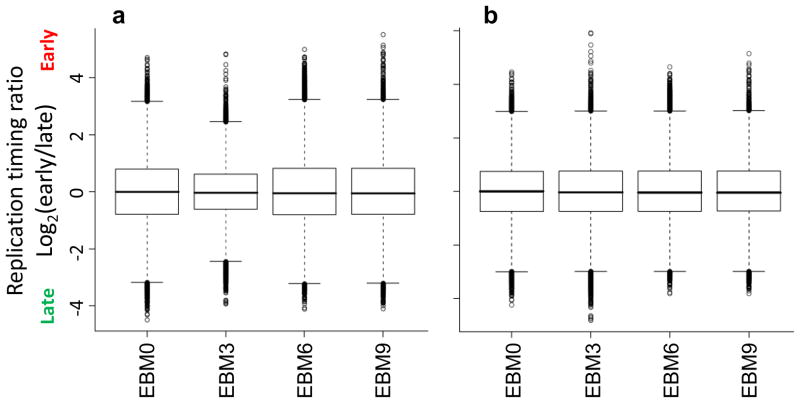
Array scanning
Perform scanning according to manufacturer recommendations, avoiding unnecessary laser exposure. Take care to align channels with respect to signal intensity frequencies, though minor differences between channels usually do not impact smoothed timing profiles after normalization.
Quality control of microarray data
Prior to analysis, the overall quality of a microarray experiment should be examined from several angles. In general, there are six qualities are important for reliable results of replication timing analyses on CGH arrays that should be verified at the corresponding Procedure steps:
Comparable signal intensity distributions for red and green channels (step 74)
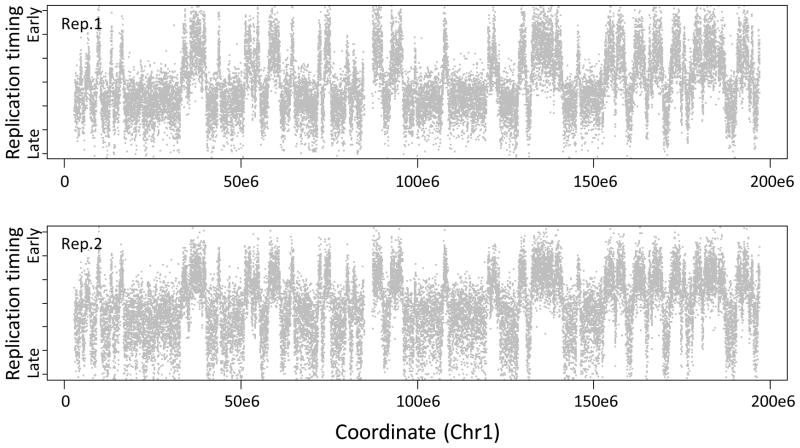
Unbiased signal ratios with respect to signal intensity (step 75)
Comparable timing value distributions between experiments (step 76)
A high overall signal-to-noise ratio of the experiment (step 84)
Lack of artifacts in raw and false-color microarray images (step 85)
High correlations between replicate experiments (step 86b)
Downstream analysis
When comparing the timing program to other genetic and epigenetic properties, differences in formats between ChIP-chip, ChIP-seq, and other approaches will require some care in processing, and even datasets from similar platforms often have idiosyncrasies that must be accounted for. In particular, take care to ensure that replication timing and other data types are compared in compatible genomic builds and equivalent cell types, and use a method of quantification consistent with the methods and goals of the studies involved.
MATERIALS
REAGENTS
CRITICAL: All solutions are made with ddH2O and stored at room temperature (22 °C) unless otherwise indicated.
Cells of interest. Cultures can be grown in any size cell culture dish, but must be in an actively dividing state for use in this protocol. A minimum of 50,000 S phase cells is required for the protocol. However, it is recommended that cultures with at least 120,000 S phase cells be used.
BrdU (5-bromo-2′-deoxyuridine)(Sigma Aldrich, B5002) Make stock solutions of 10mg/mL and 1mg/mL in ddH2O and store at −20°C.
Cell culture medium appropriate for cell type
1X Trypsin-EDTA (Mediatech 25-053-Cl)
Accutase (Innovative Cell Technologies AT104). For long term storage, store at −20°C. Thaw overnight at 4°C before use. Once thawed, store at 4°C for up to two months. Warm to room temperature before each use.
PBS. See Reagent Setup
Fetal Bovine Serum (FBS) (GIBCO 16000). Prepare 1%(vol/vol) in PBS and store at 4 °C.
DAPI (BioChemika 32670) Dissolve stock in ddH2O to final concentration of 10mg/mL. Store at −20°C protected from light.
Propidium Iodide (PI, Sigma P4179-100MG) see Reagent Setup
10mg/mL RNase A (Sigma R6513) Store at −20°C
20mg/mL Proteinase K (Amresco E195) Store at −20°C
20mg/mL Glycogen (Fermentas RO561) Store at −20°C
Isopropanol (Sigma 59304)
100% ethanol (Sigma E7023)
70% (vol/vol) ethanol.
Tris Base (Fisher Scientific BP152-5)
HCl (EMD HX0603P-5)
NaCl (Fisher Scientific BP358-1)
SDS (Invitrogen 15525-017)
EDTA (Invitrogen 15576-028)
SDS-PK buffer. See Reagent Setup
Tris-saturated Phenol (Fisher, BP226-500) (Store at −20°C). ! CAUTION Caustic and harmful if inhaled or ingested. Wear gloves and other appropriate protective equipment. Use adequate ventilation. Store at −20 °C.
Chloroform (Sigma 34854) ! CAUTION Probable carcinogen and harmful if inhaled or ingested. Wear gloves and other appropriate protective equipment. Use adequate ventilation.
Anti-BrdU antibody (BD Biosciences Pharmingen 555627) Store at 4 °C.
Ammonium acetate (Fisher Scientific A639-500) See Reagent Setup.! CAUTION Irritant. Harmful if swallowed. Wear gloves and other appropriate protective equipment. Use adequate ventilation. Store at room temperature.
Rabbit anti-mouse IgG (Sigma M-7023) Store at 4 °C.
Anti-Mouse IgG-AlexaFluor488 (Invitrogen/Molecular Probes, Cat#A-11029) Store at 4 °C.
Taq DNA Polymerase with ThermoPol Buffer (New England BioLabs, M0267)
10 μM dNTPs (Bioline, BIO-39025)
10 mg/ml Ethidium Bromide (Fisher, BP102-5)
Agarose (OmniPur, 2125)
PCR primers for BrdU-IP quality verification (steps 45–49), see Table 1
0.1M HCl/0.5% (vol/vol) Triton X-100(Sigma T9284) in ddH2O. Store at room temperature
0.1M Sodium Tetraborate, (Na2B4O7 10H2O)(Sigma S-9640), pH 8.5 in ddH2O
0.5% (vol/vol) Tween20 (Sigma P-1379)/1% (wt/vol) BSA(Fisher Scientific BP1600-1) in PBS
0.1% (vol/vol) Triton X-100(Sigma T9284) in PBS
0.5% (vol/vol) Triton X-100(Sigma T9284) in PBS
BSA (Fisher Scientific BP1600-1)
GenomePlex Complete Whole Genome Amplification Kit (Sigma WGA2)
GenomePlex WGA Reamplification Kit (Sigma WGA3)
QIAquick PCR Purification Kit (QIAGEN 28106)
NimbleGen Dual-Color DNA Labeling Kit (cat. no. 05223547001)
NimbleGen Hybridization Kit (cat. no. 05583683001)
NimbleGen Wash Buffer Kit (cat. no. 05584507001)
EQUIPMENT
Nylon mesh 37 micron (Small Parts, CMN-0040-D)
5mL round bottom polystyrene tube (Falcon 352054)
15mL round bottom tube (Falcon 2059)
FACSAria cell sorter (or comparable sorter)
Hemacytometer
Vortex Genie
Sonicator
Thermocycler
Electrophoresis apparatus
Appropriate NimbleGen Arrays and Mixers
Appropriate NimbleGen Hybridization System
Appropriate NimbleGen microarray scanner
REAGENT SETUP
1 X PBS To make 1Liter, dissolve 8g NaCl, 0.2g KCl, 1.44g Na2HPO4, 0.24g KH2PO4 in 800mL ddH20. Adjust pH to 7.4 with HCl and adjust final volume to 1 Liter. Sterilize by autoclaving. Store at room temperature.
0.2X Trypsin-EDTA To make 50mL, combine 10mL 1X Trypsin-EDTA with 40mL 1X PBS. Store at 4°C for up to one month. Warm to room temperature before each use.
1 mg/mL Propidium Iodide To make 20mL, dissolve 20 mg propidium iodide powder in autoclaved ddH2O to achieve a final volume of 20mL and filter. Store for up to one year at 4°C protected from light.
DAPI Staining Solution To make approximately 1mL, add 10μl 10% Triton X-100 and 2μl 1mg/mL DAPI to 1mL PBS. Make solution fresh before each use.
12.5μg/mL Anti-BrdU antibody Dilute antibody in 1X PBS from the stock concentration of 0.5mg/mL to a final concentration of 12.5 μg/mL. Prepare 40μl of diluted antibody for each sample and discard unused diluted antibody.
10M Ammonium Acetate To make 100mL, dissolve 77.08g ammonium acetate in 50mL ddH2O. Add ddH2O to adjust final volume to 100mL. Filter and store at room temperature.
1M Tris-HCl, pH 8.0 To make 500mL, dissolve 60.57g Tris base in 400mL ddH2O. Add HCl to adjust pH to 8.0. Add additional ddH2O to adjust the final volume to 500mL. Sterilize by autoclaving. Store at room temperature.
0.5M EDTA To make 1L, dissolve 186.1 grams disodium EDTA in 800mL ddH2O. Stir vigorously on a magnetic stirrer. Adjust the pH to 8.0 by addition of NaOH. Add ddH2O to a final volume of 1L. Sterilize by autoclaving. Store at room temperature.
1 X TE To make 1 L, add 10 mL of 1M Tris-HCl pH 8.0 to 2 mL of 0.5M EDTA pH 8.0 and adjust the final volume to 1L with autoclaved ddH2O. Store at room temperature.
Phenol-Chloroform To make 50mL, combine 25mL of tris-saturated phenol with 25mL of chloroform. Allow separation of layers before use. It is recommended to store the solution overnight before use to allow adequate separation or centrifuge solution at maximum speed for 10 minutes before use to achieve separation. Store at 4°C protected from light.
SDS-PK buffer To make 50mL, combine 34mL autoclaved ddH2O, 2.5mL 1M Tris-HCl pH 8.0, 1mL 0.5 M EDTA, 10mL 5 M NaCl and 2.5mL 10% (wt/vol)SDS. Store at room temperature. Warm to 56°C before use to completely dissolve SDS.
10X IP Buffer To make 50mL, combine 28.5mL ddH2O, 5mL 1M Sodium Phosphate pH 7.0, 14 mL 5M NaCl, and 2.5mL 10% (wt/vol) Triton X-100. Store at room temperature.
1 X IP Buffer To make 50mL, add 5mL of 10X IP Buffer to 45mL autoclaved ddH2O. Store at room temperature.
Digestion Buffer To make 50mL, combine 44mL autoclaved ddH2O, 2.5mL 1M Tris-HCl pH8.0, 1mL 0.5M EDTA, 2.5mL 10% (wt/vol)SDS. Store at room temperature.
EQUIPMENT SETUP
Sonicator: Adjust sonicator settings as needed to achieve a 250bp-2kb distribution of DNA fragment sizes. We use a waterbath-type sonicator (Heat Systems-Ultrasonics W-380) with a 2 second, 50% duty cycle and an output setting of 7 for 4 min.
PROCEDURE
BrdU labeling and staining of cells for FACS
-
1
To perform PI staining and ethanol fixation prior to sorting, follow option A; this method is the most commonly used as it allows for shipping or long-term storage, and has worked well for most mouse cell lines5,6. For cells that break or clump in ethanol, follow option B; note that a drawback of option B is that cells need to be sorted immediately following BrdU labeling. Alternatively, carry out the procedure for S/G1 sorting described in Box 1 instead of steps 1–57. This method obviates the need for BrdU-IP and whole-genome amplification (WGA), and can alleviate concerns that sorting early and late fractions of S-phase or WGA introduce a temporal bias; however, in our hands E/L fractionation has produced results equivalent to the S/G1 method, as well as sorting of additional S-phase fractions3,37.
BOX 1. Method for sorting according to S/G1phase - TIMING 1 d.
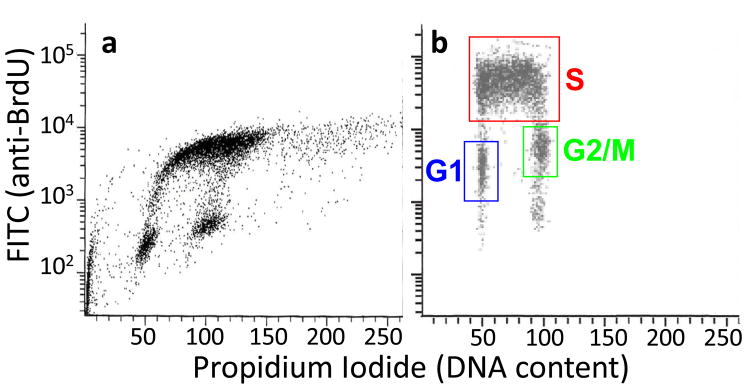
In this method, cells are sorted into two fractions, G1 phase and S phase, based on DNA content, and replication timing is derived from the 2-fold copy number increase for early vs. late replicating sequences in pure S-phase populations. DNA analysis using flow cytometry can be performed simply by the use of a single DNA-binding fluorescent dye such as PI or DAPI as originally described40. Although this method is adequate, simultaneous measurement of BrdU-labeled DNA by performing BrdU/PI double staining for cell cycle analysis, can discriminate G1 and early-S cells much more efficiently than by PI-only staining. In addition, some cell types, particularly those derived from differentiated stem cells or primary tissues, can produce debris that interferes with good S phase sorting and a short BrdU label described here can eliminate debris, which is not labeled with BrdU. The advantage of this method is that it eliminates the need for BrdU-IP and whole genome amplification steps (described in steps 13–44 and 51–57), which need to be carefully controlled. However, direct comparisons have shown that this method produces a lower signal to noise ratio than the method described in the main Procedure6.
A much shorter BrdU pulse label is used in this protocol at lower concentration, because we are only trying to identify the cells in S phase. With longer BrdU labeling times, G2/M cells become labeled. It should be noted that we originally used the standard protocol for BrdU/PI analysis provided by Becton-Dickenson, which is fine for analysis. However, we found that the high concentration of HCl in this method sheared genomic DNA to very small fragments that precluded subsequent steps of the protocol. By titrating the HCl, we found that 0.1M HCl produced the optimal compromise between good S vs. G1 separation and minimal DNA shearing.
For BrdU/PI double staining, correction of spectral overlap is critical for successful experiments. Spectral overlap exists between emission spectra of PI and FITC/AlexaFluor488 (for BrdU). Without correction, the BrdU/PI plots typically look like Fig. 2A. For this correction, the adjustment of the ratio between PI and AlexaFluor488 (or FITC) gains can significantly reduce the skewing shown in Fig. 2A. Subtraction of the FITC signal from the PI signal (=compensation) may also be required. To perform these corrections, a “BrdU-only” control is required, prepared by staining BrdU-labeled cells without the addition of PI. A “PI-only” control also helps, prepared by staining non BrdU-labeled cells for BrdU and PI. [Note: BrdU-labeled specimen stained for PI only does not reflect background signals derived from the anti-BrdU antibody and thus is not as good as unlabeled cells.] This step can be time-consuming but is critical for successful sorting. We suggest that you first adjust the gains of FSC and SSC, then adjust the PI and AlexaFluor488 gains by trial and error to get the best possible BrdU/PI plot. You may be able to obtain a reasonable BrdU/PI plot without compensation, but if not, compensate by subtracting FITC signal from PI signal. The lower the percentage subtracted, the better.
S/G1 FACS Sorting TIMING - 1 d
For adherent cells, remove cell culture medium from exponentially growing cells and replace with cell culture medium containing BrdU at a final concentration of 10 μM. For suspension cells, add BrdU to the cell culture medium at a final concentration of 10μM. In order to obviate the amplification step prior to labeling and array hybridization, start with 6 million cells. One should also prepare a small sample of non BrdU-labeled, EtOH-fixed cells for PI-only control and set aside a small number of BrdU-labeled cells for BrdU-only control.
Incubate cells for 15 minutes in a carbon dioxide incubator at 37°C, 5% CO2
-
Fix as in steps 1A, iii-x of the main Procedure.
PAUSE POINT Cells can be stored as in step 1A.
Aliquot (multiples of) 3 × 106 cells in a 1.5mL tube(s), centrifuge for 5 min., 200 × g at room temperature. Removal of supernatant is much easier with 1.5 mL tubes as the pellets are very loose.
Aspirate supernatant completely with P200. Here and elsewhere, an additional pulse spin (~3 sec) will help with discarding residual supernatant.
Loosen pellet by brief vortexing.
Add 1ml of 0.1M HCl/0.5% Triton X-100, resuspend by tapping.
Incubate 15 min at room temperature in the dark.
Centrifuge 5 min, 200 × g at room temperature, then aspirate supernatant completely.
Add 1ml of 0.1M Sodium Tetraborate and resuspend by tapping.
Centrifuge 5 min, 200 × g at room temperature, then aspirate supernatant completely.
Add 0.15 μg anti-BrdU antibody in 0.5 ml 0.5% Tween20/1% BSA/PBS and resuspend by tapping.
Incubate for 30 min at room temperature in the dark.
Centrifuge 5 min, 200 × g at room temperature, then aspirate supernatant completely.
Add 0.5 ml of 0.5% Tween20/1% BSA/PBS.
Centrifuge 5 min, 200 × g at room temperature, then aspirate supernatant completely.
Add 1 μg anti-Mouse IgG-AlexaFluor488 in 100 μl 0.5% Tween20/1% BSA/PBS and resuspend by tapping (or when 1–2 × 106 cells are used, add 0.5 μg anti-Mouse IgG-AlexaFluor488 in 50 μl).
Incubate for 30 min at room temperature in the dark.
Centrifuge 5 min, 200 × g at room temperature, then aspirate supernatant completely.
Add 0.5 ml of 0.5% Tween20/1% BSA/PBS.
Centrifuge 5 min, 200 × g at room temperature, then aspirate supernatant completely.
Resuspend the pellet in 1 ml of 5μg/ml PI in PBS (For “BrdU-only” control, just add PBS).
Transfer to a round bottom 5mL tube (i.e. Falcon 2054).
Adjust concentration to 2 × 106/ml by adding 5 μg/ml PI in PBS. For BrdU-only sample, adjust to the same concentration by adding PBS without PI.
Filter with 37 μm mesh filter.
Bring to Flow lab for sorting (on ice, dark). Resume the main Procedure at step 58.
(A) Labeling and PI staining of cells for FACS following ethanol fixation - TIMING 3.5 h
Add BrdU to cells in culture medium at a final concentration of 50μM.
Incubate cells for two hours in a carbon dioxide incubator at 37°C, 5% CO2.
For adherent cells, rinse gently with ice-cold PBS twice. For suspension cells, collect cells in a 15mL tube and proceed directly to step 1a(vi).
-
Detach adherent cells using 0.2X Trypsin-EDTA for 2–3 minutes or Accutase for 3–6 minutes.
CRITICAL STEP Incubate cells at 37 °C with the enzyme treatment and/or use gentle trituration if necessary to achieve a single cell suspension, as this is essential for accurate FACS sorting.
Add 5mL of cell culture medium (containing FBS if trypsin has been used) to the cell culture dish or flask, pipette gently, and transfer contents to a 15mL round bottom tube.
Count the number of cells collected using a hemacytometer. Collect enough cells to obtain at least 20,000–30,000 (preferably >150,000) cells in each fraction after sorting (step 2); this will generally require 0.5–1×106 cells, with more required if few cells are in S-phase. For first-time users, we recommend starting with 4×106 – 8×106 cells.
Centrifuge at approximately 200 × g for 5 minutes at room temperature.
Aspirate supernatant carefully and resuspend cells in 2.5 mL of ice-cold PBS containing 1% FBS.
-
Add 7.5 mL of ice-cold 100% ethanol dropwise while gently vortexing.
CRITICAL STEP Note that vortexing should be performed gently to avoid cell damage.
-
Seal the cap of the 15 mL tube with parafilm and mix gently but thoroughly.
PAUSE POINT If necessary, cells can be stored in the dark at −20°C indefinitely.
Resuspend the BrdU-labeled, ethanol fixed cells by tapping and inverting the tube.
Transfer 4 × 106 – 8 × 106 cells to a 5 mL polystyrene round bottom tube.
Centrifuge at approximately 200 × g for 5 minutes at room temperature.
Decant supernatant carefully
Resuspend the cell pellet in 2 mL of PBS with 1% FBS. Mix well by tapping the tube.
Centrifuge at approximately 200 × g for 5 minutes at room temperature.
Decant supernatant carefully.
Resuspend cell pellet in PBS with 1% FBS to achieve a solution of 3 × 106 cells/mL.
Add 1mg/mL propidium iodide to a final concentration of 50 μg mL.
Add 10 mg/mL RNase A to a final concentration of 250 μg/mL.
Tap the tube to mix and incubate for 20 to 30 minutes at room temperature (22°C) in the dark.
Filter cells by pipeting them through 37 micron nylon mesh into a 5 mL polystyrene round bottom tube.
Keep samples on ice in the dark and proceed directly to FACS sorting.
(B) BrdU labeling and DAPI staining of cells for FACS - TIMING 3 h
Follow steps 1A i-vii.
Aspirate the supernatant carefully.
Add 5mL of ice-cold PBS and pipette gently but thoroughly.
Centrifuge at approximately 200 × g for 5 minutes at room temperature.
Aspirate supernatant carefully.
Resuspend the cell pellet in DAPI staining solution to achieve a solution of 5 × 106 – 10 × 106 cells/mL.
Filter cells by pipeting them through 37 micron nylon mesh into a 5 mL polystyrene round bottom tube.
Keep samples on ice in the dark and proceed directly to FACS sorting.
-
2
Run sample on FACSAria cell sorter (Any comparable cell sorter can be used)
CRITICAL STEP It is very important to keep live samples chilled on ice or at 4°C during FACS analysis to avoid cell cycle progression in the absence of BrdU. Protect samples from light.
-
3
Use forward and side scatter information to select the desired population of cells to be included in the sort, and exclude doublets or cell debris.
-
4
Create a histogram that plots cell count on the y-axis and DNA content (fluorochrome intensity) on the x-axis. See Fig. 1
-
5
Select two distinct S-phase populations to be sorted into separate fractions as indicated in Fig. 1.
-
6
Sort cells into fresh 5ml round bottom tubes and keep at 4°C during the sort.
PAUSE POINT Store cells immediately on ice in the dark until all samples have been sorted.
? TROUBLESHOOTING
-
7
Centrifuge at 400 × g for 10 minutes at 4°C. Alternatively, if fewer than 150,000 cells have been collected for each fraction, proceed directly to step 9.
-
8
Decant supernatant gently, only once.
CRITICAL STEP Some residual sheath fluid can be left in the tube to prevent losing the cell pellet, which can easily detach from the tube during this step.
-
9
Add 1 mL of SDS-PK buffer containing 0.2mg/mL Proteinase K and 0.05mg/mL glycogen for every 100,000 cells collected and mix vigorously by tapping the tube.
-
10
Incubate samples in a 56°C water bath for 2 hours.
-
11
Mix sample thoroughly and aliquot 200μl, equivalent to approximately 20,000 cells, per 1.5 mL tube. PAUSE POINT Samples can be stored for at least 6 months at −20°C in the dark before use.
-
12
Add 200μl SDS-PK buffer with 0.05mg/mL glycogen to each aliquot and proceed directly to BrdU immunoprecipitation.
Figure 1.

A typical cell cycle profile for a mammalian fibroblast population obtained during FACS analysis by plotting cell count vs. DNA content. In this example, cellular DNA was stained with PI, so DNA content is represented by PI intensity. A G1 peak, representing cells with 2N DNA content, and a G2/M peak, representing cells that have undergone replication and therefore possess a 4N DNA content are labeled. The area between these two peaks is representative of cells in S phase, and can be sorted into two fractions, as indicated here, to obtain early and late S phase samples.
BrdU immunoprecipitation - TIMING 2 d
-
13
Extract once with phenol-chloroform, collecting the upper phase in a 1.5mL tube.
-
14
Extract once with chloroform, collecting the upper phase in a 1.5mL tube.
-
15
Add 1 volume of isopropanol and mix well.
-
16
Store at −20°C for 20 minutes. PAUSE POINT Samples can be stored in the dark at −20°C overnight.
-
17
Centrifuge at 16000 × g for 30 minutes at 4°C.
-
18
Discard the supernatant and add 750μl 70% ethanol to the pellet
-
19
Centrifuge at 16,000 × g for 5 minutes at 4°C, then remove all ethanol and let the pellet dry.
-
20
Resuspend the pellet in 500μl 1x TE.
PAUSE POINT If necessary, the pellet can be stored overnight at 4°C.
-
21
Sonicate DNA to an average size of ~0.7–0.8 kb. Settings required for a 250 bp to 2 kb range should be determined empirically for each sonicator type. See Equipment Setup.
-
22
Incubate sample at 95°C for 5 minutes to heat denature the DNA.
-
23
Cool sample on ice for 2 minutes.
-
24
Add 60μl of 10X IP Buffer to a clean 1.5mL tube.
-
25
Add the denatured DNA from step 22 to the tube from step 24.
-
26
Add 40μl of 12.5 μg/ml anti-BrdU antibody.
-
27
Incubate for 20 minutes at room temperature with constant rocking.
CRITICAL STEP Cover tubes with foil to keep samples in the dark.
-
28
Add 20μg of rabbit anti-mouse IgG.
CRITICAL STEP Cover tubes with foil to keep samples in the dark.
-
29
Incubate for 20 minutes at room temperature with constant rocking.
-
30
Centrifuge at 16,000 × g for 5 minutes at 4°C
-
31
Remove supernatant completely.
CRITICAL STEP If pellet becomes loose, then briefly centrifuge sample again in order to completely remove the supernatant without disturbing the pellet. Several centrifugations may be necessary to completely remove supernatant.
-
32
Add 750μl of 1X IP Buffer that has been chilled on ice.
-
33
Centrifuge at 16,000 × g for 5 minutes at 4°C.
-
34
Remove supernatant completely, as in step 31.
-
35
Resuspend the pellet in 200μl digestion buffer with freshly added 0.25mg/mL proteinase K. Incubate samples overnight at 37°C.
-
36
Add 100μl of fresh digestion buffer with freshly added 0.25mg/mL proteinase K.
-
37
Incubate samples for 60 minutes at 56°C.
-
38
Extract once with phenol-chloroform, collecting the upper phase in a 1.5mL tube.
-
39
Extract once with chloroform, collecting the upper phase in a 1.5mL tube.
-
40
Add 0.625μl of 20mg/mL glycogen, 100μl of 10 M ammonium acetate, and 750μl of 100% ethanol and mix well.
-
41
Store at −20°C for 20 minutes. PAUSE POINT Samples can be stored in the dark at −20°C indefinitely.
-
42
Centrifuge at 16,000 × g for 30 minutes at 4°C.
-
43
Remove supernatant, rinse pellet with 70% ethanol and dry.
-
44
Resuspend the pellet in 80μl 1X TE (for a final concentration of 250 cell eq./μL).
PAUSE POINT Store DNA at 4°C for up to one month, or at −20°C for longer storage.
PCR method for quality control of BrdU-immunoprecipitation - TIMING 5 h
-
45
Prepare enough PCR master-mix to screen all IP samples with each primer set listed in Table 1. An example PCR mix is listed in the table below. Mitochondrial primer sets should be used at 1.0 μM concentration instead of 0.5 μM; add 0.63 μL F/R 20μM combined primers and adjust nuclease-free water accordingly.
Component Amount per reaction (μL) Final 10X Taq buffer 1.25 1X 10mM dNTPs 0.25 0.2 mM 20 U/μL Taq Polymerase 0.06 1.2 U F/R 20 μM combined primers 0.31 0.5 μM Nuclease free water to 12.5 -
46
Aliquot 11.5 μL master-mix per tube and add 1 μL IP sample.
-
47
Run samples in thermocycler with the following conditions:
Cycle number Denature Anneal Extend 1 94°C, 2 min 2–39 94°C, 45 s 60°C, 45 s 72°C, 2 min 40 72°C, 5 min -
48
Add 2.5 μL 6x loading dye to every 12.5 μL reaction and load 6 μL onto 1.5% agarose gel. Run the gel at 125V for 16 min.
-
49
Score each IP based on anticipated enrichment of amplicon DNA (see Experimental Design). Multiple samples from the same cell type should amplify consistently, with enrichment consistent with genes of known replication timing for the given cell type.
CRITICAL STEP Before proceeding, verify sample quality with corresponding primer sets listed in Table 1.
? TROUBLESHOOTING
-
50
If several IPs of the same sample and S-phase fraction pass the screening, pool equal amounts of each IP to a final volume of 50 μL (e.g. If two IP pass, combine 25 μL of each to pool).
Whole genome amplification - TIMING 8 h
-
51
Precipitate DNA fractions by adding 1.25 μL 2 mg/mL glycogen, 20 μL 10M ammonium acetate and 150 μL ethanol to each 50 μL IP sample (If pooling multiple samples, 50 μL total volume should still be used). Mix well, let stand at −20°C for 20 min, then centrifuge for 30 min at max speed at 4°C.
-
52
Rinse pellets with 70% ethanol, air dry and resuspend in 10 μL nuclease free water.
-
53
Transfer the 10μL samples to 0.2 mL PCR tubes and carry out whole genome amplification using an appropriate method or kit. In our hands, the GenomePlex Complete Whole Genome Amplification Kit has worked well, starting from the Library Preparation step (i.e., skipping Fragmentation)39.
-
54
Purify entire WGA products using an appropriate PCR purification kit, such as QIAquick. Elute in 30 μL nuclease free water pre-warmed to 65°C and determine concentration using Nanodrop.
-
55
Dilute WGA samples to appropriate concentration (we use 1 μL DNA of 20 ng/μL) and, if necessary to obtain sufficient material for the chosen array platform, perform a second round of whole genome amplification. We follow the GenomePlex WGA Reamplification Kit Reamplification Procedure A.
-
56
Purify entire reamplified WGA products as in step 54.
-
57
Screen purified final products using the PCR method described in steps 46–49.
PAUSE POINT Samples can be stored in the dark at −20C for up to one month.
? TROUBLESHOOTING
Labeling and hybridizing - TIMING 1–3 d
-
58
Differentially label reamplified early and late WGA DNA fractions with Cy3 and Cy5 dyes according to the method most appropriate for the chosen array platform. We follow the Sample Labeling Instructions for NimbleGen Dual-Color DNA Labeling Kit.
-
59
Hybridize samples to array(s) using the corresponding method or kit. We use the NimbleGen Hybridization Kit.
-
60
Following Hybridization, wash array(s) as needed. We perform this step as directed using NimbleGen Wash Buffer Kit.
-
61
Scan array(s) using appropriate microarray scanner and software package. We use NimbleGen scanner GenePix 4000B and the accompanying NimbleGen Arrays User’s Guide, CGH Analysis v5.1. Newer equipment is accompanied with a newer version of the User’s Guide and operated slightly differently. For NimbleGen arrays, raw images should be saved as .tif files, and two .pair files (one each for Cy3 and Cy5 channels) will be created per experiment.
Normalization of raw datasets TIMING - 1 d
-
62
If necessary, install R from http://www.r-project.org/. Create RGL (Red Green List) files from the original NimbleGen .pair files as described in steps 63–68. These files contain columns for both red (Cy5) and green (Cy3) channel signal intensities; example pair files used in step 65 and throughout are available in Supplementary Data 1.
-
63
Set the working directory using the command ‘setwd’ in the R console to specify the appropriate filepath. Here and in later steps, the “>” symbol denotes the R prompt at the beginning of a line and should be omitted when typing the command.
> setwd(“D:\\RT project\\Raw datasets”)
-
64
Read the first five rows of data from the raw data files, and determine the data type of each column using the sapply() function:
> tab5rows <- read.delim(“318990_4L1210LymphoblastP1_532.pair”, header = T, nrows = 5, skip=1)
> classes <- sapply(tab5rows, class)
CRITICAL STEP: When reading large tables in R, such as .pair files, explicitly noting the number of rows and data type of each column as illustrated here and in step 65 will save a substantial amount of memory and calculation time. Occasionally, the sapply() function will set the genomic position columns of large datasets as an integer type, which lacks the memory space to store large numbers. If so, set the type manually with > classes[x] = “numeric” (where x is the column number containing position information) after creating the classes variable.
-
65
Read the raw datasets into memory. Note that variable names and filenames may be substituted here and elsewhere, as appropriate. The “nrows” parameter can be a modest overestimate; the correct number of rows will be present in the final table, but an estimate allows the system to allocate the correct amount of memory.
> mLymph1Cy3 <- read.delim(“L1210LymphoblastR1_532.pair”, header=T, nrows=390000, comment.char = ““, colClasses=classes, skip=1)
> mLymph1Cy5 <- read.delim(“L1210LymphoblastR1_635.pair”, header=T, nrows=390000, comment.char = ““, colClasses=classes, skip=1)
> mLymph2Cy3 <- read.delim(“L1210LymphoblastR2_532.pair”, header=T, nrows=390000, comment.char = ““, colClasses=classes, skip=1)
> mLymph2Cy5 <- read.delim(“L1210LymphoblastR2_635.pair”, header=T, nrows=390000, comment.char = ““, colClasses=classes, skip=1)
-
66
Extract the Cy3 and Cy5 channel signal intensities from the raw datasets, e.g.:
> mLymph1 <- data.frame(S_Cy5=mLymph1Cy5[,10],S_Cy3=mLymph1Cy3[,10])
> mLymph2 <- data.frame(S_Cy5=mLymph2Cy5[,10],S_Cy3=mLymph2Cy3[,10])
-
67
Write the columns extracted in step 66 to separate RGL files for normalization:
> write.table(mLymph1, file=“318990_4L1210LymphoblastP1.rgl.txt”, row.names=F, quote=F, sep=“\t”, eol=“\r\n”)
> write.table(mLymph2, file=“319048_4L1210LymphoblastP1-2.rgl.txt”, row.names=F, quote=F, sep=“\t”, eol=“\r\n”)
-
68
Create a “targets” text file that describes the target files for normalization. We will name this file “T.txt” (See Supplementary Data 2 for an example targets file). Note that, to be read correctly, the file should be tab-delimited and contain only one carriage return at the end of the final line. Place this file in the same directory as the raw .pair files and .rgl files generated above.
-
69
Install a current version of the LIMMA package according to the instructions at http://bioinf.wehi.edu.au/limma/, or using the command line interface:
> source(“http://www.bioconductor.org/biocLite.R”)
> biocLite(“limma”)
> biocLite(“statmod”)
-
70
Perform loess and scale normalization using LIMMA as described in steps 71–73, and verify the results as described in steps 74–85. This process is more straightforward than many two-color normalization methods, since NimbleGen arrays do not have print tip groups, spot background areas, or mismatch spots that must be accounted for. Loess normalization (normalizeWithinArrays) corrects the internal dependence of red-green ratios on their intensity independently for each array, and is examined further in step 74 and step 75. Scale normalization (normalizeBetweenArrays) equalizes the distribution of timing values between multiple samples for comparisons, and can be verified in step 76.
-
71
Load the limma package, and read the raw datasets listed in the file created in step 68. This will generate a MAlist-type data object, r, that stores the raw samples before normalization:
> library(limma)
> t = readTargets(“T.txt”, row.names=“Name”)
> r = read.maimages(t, source=“generic”,columns=list(R=“S_Cy5”, G=“S_Cy3”))
-
72
Perform loess normalization. This will generate a second MAlist-type data object, MA.l, that stores the samples after within-array normalization.
> MA.l = normalizeWithinArrays(r, method=“loess”)
-
73
Perform scale normalization. This will generate a third MAList object, MA.q, that stores the samples after between-array normalization. As with ChIP-chip methods13,14, this type of scale normalization may not be appropriate for examining subsets of the genome where large unbalanced timing changes are expected (e.g., timing of the X chromosome before and after inactivation), but is ideal for whole-genome analyses.
> MA.q = normalizeBetweenArrays(MA.l, method=“scale”)
-
74
Check the distribution of spot intensities for red and green channels after each stage of normalization (Fig. 3). These distributions should be fairly well-aligned, and have tails with high signal values. Experiments in which signal intensity drops off more sharply will often show higher levels of noise in the final dataset. (Here and in subsequent steps, text following the “#” symbol represents non-executed comments.)
> plotDensities(r)
# Raw data
> plotDensities(MA.l)
# After within-array normalization
> plotDensities(MA.q)
# After between-array normalization
-
75
Check the dependence of timing ratios on signal intensities by creating MA plots (Fig. 4). Points will often be skewed to low Cy5/Cy3 ratios at low intensities due to photobleaching of Cy5, but should be corrected after within-array loess normalization in LIMMA. This bias is the most common artifact for NimbleGen arrays, but other types can also be diagnosed with MA plots43.
> plotMA(r, array=1)
# Raw data, replicate 1
> plotMA(MA.l, array=1)
# After within-array normalization
-
76
Verify that the distribution replication timing values is equivalent across experiments after normalization by creating boxplots of Cy5/Cy3 ratios for each experiment (Fig. 5). These distributions may be slightly different before normalization (and after within-array normalization), but 1st and 3rd quartiles (the box boundaries) of all experiments should be equal after between-array normalization.
> boxplot(MA.l$M~col(MA.l$M), names=colnames(MA.l$M))
> boxplot(MA.q$M~col(MA.q$M), names=colnames(MA.q$M))
-
77
Create an intermediate file containing the normalized datasets by typing, e.g,:
> write.table(MA.q$M, file=“Loess_mLymph_112909.txt”, quote=F, row.names=F, sep=“\t”)
This tab-delimited text file will be further processed in steps 79–85 to sort and average the normalized datasets and check other quality control measures.
-
78
Remove the other objects from memory:
> rm(r, MA.l, MA.q, mLymph1Cy3, mLymph1Cy5, mLymph2Cy3, mLymph2Cy5, mLymph1, mLymph2); gc(reset=T)
Or, to remove all objects:
> rm(list=ls())
-
79
Assign position and chromosome information to the normalized datasets. This can be accomplished using the original .pair files, which typically contain this information in columns “POSITION” and “SEQ_ID” respectively (option A). Some data formats, such as HD2 triplex arrays, contain a different format of SEQ_ID column with chromosome and chromosome endpoints combined (e.g., “chr11:1-134452384”), or no SEQ_ID column. In these cases, extract chromosome labels from the PROBE_ID column (option B):
Figure 3.
Distribution of signal intensities before and after normalization. Panels depict the distribution of Cy5 (red) and Cy3 (green) signal values before normalization (a), after within-array normalization (b), and after between-array normalization (c) in LIMMA. Since replication timing is a relative property, equivalent amounts of DNA are transcribed before and after the middle of S-phase, allowing distributions to be transformed to a common scale for each channel (b) and array (c).
Figure 4.
Dependence of timing ratios on signal intensity. MA plots from LIMMA illustrate the relationship between red/green ratios (y-axis) and signal intensity (x-axis) before (a) and after (b) within-array normalization. The skew of low-intensity data points toward Cy3 (here, late) values is a common characteristic of two-color arrays, and is corrected after normalization.
Figure 5.
Verification of scale normalization between datasets. Exemplary boxplots of timing values before (a) and after (b) normalization between arrays, for a 9-day differentiation protocol with 3-day intermediates5. Modest differences in the distribution of timing values (with box boundaries representing the 1st and 3rd quartiles) are equalized after scaling.
A.) Copy position and chromosome columns from original .pair files
-
Read the intermediate file created in step 77:
> tab5rows = read.table(“Loess_mLymph_112909.txt”, header = T, nrows = 5)
> classes = sapply(tab5rows, class)
> RT = read.table(“Loess_mLymph_112909.txt”, header=T, nrows=389306, comment.char = ““, colClasses=classes)
-
Next, read the original .pair file containing POSITION and SEQ_ID columns:
> tab5rows = read.delim(“L1210LymphoblastP1_635.pair”, header = T, nrows = 5, skip=1)
> classes = sapply(tab5rows, class)
> a = read.delim(“L1210LymphoblastP1_635.pair”, header=T, nrows=389306, comment.char = ““, colClasses=classes)
-
Finally, remove unmapped probes from the files loaded in steps 79a(i) and 79a(ii) and assign position and chromosome information to the normalized datasets:
> RT = subset(RT, a$POSITION != 0)
> a = subset(a, a$POSITION != 0)
> RT$CHR = a$SEQ_ID; RT$POSITION = a$POSITION
B.) Parse position and chromosome information from PROBE_ID column
Load the normalized and .pair files as outlined in steps 79a(i) and 79a(ii).
-
Split the PROBE_ID column into the elements preceding and following “FS”; for example, “CHR12FS006244334” will become “CHR12” and “006244334”.
> x = strsplit(as.character(a$PROBE_ID), “FS”)
> y = unlist(x)
# chr [1:770156] “CHR01” “003001832” …
-
Separate the odd- and even-numbered indices of this object into separate columns, and convert the position strings to numeric values.
> y1 = y[c(TRUE, FALSE)]
# chr [1:385078] “CHR01” “CHR01” …
> y2 = y[c(FALSE,TRUE)]
# chr [1:385078] “003001832” “003018759”…
> y2 = as.numeric(y2)
# num [1:385078] 3001832 3018759 …
-
Finally, assign the position and chromosome information to the normalized dataset:
> RT = data.frame(CHR=y1, POSITION=y2, RT, stringsAsFactors=F)
-
80
Sort datasets by chromosome and position. This will ensure that the plotting and autocorrelation checks in steps 81 and 84 are accurate, and is required for most downstream analysis. By the default sorting method, the order of mouse chromosomes will be 1, 10–19, 2–9, X, then Y. This order itself is unimportant, but should be consistent across experiments to prevent errors in downstream analysis.
> RT = RT[order(RT$CHR, RT$POSITION),]
-
81
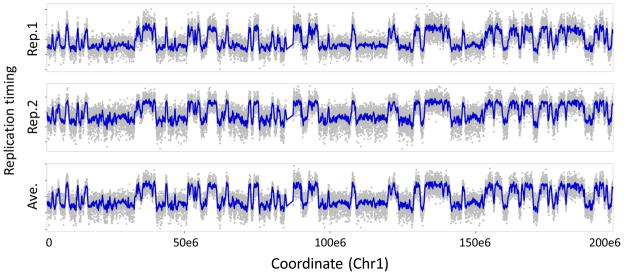
Plot timing values across a chromosome (Fig. 6). This serves to verify the orientation for early/late domains, as well as the overall technical quality of the experiments. Check the dataset structure using “str(RT)” for the correct column numbers to plot, and adjust the y-axis span (“ylim”) as needed.
? TROUBLESHOOTING
> RTb = subset(RT, RT$CHR == “chr1”)
# Create a subset of timing values in chromosome 1
> par(mar=c(3.1,4.1,1,1),mfrow=c(2,1))
# Set plot margins; include two rows in layout
> plot(RTb[,1]~RTb$POSITION,pch=19,cex=0.2,col=“grey”,ylim=c(−3,3))
# Plot replicate 1
> plot(RTb[,2]~RTb$POSITION,pch=19,cex=0.2,col=“grey”,ylim=c(−3,3))
# Plot replicate 2
-
82
Using known regions of early or late replication, verify that the timing values are properly oriented. If not, reverse them by multiplying the appropriate data columns by −1:
> RT[,1] = RT[,1] * -1
-
83
Rename datasets and average replicates as desired, then write a finalized file containing normalized data to the current working directory (see step 63); for example,
> names(RT)[1:2] = c(“mLymphR1”, “mLymphR2”)
> RT$mLymphAve = (RT[,1] + RT[,2])/2
> write.table(RT, “ LoessScale+CHRPOS_mLymph_112909.txt”, row.names=F, quote=F, sep=“\t”)
-
84
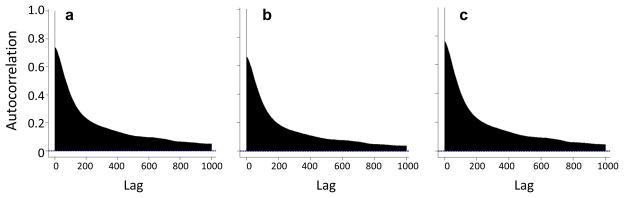
For each dataset, determine the autocorrelation function (ACF), which describes the correlation between neighboring data points as a function of their genomic distance (Fig. 7). Since nearby loci should replicate almost synchronously, the ACF is a useful measure of overall data quality. High-quality datasets will have a correlation between nearest neighbor timing values of R = 0.60–0.80. This measure of signal-to-noise ratio will improve as more replicates with equivalent states are averaged.
> acf(RT[,1],lag=1000)$acf[2]
# Replicate 1: R = 0.742
> acf(RT[,2],lag=1000)$acf[2]
# Replicate 2: R = 0.665
> acf(RT$mLymphAve, lag=1000)$acf[2]
# Averaged 1 and 2: R = 0.762
? TROUBLESHOOTING
-
85
To check for spatial artifacts, examine the original .tif images (Fig. 8) for common characteristics of regional bias, such as streaks, blank regions, or overabundance of either channel in any region of the array44 Note that the “rtiff” package may first need to be installed as in step 72. Since most probes on tiling microarray designs are randomly distributed with respect to genomic location, spatial artifacts in the scanned images should not much affect timing values in any particular location in the genome, but may reduce the overall signal to noise ratio of the experiment if they cover a substantial portion of the array.
> library(rtiff)
> Cy5 = readTiff(“318990_3MEFfemale_532.tif”)
> plot(Cy5)
Figure 6.
Replication timing values across chromosome 1. For each replicate, individual log2(Cy5/Cy3) probe intensities are plotted in grey (y-axis) against their position on chromosome 1 (x-axis). Due to photobleaching of Cy5 diagnosed in step 75, timing is skewed toward early values in replicate 1 (top) and late values in replicate 2 (bottom), demonstrating the practical advantages of dye-swap replicates.
Figure 7.
Autocorrelation functions for two replication timing experiments and their average. Correlation values (y-axis) decline as a function of genomic distance between data points (with ‘lag’ on the x-axis representing the separation between probes), but should start above 0.6 for high-quality datasets and improve upon averaging replicates.
Figure 8.
A typical NimbleGen microarray image after a successful experiment. The lighter points in a grid pattern are control features that aid with spot alignment.
Static properties of the timing program in a given cell type TIMING - 3 h
-
86
After normalization, choose among several common options to analyze the characteristics of timing datasets. Although optional, each method is complementary and useful for a wide range of downstream analysis. To derive an overall timing profile from noisier raw data points, apply a loess smoothing function (option A). Use a correlation metric, generally after loess smoothing, to determine the overall levels of similarity among two or more datasets (option B). Perform segmentation (option C) to define the boundaries of replication domains and determine their average timing.
A.) Loess smoothing
-
Apply loess smoothing to each chromosome as outlined below (Fig. 9). For human and mouse datasets, we perform smoothing with a bandwidth of 300 kilobases; other systems may have different optimal smoothing spans that should be determined empirically using the smallest span that reproduces most of the features between replicate profiles.
> chrs = levels(RT$CHR); str(chrs)
# Create a list of all chromosomes
> AllLoess = NULL
# Initialize a variable to store all loess-smoothed data
> for (chr in chrs) {# For each chromosome,
> RTl = NULL
# Create a variable to store loess-smoothed values
> RTb = subset(RT, RT$CHR == chr)
# Subset the dataset to a single chromosome
> lspan = 300000/(max(RTb$POSITION)-min(RTb$POSITION))
# Set smoothing span
> cat(“Current chromosome: “, chr, “\n”)
# Output current chromosome to console
> RTla = loess(RTb$ mLymphR1~ RTb$POSITION, span = lspan)
# Smooth dataset 1
> RTlb = loess(RTb$mLymphR2~ RTb$POSITION, span = lspan)
# Smooth dataset 2
> RTlc = loess(RTb$mLymphAve ~ RTb$POSITION, span = lspan)
# Smooth dataset 3
> RTl = data.frame(CHR=RTb$CHR, POSITION=RTb$POSITION, RTla$fitted, RTlb$fitted,
# Combine the datasets for the current chromosome
> AllLoess = rbind(AllLoess, RTl)
# Combine current chromosome with overall dataset
> }
# End for loop
> x = as.data.frame(AllLoess)
# Reformat the smoothed datasets as a data frame
-
Rename the loess-smoothed datasets as desired, and save these to a tab-delimited text file. Note that column names within a data frame cannot begin with a number.
> names(x)[3:5] = c(“x300smo_mLymphR1”, “x300smo_mLymphR2”, “x300smo_mLymphAve)
> write.table(x, “300kb_LoessSmo_mLymph_112909.txt”, row.names=F, quote=F, sep=“\t”)
-
Plot the results of loess smoothing as follows (Fig. 9). The “mfrow” parameter may be adjusted for different numbers of datasets.
> RTc = subset(RT, CHR == “chr1”)
# Subset of raw timing data in chr1
> LSc = subset(LS, CHR == “chr1”)
# Subset of smoothed data in chr1
> par(mar=c(2.2,5.1,1,1), mfrow=c(3,1), col=“grey”, pch=19, cex=0.5, cex.lab=1.8, xaxs=“i”)
> plot(RTc$mLymphR1~RTc$POSITION, ylab=“mLymph R1”, xaxt=“n”)
# Plot raw data points
> lines(LSc$x300smo_mLymphR1~LSc$POSITION, col=“blue3”, lwd=3)
# Overlay loess line
> plot(RTc$mLymphR2~RTc$POSITION, ylab=“mLymph R2”, xaxt=“n”)
> lines(LSc$x300smo_ mLymphR2~LSc$POSITION, col=“blue3”, lwd=3)
> plot(RTc$mLymphAve~RTc$POSITION, xlab=“Coordinate (bp)”, ylab=“mLymph ave”)
> lines(LSc$x300smo_ mLymphAve~LSc$POSITION, col=“blue3”, lwd=3)
Figure 9.
Raw (gray) and loess-smoothed (blue) replication timing values along chromosome 1.
B.) Correlations between datasets
-
Once the technical quality of the array data is established, compare biological replicate experiments to determine the relative level of biological similarity between samples. When comparing different cell types, to isolate biological rather than array quality differences, we typically use loess-smoothed averaged-replicate data, rather than individual, raw or normalized data:
> cor(x[,c(4:6)]
Rep1 Rep2 Ave Lymphoblast Rep1 1.000 0.978 0.995 Lymphoblast Rep2 0.978 1.000 0.994 Lymphoblast Ave 0.995 0.994 1.000 The cor() function defaults to Pearson correlation, but other methods are available (see ?cor in R). If missing values are present, add “na.rm=T” to remove them.
C.) Segmentation
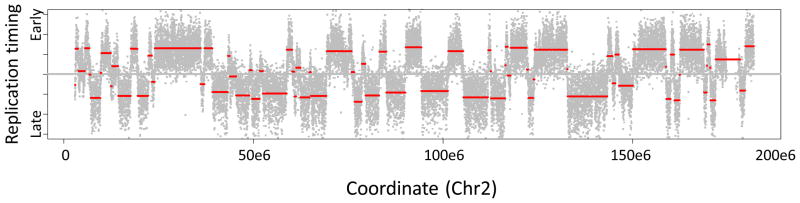
Perform circular binary segmentation as outlined in steps 86c(ii)-(iv) (Fig. 10). Biologically, these segments (or “replication domains”) appear to correspond to domains of coordinately regulated, synchronously firing origins that may be part of replication factories. We perform segmentation as follows using the DNACopy algorithm designed by Venkatraman et al.45, which performs favorably compared to alternatives for CGH copy number analysis46–48.
-
First, load the DNAcopy package and prepare a CNA (copy number array) object for segmentation:
> library(DNAcopy)
> mLymph = CNA(RT$mLymphAve, RT$CHR, RT$POSITION, data.type=“logratio”, sampleid = “mLymph”)
-
Next, segment the CNA object with the desired parameters. The parameters shown are those we have used for analysis of mouse and human timing datasets with autocorrelations near 0.83,6; data of different quality or in different formats may require these to determined empirically.
> Seg.mLymph = segment(mLymph, nperm=10000, alpha=1e-15, undo.splits=“sdundo”, undo.SD=1.5, verbose=2)
-
Examine the resulting segmentation object ‘Seg.mLymph’, which contains the raw data and segmentation breakpoints assigned by circular binary segmentation49. The number of segments assigned can be determined using str(Seg.mLymph$output) and visualized using various functions built into DNAcopy (Fig. 10).
> par(ask=T,mar=c(3.1,4.1,1,1))
# Set figure margins; ask before replotting
> plot(Seg.mLymph, plot.type=“c”)
# Plot each chromosome separately
> plot(Seg.mLymph, plot.type=“s”)
# Plot overview of all chromosomes
> plot(subset(Seg.mLymph,chromlist=“chr2”), pch=19, pt.cols=c(“gray”,”gray”), xmaploc=T, ylim=c(− 3,3))
# Plot a single chromosome with alternate format
-
Create a tab-delimited text file containing segment endpoints and average replication timing values for each segment. The file will be written to the current working directory (see step 63).
> write.table(Seg.mLymph$output, row.names=F, quote=F, sep=“\t”)
-
After segmentation, calculate the sizes of replication domains from the segmented dataset to examine average sizes for domains with early or late timing.
> Lymph = Seg.mLymphR1$output
# Extract domain information
> Lymph$size = Lymph$loc.end - Lymph$loc.start
# Calculate domain sizes
> LymphEarly = subset(Lymph, Lymph$seg.mean > 0)
# Create subset of early domains
> LymphLate = subset(Lymph, Lymph$seg.mean < 0)
# Create subset of late domains
> boxplot(LymphEarly$size, LymphLate$size)
# Distribution of early/late domain sizes
Figure 10.
Raw data (gray) overlaid with segmented timing domains (red) along chromosome 2, as defined by circular binary segmentation45
Dynamic changes in the timing program TIMING - 3 h
-
87
To examine changes in the replication program during differentiation, use one or several of the methods in this step to leverage the segmentation and loess smoothing methods introduced in step 86. Since no single method is sufficient to fully describe the type, degree, and distribution of timing changes during development, we cover several complementary ways to measure these properties and explore the relationships between cell types. These include: A) The amount of the genome changing replication timing (percent changes analysis); B) The degree and relationships of RT changes between cell types (clustering approaches); and C) The properties of domains that change timing upon differentiation (switching domain analysis)
A.) Percent changes analysis
-
Determine the amount of the genome with differential timing between two or more cell types using an arbitrary, percentile, or significance-based cutoff for RT changes. We recommend scaling datasets to equivalent ranges and applying an empirical cutoff for changes verifiable by PCR to quantify these genome-wide, as shown here. As most methods for quantifying timing changes are sensitive to scale differences, datasets should be first scaled and normalized together in LIMMA (see steps 62–76).
> RTd1 = RT$mLymphR1 - RT$mLymphR2
# Calculate timing differences between datasets
> mLength = length(RTd1)
# Determine total number of probes
> s = 0.67
# Set cutoff for significant changes
> sum(abs(RTd1)>s)/mLength
# Percentage changing, R1 vs. R2
> sum(RTd1 < −s)/mLength
# Early to Late changes: 1.6% of all probes
> sum(RTd1 > s)/mLength
# Late to Early changes: 1.3% of all probes
B.) Clustering approaches
Perform clustering to aggregate experiments with similar timing patterns. For k-means clustering we have used the programs Cluster50 and TreeView (http://rana.lbl.gov/EisenSoftware.htm) and refer readers to their corresponding guides. For hierarchical clustering, we use the “pvclust” package in R51 to compute clusters based on the stability of connections between cell types and ascribe p-values to each node.
-
To improve the precision of individual RT measurements and lessen the considerable computational expense of most clustering algorithms, average individual timing values into larger windows prior to clustering. We typically average datasets in windows of approximately 200 kb.
> mLymph.R1 = NULL; mLymph.R2 = NULL
# Initialize variables to store averaged data
> nWin = 35
# 5.8kb median probe spacing * 35 = 203kb
> mLength = nrows(RT)/nWin
# Calculate number of windows
> for (x in 1:mLength) {# For each potential window,
> z1 = x * nWin
# Determine probe number at window start
> z2 = (x+1) * nWin
# Determine probe number at window end
> mLymph.R1[x] = mean(RT$mLymphR1[z1:z2])
# Average replicate 1 across window
> mLymph.R2[x]= mean(RT$mLymphR2[z1:z2])
# Average replicate 2 across window
> cat(“Current window: “, x, “/”, mLength, “\n”)
# Write the current window to the console
> }
# End for loop
> RTWind = data.frame(mLymph.R1, mLymph.R2)
# Write the results to a new data frame
-
Load the pvclust51 package and use its corresponding function to cluster datasets using multiscale bootstrap resampling, which will assign p-values to each node in the hierarchical clustering dendrogram. See ?pvclust after loading the package for additional options and settings.
> library(pvclust)
> cluster.bootstrap <- pvclust(RTWind, nboot=1000, method.dist=“abscor”)
-
Plot the cluster dendrogram as performed below.
> plot(cluster.bootstrap)
# Plot overall dendrogram
> pvrect(cluster.bootstrap)
# Outline datasets that cluster at a significant level
CRITICAL STEP Take care when interpreting the results of hierarchical clustering, as: a wide variety of topologies are possible for a single dendrogram, since any node can be flipped horizontally without changing the connections between clusters; agglomerative clusters can change substantially when new experiments are added, and; the exact connections produced (though usually not the overall structure of the dendrogram) often change for different clustering algorithms or distance metrics.
C.) Properties of RT switching domains
-
Perform segmentation on the differences between timing profiles to define the boundaries of domains that switch to earlier or later replication (switching domains) and analyze the properties of genetic and epigenetic elements within them. To compute these domains, first subtract the normalized (not loess smoothed) values of the two experiments to be compared, and create a CNA object in a similar fashion to step 86c(ii).
> dRT = CNA(RT$NPCave-RT$ESCave, RT$CHR, RT$POSITION, data.type=“logratio”, sampleid=“NPC-ESC dRT”)
-
Next, segment the resulting object, calculate domain sizes, and write the segments to a tab-delimited text file,
> Seg.dRT = segment(dRT, nperm=10000, alpha=1e-15, undo.splits = “sdundo”, undo.SD=1.5, verbose=2); dRTdom = Seg.dRT$output
> dRTdom$size = dRTdom$loc.end - dRTdom$loc.start
> write.table(dRTdom, “Switching segments, mNPC vs. mESC.txt”, row.names=F, quote=F, sep=“\t”)
-
Identify domains with the largest timing changes in either direction, as well as domains with stable timing between datasets, using cutoffs from the quantile() function.
> quantile(dRTdom$seg.mean, probs = c(0.05, 0.95))
# Top 5% of changes to early/late
> quantile(dRTdom$seg.mean, probs = c(0.40, 0.60))
# Middle 20% of smallest changes
> LtoEdom = subset(dRTdom, dRTdom$seg.mean > 1.28552)
# Isolate late-to-early domains
> EtoLdom = subset(dRTdom, dRTdom$seg.mean < −1.32328)
# Isolate early-to-late domains
> middleDom = subset(dRTdom, dRTdom$seg.mean > −0.14808 & dRTdom$seg.mean < 0.23698)
# Isolate non-switching domains
> boxplot(middleDom$size, LtoEdom$size, EtoLdom$size)
# Plot distributions of domain sizes
Comparison and alignment to outside datasets TIMING - 6 h
-
88
Choose among several alternative approaches to compare the timing program to the vast array of genome-wide or gene-centric data made available through initiatives such as ENCODE52–54 and public repositories like GEO55,56. To study gene-level regulation, assign replication timing values (option A) and epigenetic marks (option B) to lists of RefSeq (http://www.ncbi.nlm.nih.gov/RefSeq/) or other gene locations. For domain-level analysis, average values within the boundaries of replication domains (option C).
A.) Assignment of replication timing values to gene promoters
Assign loess-smoothed timing values to gene promoters as outlined below. Although the main purpose of loess is to derive an overall smoothed replication timing profile, the smoothed data object produced can be interrogated at any set of genomic coordinates, making it especially valuable for comparing datasets from different array platforms and coordinates. Using this approach, we assign timing data to NCBI’s RefSeq gene promoter locations as follows:
Begin by loading the required datasets; these include a table of RefSeq gene locations for the desired species (found at http://www.ncbi.nlm.nih.gov/RefSeq/), and a list of smoothed replication timing values created in step 86 and loaded as in step 65.
-
Next, create a list of chromosomes to be analyzed and variables to store the data in each chromosome.
> chrs = levels(RefSeq$CHR)
# Create a list of chromosomes to be analyzed
> AllSm = NULL
# Variable to store smoothed data for all chromosomes
> ChrSm = NULL
# Variable to store smoothed data for one chromosome
-
Run the following loop to calculate replication timing values at transcription start sites of RefSeq genes. Advanced R users may substitute an appropriately reformatted function if desired, and the approach below may be used generically to apply values from any data type regulated on large scales (relative to array probe density) to any list of genomic coordinates.
> for(chr in chrs) {# For each chromosome,
> RTc = subset(RT, CHR == chr)
# Create subset of timing values in the chromosome
> RSc = subset(RefSeq, CHR == chr)
# Create subset of RefSeq genes in the chromosome
> cat(“Current chromosome: “, chr, “\n”)
# Output current chromosome to console
> lspan = 300000/(max(RTc$POSITION)-min(RTc$POSITION))
# Set smoothing span
> smLym1 = loess(RT$mLymphR1 ~ RT$POSITION, span = lspan)
# Smooth dataset 1
> smLym2 = loess(RT$mLymphR2 ~ RT$POSITION, span = lspan)
# Smooth dataset 2
> smLym3 = loess(RT$mLymphAve ~ RT$POSITION, span = lspan)
# Smooth dataset 3
> Lym1 = predict(smLym1, RSc$TSS)
# Predict (interpolate) values at transcription start sites
> Lym2 = predict(smLym2, RSc$TSS)
# Predict values for dataset 2
> Lym3 = predict(smLym3, RSc$TSS)
# Predict values for dataset 3
> ChrSm = data.frame(CHR=chr,POSITION= RSc$TSS, Lym1, Lym2, Lym3)
> AllSm = rbind(AllSm, ChrSm)
# Combine information for all experiments/chromosomes
> }
# End for loop
-
As in steps 78–79, write the results of analysis to an external file before unloading the data from memory:
> write.table(AllSm, “Mouse lymphoblast RT at RefSeq gene positions.txt”, quote=F, sep=“\t”)
B.) Assignment of histone and other epigenetic marks to gene promoters
Assign epigenetic and other datasets to gene promoters using steps 88b(ii)–(v). Unlike replication timing, values from epigenetic datasets are often too sparse relative to their unit of regulation to apply the method in option 88a. For this example, we assign values from a generic genomewide ChIP-seq experiment to windows +500 to −2500 bases from RefSeq gene promoters.
As in option A, first load the required datasets as described in steps 65 and 88a(ii). Two files are required: One with columns describing the genomic coordinate, orientation (+/−), and chromosome of each gene (read into a variable named “RefSeq”), and another with the coordinate, chromosome, and data value for each mark (read into variable “Marks”).
-
Create a list of chromosomes to be analyzed and variables to store the assigned values:
> chrs = levels(Marks$CHR); AllGenes = NULL; AllHist = NULL
-
Run the following loop to assign values near transcription start sites to RefSeq genes. We generally set the apply function to assign the highest value within the promoter window to the gene; other approaches include averaging the number of reads within the body of genes57, individually analyzing equally spaced bins across open reading frames58, and assessing promoters with significant binding above background59. Bear in mind that the transcription start site may not be the best target for all modifications; indeed, for H3K36me3 marking transcription elongation, values at the transcription endpoint or exon 5′ ends may better represent overall enrichment60.
> for (chr in chrs) {# For each chromosome,
> RSc = subset(RefSeq, CHR == chr)
# Create subset of RefSeq genes in the chromosome
> MKc = subset(Marks, CHR == chr)
# Create subset of mark values in the chromosome
> for(m in 1:nrow(RSc)) {# For each gene in the chromosome,
> if(RSc[m,]$Strand == “+”) {# If the gene is in the forward orientation,
> RTcSub = subset(RTc, (RTc$Start < RSc[m,]$txStart +500) & (RTc$Start>
RSc[m,]$txStart - 2500))
# Collect values from txStart +500 to −2500bp
> AllHist = rbind(AllHist, apply(RTcSub, 2, max)[3:12])
# Assign max value to gene
> AllGenes = rbind(AllGenes, RSc[m,]$Gene)
# Combine with overall list
> }
# End if
> if(RSc[m,]$Strand == “−”) {# If the gene is in the reverse orientation,
> RTcSub = subset(RTc, (RTc$Start < RSc[m,]$txEnd +2500) & (RTc$Start >
RSc[m,]$txEnd - 500))
# Collect values from txEnd +2500 to −500 bp
> AllHist = rbind(AllHist, apply(RTcSub, 2, max)[3:12])
# Assign max value to gene
> AllGenes = rbind(AllGenes, RSc[m,]$Gene)
# Combine with overall list
> }
# End if
> cat(“Chromosome:”, chr, “Gene:”, m, “/”, nrow(RSc), “\n”)
# Output current gene
> }
# End gene loop
>}
# End chromosome loop
-
Finally, similarly to previous steps, combine the gene and epigenetic mark information into a single table and output as tab-delimited text.
> OutFile = data.frame(cbind(AllGenes, AllHist), stringsAsFactors=F)
> write.table(OutFile, file=“Histone modifications at RefSeq gene positions.txt”, row.names=F, quote=F, sep=“\t”)
C.) Integration of epigenetic mark values over replication domains
Use the method below to correlate domainwide replication timing values and the average level of epigenetic marks within timing domains segmented in step 87C (for static timing domains) or 88C (for domains that switch timing). Given that the magnitude of correlations between genetic properties generally increases when measured in larger windows, it is important to quantify these relationships in windows consistent with biologically regulated unit sizes.
-
Read the replication domains created in step 87C or 88C into variable “Seg.RT”.
> Seg.RT = read.table(“Lymph-1 segments.txt”,header=T)
-
Create a list of chromosomes and variables in which to store average epigenetic mark and timing values.
> chrs = levels(Seg.RT$chrom)
> MarksData = NULL; RTData = NULL
-
Run the loop below to assign the average values of one or multiple epigenetic datasets to each replication domain, or modify as needed.
> dom = 0
# Initialize domain number to 0
> for(chr in chrs) {# For each chromosome,
> Seg.RTb = subset(Seg.RT, Seg.RT$chrom == chr)
# Get timing domains in chromosome
> MarksB = subset(Marks, Marks$CHR == chr)
# Get mark data in chromosome
> for (i in 1:dim(Seg.RTb)[1]) {# For each domain,
> cat(“Current chr:”, chr, “Domain:”, dom, “\n”)
# Output current domain
> MarksD = subset(MarksB, MarksB$Start > Seg.RTb[i,]$loc.start & MarksB$Start <
Seg.RTb[i,]$loc.end)
# Find subset of marks in domain
> MarksD = MarksD[,3:12]
# Exclude chr/pos from mark data
> MarksD[,1:10] = MarksD[,1:10] - MarksD[,1]
# Subtract control values, if needed
> MarksData = rbind(MarksData, apply(MarksD,2, “mean”))
# Average mark data in domain
> dom = dom + 1
# Increment domain number
> }
# End domain loop
> }
# End chromosome loop
-
Lastly, find the correlations between domainwide replication timing and each type of epigenetic mark, and create scatterplots to visualize these relationships.
> cor(Seg.RT$seg.mean, data.frame(MarksData))
> plot(Seg.RT$seg.mean, data.frame(MarksData)[1])
TIMING
Steps 1–12, BrdU Labeling and FACs sorting: ~5–6 hours
Steps 13–44, BrdU Immunoprecipitation: ~2–3 days
Steps 45–50, PCR Assay: ~4–6 hours
Steps 51–57, WGA: ~5 hours
Box 1, S/G1 FACS Sorting ~1 day
Step 58, Dye Labeling: ~3–4 hours
Step 59, Hybridization: ~1 hour plus hybridization time
Steps 60–61, Washing and Scanning: ~1–2 hours
Steps 62–85, Normalization: ~1 day
Step 86, Static properties: ~3 h
Step 87, Dynamic properties: ~3 h
Step 88, Outside datasets: ~6 h
? TROUBLESHOOTING
Troubleshooting advice can be found in Table 2.
TABLE 2.
Troubleshooting table.
| Step | Problem | Possible Reason | Solution |
|---|---|---|---|
| 6 | Cell aggregation or debris accumulation prevents accurate cell sorting | Failure to achieve single cell suspension with certain problematic cell types | Incubate with enzyme treatment, such as Tryspin-EDTA or accutase, for a longer period of time. Use gentle trituration to ensure that cell aggregates are broken apart prior to fixation and/or sorting. Occasional pausing and filtering of cell samples during FACs may help. |
| Vortexing during ethanol fixation was too harsh | Use the lowest vortex setting available while adding ethanol dropwise | ||
| 49 | Inconsistent PCR bands between aliquots of the same sample | Contamination between fractions during FACS, likely due to problems in cell fixation | Switch from PI staining (with fixation) to DAPI staining (without fixation) |
| Inconsistent number of cells aliquoted to each tube | Mix contents thoroughly before aliquoting and freezing for storage. Aliquot 20,000 cells while the samples are hot, to avoid pipetting errors due to SDS formation in the solution | ||
| BrdU labeling time was insufficient | Incubate growing cells with BrdU for a longer period of time. Cells with longer S-phases require longer BrdU incubation times. | ||
| Varying efficiency of BrdU-immunoprecipitation between samples caused by loss of DNA-protein pellet. | Use caution when removing supernatant from the loose DNA-protein pellet. Centrifuge the sample multiple times as needed to remove supernatant without disturbing the pellet. | ||
| 57 | Samples do not pass screening | Bias created during WGA | Increase the amount of starting material for WGA. For instance, start with 100 ul of IP sample pool instead of 50 ul at step 52. |
| 81 | Skew towards early or late values | Bias created during WGA or labeling, or excessive photobleaching during scanning. | Check early vs. late WGA yields, and avoid multiple scans of the array |
| 84 | Low autocorrelation (high level of noise) | Values are not properly sorted by chromosomal location | Ensure that chromosome and position columns are properly assigned to experimental values, and sorted as in step 80 |
| Low signal intensity (step 74) | Check yield after labeling and amplification steps, as well as scanner settings. | ||
| 86c(v) | Large difference in domain numbers between similar datasets | Sensitivity of segmentation algorithms to differences in data quality | Either adjust the parameter undo. SD (using similar autocorrelation-level datasets as a guide), or add Gaussian noise to higher-quality datasets to equalize their acf (step 84) prior to segmentation. |
ANTICIPATED RESULTS
Our research has shown that the described method is a powerful tool for genome-scale analysis of replication timing. However, meaningful data analysis is dependent on the quality of available data. Therefore, measures should be taken throughout the protocol to ensure that each phase of the procedure produces quality starting material for subsequent phases. Anticipated results for various steps of the protocol are described here. Typical FACS plots demonstrating successful DNA content analysis and indicating appropriate S-phase fractions to be collected are shown in Figure 2.
Figure 2.
2D Cell Cycle Sorting for S and G1 phases. Cells labeled with BrdU and stained as described in Box 1, and then analyzed on a FACS. A. A typical non-corrected BrdU/PI plot. Note how the plot is skewed to the right due to spectral overlap. B. A corrected BrdU/PI plot. Sorting windows for nicely separated G1, S and G2/M phases of the cell cycle are indicated.
Following cell sorting and BrdU-immunoprecipitation, marker genes with known relative replication timing (Table 1) should be amplified by PCR for multiple IP samples and detected by electrophoresis on an agarose gel. Among the mouse sequences listed in Table 1, mitochondrial DNA replicates throughout the cell cycle61 and will typically be equally represented in early and late S-phase fractions. Alpha-globin, Pou5f1 and Mmp15 are generally early replicating markers, whereas beta-globin, Zfp42, Dppa2, Ptn, Mash1, and Akt3 are generally late replicating markers. Note that some genes switch replication timing at some point during development; for instance, Zfp42 and Dppa2 are early replicating in ESCs but late replicating in all somatic cell types examined to date. Therefore, consistency across multiple samples from the same cell type is usually the most reliable way to assess the quality of IP samples. Among the human sequences listed in Table 1, mitochondrial DNA is equally represented in early and late S-phase fractions, while alpha-globin, MMP15 and BMP1 are generally early replicating markers. PTGS2, NETO1, SLITRK6, ZFP42 and DPPA2 are generally late replicating. High quality immunoprecipitation reactions show consistency in the relative amount of BrdU-labeled DNA in respective S-phase fractions between samples of the same cell type. This PCR analysis should be performed again directly following whole genome amplification in order to ensure that no bias has been introduced during this step of the procedure. If no bias is detected, 4–8μl of purified WGA3 DNA should be run on a 1.5% agarose gel in order to determine its quality. Quality DNA will range in size from 100–1,000 bp, with an average size of about 400bp. Additionally, WGA3 DNA should have an A260/A80 value greater than or equal to 1.8 and an A260/A230 value greater than or equal to 1.9 in order to function as high quality starting material for the labeling reaction. NimbleGen Arrays User’s Guide, CGH Analysis should be consulted for anticipated results of the hybridization and scanning procedures.
After a successful experiment, domains of coordinate replication timing (replication domains) will be clearly visible in the raw data after plotting these across a chromosome (Figure 6). Less successful experiments will have autocorrelation values below 0.6 (Figure 7), and visibly higher levels of noise, limiting the resolution of smaller replication domains. Further, low signal in MA plots (Figure 4) and signal intensity distributions (Figure 3) will also often present with low autocorrelation, and may indicate a low volume of Cy-labeled DNA or problems with scanning. If several replicate experiments were performed, these should have high (>0.90) correlations between loess-smoothed timing values (step 86B).
Supplementary Material
Acknowledgments
We thank Juan Carlos Rivera Mulia, Athena Rycyk, and the anonymous reviewers for helpful comments on the manuscript. Research in the Gilbert lab is funded by NIH grants GM083337 and GM085354.
Footnotes
AUTHOR CONTRIBUTIONS
D.G. and I.H. conceived the study and designed the experiments. T.R. and I.H. devised the computational methods. T.R., D.B., B.P., and D.G. wrote the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare that they have no competing financial interests.
References
- 1.Pope BD, Hiratani I, Gilbert DM. Domain-wide regulation of DNA replication timing during mammalian development. Chromosome Research. 2010;18:127–36. doi: 10.1007/s10577-009-9100-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yaffe E, et al. Comparative analysis of DNA replication timing reveals conserved large-scale chromosomal architecture. PLoS Genetics. 2010;6:e1001011. doi: 10.1371/journal.pgen.1001011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ryba T, et al. Evolutionarily conserved replication timing profiles predict long-range chromatin interactions and distinguish closely related cell types. Genome Research. 2010;20:761–70. doi: 10.1101/gr.099655.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gilbert DM, et al. Space and Time in the Nucleus: Developmental Control of Replication Timing and Chromosome Architecture. Cold Spring Harbor symposia on quantitative biology. 2010 doi: 10.1101/sqb.2010.75.011. [DOI] [PubMed] [Google Scholar]
- 5.Hiratani I, et al. Genome-wide dynamics of replication timing revealed by in vitro models of mouse embryogenesis. Genome Research. 2010;20:155–69. doi: 10.1101/gr.099796.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hiratani I, et al. Global reorganization of replication domains during embryonic stem cell differentiation. PLoS Biology. 2008;6:e245. doi: 10.1371/journal.pbio.0060245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schwaiger M, et al. Heterochromatin protein 1 (HP1) modulates replication timing of the Drosophila genome. Genome research. 2010 doi: 10.1101/gr.101790.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schwaiger M, et al. Chromatin state marks cell-type- and gender-specific replication of the Drosophila genome. Genes & Development. 2009;23:589–601. doi: 10.1101/gad.511809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schübeler D, et al. Genome-wide DNA replication profile for Drosophila melanogaster: a link between transcription and replication timing. Nature Genetics. 2002;32:438–42. doi: 10.1038/ng1005. [DOI] [PubMed] [Google Scholar]
- 10.Lee T-J, et al. Arabidopsis thaliana chromosome 4 replicates in two phases that correlate with chromatin state. PLoS Genetics. 2010;6:e1000982. doi: 10.1371/journal.pgen.1000982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koren A, Soifer I, Barkai N. MRC1-dependent scaling of the budding yeast DNA replication timing program. Genome Research. 2010;20:781–90. doi: 10.1101/gr.102764.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Raghuraman MK, Brewer BJ. Molecular analysis of the replication program in unicellular model organisms. Chromosome research: an international journal on the molecular, supramolecular and evolutionary aspects of chromosome biology. 2010;18:19–34. doi: 10.1007/s10577-009-9099-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hayashi M, et al. Genome-wide localization of pre-RC sites and identification of replication origins in fission yeast. The EMBO journal. 2007;26:1327–39. doi: 10.1038/sj.emboj.7601585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karnani N, Taylor CM, Dutta A. Microarray analysis of DNA replication timing. Methods in molecular biology (Clifton, NJ) 2009;556:191–203. doi: 10.1007/978-1-60327-192-9_14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Farkash-Amar S, Simon I. Genome-wide analysis of the replication program in mammals. Chromosome Research. 2009 doi: 10.1007/s10577–009–9091–5. [DOI] [PubMed] [Google Scholar]
- 16.Sasaki T, et al. The Chinese hamster dihydrofolate reductase replication origin decision point follows activation of transcription and suppresses initiation of replication within transcription units. Molecular and cellular biology. 2006;26:1051–62. doi: 10.1128/MCB.26.3.1051-1062.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gilbert DM. Replication origin plasticity, Taylor-made: inhibition vs recruitment of origins under conditions of replication stress. Chromosoma. 2007;116:341–7. doi: 10.1007/s00412-007-0105-9. [DOI] [PubMed] [Google Scholar]
- 18.Anglana M, et al. Dynamics of DNA replication in mammalian somatic cells: nucleotide pool modulates origin choice and interorigin spacing. Cell. 2003;114:385–94. doi: 10.1016/s0092-8674(03)00569-5. [DOI] [PubMed] [Google Scholar]
- 19.Gilbert DM. Evaluating genome-scale approaches to eukaryotic DNA replication. Nature reviews. Genetics. 2010 doi: 10.1038/nrg2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gilbert DM. Temporal order of replication of Xenopus laevis 5S ribosomal RNA genes in somatic cells. Proceedings of the National Academy of Sciences of the United States of America. 1986;83:2924–8. doi: 10.1073/pnas.83.9.2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gilbert DM, Cohen SN. Bovine papilloma virus plasmids replicate randomly in mouse fibroblasts throughout S phase of the cell cycle. Cell. 1987;50:59–68. doi: 10.1016/0092-8674(87)90662-3. [DOI] [PubMed] [Google Scholar]
- 22.Hansen RS, et al. Association of fragile X syndrome with delayed replication of the FMR1 gene. Cell. 1993;73:1403–9. doi: 10.1016/0092-8674(93)90365-w. [DOI] [PubMed] [Google Scholar]
- 23.Yokochi T, et al. G9a selectively represses a class of late-replicating genes at the nuclear periphery. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:19363–8. doi: 10.1073/pnas.0906142106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu J, et al. G2 phase chromatin lacks determinants of replication timing. The Journal of cell biology. 2010;189:967–80. doi: 10.1083/jcb.201002002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pollack JR, et al. Genome-wide analysis of DNA copy-number changes using cDNA microarrays. Nature Genetics. 1999;23:41–6. doi: 10.1038/12640. [DOI] [PubMed] [Google Scholar]
- 26.Acevedo LG, et al. Genome-scale ChIP-chip analysis using 10,000 human cells. BioTechniques. 2007;43:791–7. doi: 10.2144/000112625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Statistical Applications in Genetics and Molecular Biology. 2004;3:3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 28.Yang YH, et al. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Research. 2002;30:e15. doi: 10.1093/nar/30.4.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bolstad BM, et al. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–93. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 30.Core RD. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing Vienna Austria ISBN. 2008;3:2673. [Google Scholar]
- 31.Gentleman RC, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biology. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ihaka R, Gentleman R. R: A Language for Data Analysis and Graphics. Journal of Computational and Graphical Statistics. 1996;5:299. [Google Scholar]
- 33.Spector P. Data Manipulation with R. Vol. 154. Springer Publishing Company, Incorporated; 2008. [Google Scholar]
- 34.Chambers JM. Software for Data Analysis: Programming with R. Vol. 498. Springer Publishing Company, Incorporated; 2008. [Google Scholar]
- 35.Crawley MJ. The R Book. Vol. 950. Wiley; 2007. [Google Scholar]
- 36.Wettenhall JM, Smyth GK. limmaGUI: a graphical user interface for linear modeling of microarray data. Bioinformatics. 2004;20:3705–6. doi: 10.1093/bioinformatics/bth449. [DOI] [PubMed] [Google Scholar]
- 37.Hansen RS, et al. Sequencing newly replicated DNA reveals widespread plasticity in human replication timing. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:139–44. doi: 10.1073/pnas.0912402107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pombo A, Gilbert DM. Nucleus and gene expression: the structure and function conundrum. Current Opinion in Cell Biology. 2010 doi: 10.1016/j.ceb.2010.04.010. [DOI] [PubMed] [Google Scholar]
- 39.Geen OH, et al. Comparison of sample preparation methods for ChIP-chip assays. BioTechniques. 2006;41:577–80. doi: 10.2144/000112268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Woodfine K, et al. Replication timing of the human genome. Human Molecular Genetics. 2004;13:191–202. doi: 10.1093/hmg/ddh016. [DOI] [PubMed] [Google Scholar]
- 41.Peng S, et al. Normalization and experimental design for ChIP-chip data. BMC Bioinformatics. 2007;8:219. doi: 10.1186/1471-2105-8-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gottardo R. Modeling and analysis of ChIP-chip experiments. Methods in Molecular Biology. 2009;567:133–43. doi: 10.1007/978-1-60327-414-2_9. [DOI] [PubMed] [Google Scholar]
- 43.Peng S, et al. Normalization and experimental design for ChIP-chip data. BMC Bioinformatics. 2007;8:219. doi: 10.1186/1471-2105-8-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reimers M, Weinstein JN. Quality assessment of microarrays: visualization of spatial artifacts and quantitation of regional biases. BMC Bioinformatics. 2005;6:166. doi: 10.1186/1471-2105-6-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Venkatraman ES, Olshen AB. A faster circular binary segmentation algorithm for the analysis of array CGH data. Bioinformatics. 2007;23:657–63. doi: 10.1093/bioinformatics/btl646. [DOI] [PubMed] [Google Scholar]
- 46.Dellinger AE, et al. Comparative analyses of seven algorithms for copy number variant identification from single nucleotide polymorphism arrays. Nucleic Acids Research. 2010 doi: 10.1093/nar/gkq040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Willenbrock H, Fridlyand J. A comparison study: applying segmentation to array CGH data for downstream analyses. Bioinformatics. 2005;21:4084–91. doi: 10.1093/bioinformatics/bti677. [DOI] [PubMed] [Google Scholar]
- 48.Lai WR, et al. Comparative analysis of algorithms for identifying amplifications and deletions in array CGH data. Bioinformatics. 2005;21:3763–70. doi: 10.1093/bioinformatics/bti611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Olshen AB, et al. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics. 2004;5:557–72. doi: 10.1093/biostatistics/kxh008. [DOI] [PubMed] [Google Scholar]
- 50.Eisen MB, et al. Cluster analysis and display of genome-wide expression patterns. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:14863–8. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Suzuki R, Shimodaira H. Pvclust: an R package for assessing the uncertainty in hierarchical clustering. Bioinformatics. 2006;22:1540–2. doi: 10.1093/bioinformatics/btl117. [DOI] [PubMed] [Google Scholar]
- 52.The ENCODE (ENCyclopedia Of DNA Elements) Project. Science (New York, NY) 2004;306:636–40. doi: 10.1126/science.1105136. [DOI] [PubMed] [Google Scholar]
- 53.Birney E, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rosenbloom KR, et al. ENCODE whole-genome data in the UCSC Genome Browser. Nucleic Acids Research. 2010;38:D620–5. doi: 10.1093/nar/gkp961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic acids research. 2002;30:207–10. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Barrett T, et al. NCBI GEO: archive for high-throughput functional genomic data. Nucleic acids research. 2009;37:D885–90. doi: 10.1093/nar/gkn764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–37. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 58.Salcedo-Amaya AM, et al. Dynamic histone H3 epigenome marking during the intraerythrocytic cycle of Plasmodium falciparum. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:9655–60. doi: 10.1073/pnas.0902515106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Guenther MG, et al. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hon G, Wang W, Ren B. Discovery and annotation of functional chromatin signatures in the human genome. PLoS Computational Biology. 2009;5:e1000566. doi: 10.1371/journal.pcbi.1000566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Aladjem MI, et al. Replication initiation patterns in the beta-globin loci of totipotent and differentiated murine cells: evidence for multiple initiation regions. Molecular and Cellular Biology. 2002;22:442–52. doi: 10.1128/MCB.22.2.442-452.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.