Abstract
Precursors undertaking T-cell development shed their access to other pathways in a sequential process that begins before entry into the thymus and continues through many cell cycles afterwards. This process involves three levels of regulatory change, in which the cells’ intrinsic transcriptional regulatory factors, expression of signaling receptors like Notch1, and expression of distinct homing receptors separately contribute to confirmation of T-cell identity. Each alternative potential has a different underlying molecular basis which is neutralized, then permanently silenced through different mechanisms in early T-cell precursors. This regulatory mosaic has notable implications for the hierarchy of relationships linking T lymphocytes to other hematopoietic fates.
Commitment versus potential
Multipotent hematopoietic cells gain access to the T-cell developmental pathway, and then confirm that choice of fate by “burning their bridges” to other pathways. The process involves a continuing dialogue between the differentiating cell and the signals coming from its environment. Best understood of these signals are effects of Notch1 engagement with Delta ligands provided in the environment of the thymus, which are needed for T-cell specification throughout the lineage choice process, and only become dispensable after T-cell fate is confirmed (1-3). In vivo, expression of Notch ligand with supportive cytokines such as IL-7 and Kit ligand gives the thymus its T-cell inductive activity (3). However, the cells that begin the T cell program initially have access to other options. These can be revealed if the cells are removed from the thymic environment and challenged with different environmental signals. Only after they lose the ability to make these alternative responses are the cells committed.
Commitment is the cell-intrinsic regulatory transformation through which a cell's alternative potentials are eliminated. “Potential” is not the same as the default fate that a cell adopts in an undisturbed condition. “Fate” is the intersection between the cell's potential and the permissive or nonpermissive circumstances in which a cell finds itself. For a typical uncommitted cell, the fates its progeny actually adopt in vivo may be only a slice of the uncommitted cell's full potential, because environmental conditions are limiting. As the cell progresses toward commitment, its potential shrinks. At the point where potential rather than environment becomes limiting for fate, the cell is committed. Interestingly, one of the few cases where a “fate” assay approaches a “potential” assay is in the case of hematopoietic stem cells, where extremely long assay times are conventionally used, extremely large numbers of progeny are generated before the endpoint, and the intermediates include highly motile cells that can sample an unusually large number of in vivo environments before differentiating as scored at the assay endpoint. However, this exception proves the rule that multipotent cells can only show the subset of their potentials that their environments support.
If fate is different from potential, then why study potential at all (4,5)? Potential reveals what the regulatory machinery of the cell itself, transcription factors and signal receptors, contributes to its identity. The commitment process establishes an irreversible, portable, unconditional identity by actively narrowing a cell's range of potentials. Exclusion of any particular fate can occur by repression either of a key transcription factor or of a signaling receptor that would be needed to induce that pathway in permissive conditions. For example, T cell potential itself depends on Notch1 expression. During B-lineage commitment, T-cell potential is excluded by Pax5's silencing the expression of Notch1 (6). Ultimately, commitment to a lineage is the readout for the gene regulatory network that links positive differentiation in one pathway with the controlled silencing of particular transcription factors and signaling receptors needed for other pathways.
Normally developmental potentials of two populations are compared by testing cells under uniform conditions that are clearly permissive for the fates being assayed. However, there are caveats. When hematopoietic precursors are adoptively transferred intravenously, the protocol in effect “hands over” to the grafted cells themselves the ultimate choice of the environment into which they will lodge. The choice is mediated by adhesion molecules and chemokine receptors on the cells that can bias migration to particular microenvironments, and these “traffic control” molecules are themselves developmentally regulated. For example, the chemokine receptors CCR9 and/or CCR7 are needed for hematopoietic precursors to enter the thymus; double mutants are dramatically impaired as T-cell precursors in vivo (7-9). The abilities of cells to home predictably to particular environments in vivo are biologically important and especially relevant for therapeutic use of uncommitted cells in clinical practice (5). However, these traffic control molecules are not needed under permissive conditions to program T-cell identity itself (10).
Thus, changes in cell potential need to be explained by mechanisms operating at three distinct levels. At the cell-intrinsic level, potential is defined by transcription factors and possible epigenetic constraints. At the opposite extreme, in cases where the environment is not experimentally controlled, the cell's opportunities may be biased by its migration preferences. Between these levels, at the cell/environment interface, triggering of potential depends on expression of signaling receptors that modify transcription factor activity in response to inductive environmental signals.
Stepwise commitment of T-cell precursors
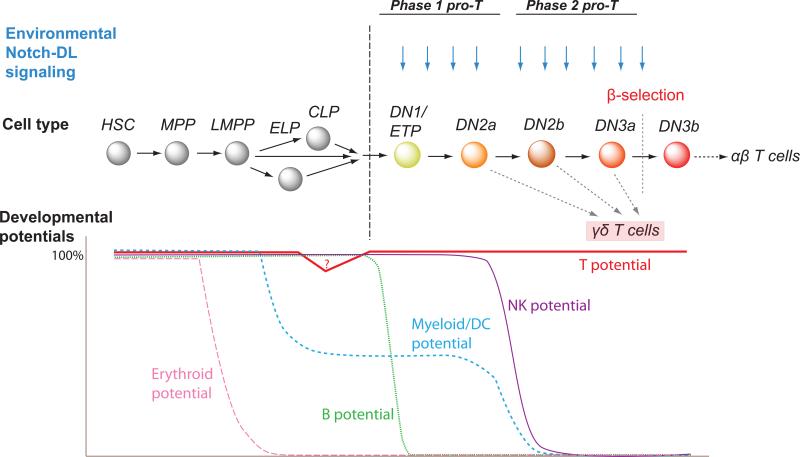
Hematopoietic stem cells begin with access to 10 or more distinguishable fates, including erythroid and megakaryocytic, macrophage, neutrophilic granulocyte, eosinophil, basophil, mast cell, conventional dendritic cell, plasmacytoid dendritic cell, natural killer (NK) cell, T lymphocyte, and B lymphocyte as well as their subtypes. In the T cell pathway, there is an ordered loss of access to these fates. Thus, commitment emerges as the end of a sequential process of lineage exclusions, summarized in Fig. 1. Before arrival in the thymus, precursors have already lost the ability to make erythroid cells and megakaryocytes, but the most immature cells in the thymus clearly retain access to dendritic cell, natural killer cell, macrophage and granulocyte, and probably also mast-cell and B cell potential in addition to T-cell potential. The B cell potential is rapidly extinguished (11). In fetal mouse T-cell development, B potential is lost even before the cells arrive in the thymus (12,13). However, NK, DC, and even macrophage and granulocyte (“myeloid”) potential remain evident until a later intrathymic stage, in both fetal and postnatal cases, if the cells are tested under permissive conditions. These NK and DC potentials then disappear at a distinct, later transition between “DN2” (c-Kit+ CD44+ CD25+) and “DN3” (c-Kitlow CD44- CD25+) (14-18), more finely mapping between the “DN2a” (c-Kit++) and “DN2b”(c-Kit+) stages (19,20). Single cells in the DN2 (DN2a) stage can still give rise to clones including T cells as well as NK cells, macrophages, or dendritic cells, or even granulocytes (15,16,18,21,22); in contrast, cells one stage later (DN2b stage) can no longer do this (19,20). This last step of alternative fate exclusion completes T lineage commitment.
Figure 1. Exclusion of different hematopoietic fate alternatives on the approach to T-cell identity.
The figure shows the changes in developmental potential described in the text, in the context of a likely sequence of hematopoietic precursors from bone marrow to thymic entry (vertical dashed line) through T-lineage commitment. Graphs below the stages shown represent levels of potential remaining at each stage for erythroid, myeloid/DC, B, and NK pathway alternative. In each case, “potential” is defined as an ability of a normal, unmodified cell to differentiate to a given fate when the cell is simply transferred to a permissive microenvironment. The uncertainty about different precursor types that may seed the thymus is shown as dual paths leading to thymic entry. A dip in “T cell potential” below the CLPs represents a subset of CLPs with reduced but detectable T cell potential. The apparent conditional decrease in myeloid potential seen for many prethymic precursors when assayed in vivo is depicted as a partial decrease of myeloid potential before the thymus, followed by a plateau of continuing potential until this fate is excluded by cell-intrinsic mechanisms at a later stage within the thymus. Cell types: HSC, hematopoietic stem cell; MPP, multipotent precursor; LMPP, lymphoid-primed multipotent precursor; ELP, early lymphoid precursor; CLP, common lymphoid precursor; DN1/ETP, c-Kit++ CD44+ CD25- DN cells (CD4- CD8- TCR- Lin- early T cells); DN2a, c-Kit++ CD44+ CD25+ DN cells; DN2b, c-Kit+ CD44+ CD25+ DN cells; DN3a, c-Kitlow CD44low CD25+ DN cells; DN3b, c-Kitlow CD44low CD25+ DN cells undergoing CD27 and CD28 upregulation and size increase triggered by β-selection. Later stages of αβ T cell development, not shown, include differentiation to CD4+ CD8+ cells after β-selection followed by selection of a few chosen survivors into CD4+ or CD8+ fates.
T-lineage gene expression is fully activated by the DN3 stage (20). The genes required for T-cell receptor (TCR) expression and function are turned on asynchronously, depending on gene-specific combinations of inputs from the transcription factors that are needed for early T-cell development, including GATA-3, TCF-1, basic helix-loop-helix (bHLH) E proteins E2A and HEB (Tcf12), Runx/CBFβ complexes, and the Notch-activated transcription factor RBPJκ (CSL) (23). In detail, the induction of T-cell genes still remains poorly explained. None of the essential factors seems dominant in the way that GATA-1 is for erythroid genes (24). However, these factors also participate individually in aspects of lineage exclusion, as described below.
Notch signaling in B and myeloid lineage exclusion
As long as immature thymocytes remain undisturbed in the thymus, their prevalent fate will normally be to generate T-cell progeny, with few NK, dendritic or myeloid progeny, and no B-cell progeny (25,26). Notch signaling makes this in vivo environment non-permissive for display of B, myeloid, and NK alternative potentials. Notch can block these alternative pathways even if the cells are forced to express ectopic transcription factors of other lineages that would otherwise impose lineage conversion (18,27-29). Thus, it is only when cells are removed from Notch signaling that distinct levels of restriction are seen. B lineage repression becomes independent of Notch signaling within a few cell divisions after thymic entry (11), when the cells are still in the initial “early T-cell precursor” (ETP/DN1, c-Kit++ CD44+ CD25-) compartment (Fig. 1), although Notch is still needed at that stage to keep myeloid potential at bay. Myeloid and dendritic lineage exclusions only become Notch independent later, at the DN2b stage (19,20) during commitment.
“Regulatory bridges” to myeloid and dendritic potential
Hematopoietic precursors, both in the bone marrow and in the early intrathymic stages, simultaneously express some levels of transcription factors that participate in disparate fates, as well as many of the target genes associated with these contradictory pathways (30,31). Indeed, early T cell precursors share certain transcription factors with the ensembles that guide differentiation of B and myeloid cells. T cells share with B cells the use of E proteins, Myb, Runx1/CBFβ, and Ikaros, and both initially share with myeloid cells the use of the ETS-family factor PU.1. In the myeloid case, potential appears to be controlled through effects on the shared factors that make a “bridge” from the T-cell pathway.
Myeloid potential reflects activity of the bZIP transcription factor C/EBPα together with the Ets-family factor PU.1 (32), which is also essential for generation of lymphoid-primed multipotent precursors (33,34). These play roles in virtually all myeloid cells, can upregulate each other (35), and together can convert even fibroblasts directly into macrophages (36). Myeloid growth factor receptor signaling from the environment can promote myeloid differentiation in part by post-translationally enhancing PU.1 activity (37-39). Since the myeloid growth factor receptors themselves are among the major target genes of PU.1 activation (40,41), PU.1 enables environmental myelopoietic growth factors to activate a positive feedback circuit to drive myeloid development. Both “myeloid” factors are present, or inducible, at stages when myeloid potential of thymocytes is highest. Prethymic precursors as well as ETP and DN2a pro-T cells all express moderately high levels of PU.1 (20,34). As the cells undergo commitment PU.1 is downregulated, with expression decreased by about a hundredfold by the time cells reach the DN3-stage checkpoint (20). Although C/EBPα expression is low in ETPs and silenced soon afterwards, lineage-tracing experiments also confirm that many T cells are indeed generated from C/EBPα+ precursors as well (42). Adding back either PU.1 or C/EBPα to committed, DN3 stage thymocytes restores myeloid potential; in the absence of Notch signals either factor can promote the reactivation of the other (27,28,43,44). These factors thus provide a clear way to understand why the ETP and DN2a thymocytes can still differentiate into myeloid cells, whereas DN2b and later stage cells normally cannot.
PU.1 is also crucial for development of almost all types of dendritic cells, in part due to its regulation of the growth factor receptor Flt3 (41). Although conventional and plasmacytoid dendritic cells develop from different programs that diverge sharply in their use of E proteins, both depend on PU.1 for generation (41) and require expression thereafter of PU.1-subfamily ETS factors, PU.1 or SpiB respectively (45). The ability of PU.1 and SpiB to support DC development can even be detected in the presence of some continuing Notch signaling (43) (M. M. Del Real and E. V. R., unpublished results), when C/EBP factors are not activated. Thus, it is not surprising that dendritic cell potential in thymocytes also closely tracks the natural expression of PU.1.
Lymphoid vs. myeloid lineage choice machinery
C/EBPα expression in thymocytes is limited by Notch signaling (46), possibly through the Notch-induced repressor Hes1 (47). It probably begins to be inhibited soon after thymic entry, since Notch targets including Hes1 are active in pro-T cells throughout the ETP to DN3a stages. Recent data from our own lab's genome-wide epigenetic mapping studies confirm that repression machinery has already begun to be recruited to the Cebpa gene, based on H3K27me3 histone marks, as early as the ETP/DN1 stage (J. Zhang, A. Mortazavi, B. Williams, B. J. Wold, E.V.R., unpublished); however, as noted above, it can still be reactivated at a later stage if PU.1 levels are high and Notch signals are removed. Under normal conditions, its expression limit may be set by the increasing dependence of pro-T cells on Notch signals for viability as they pass the DN2a to DN2b transition, discussed below (20).
The mechanism that represses PU.1 between the DN2a and DN3 stages is different. Two cis-regulatory regions of the Sfpi1 (PU.1) gene appear to mediate repression: a multifunctional Upstream Regulatory Element (48,49), and a recently described, cell-type specific silencer (50). Runx1/CBFβ complexes appear to repress at both elements. Two other factors may also contribute to PU.1 repression, namely TCF-1 (encoded by the Tcf7 gene) and Gfi1, both of which can work through sites near the Upstream Regulatory Element (51,52). Finally, GATA-3 appears to antagonize PU.1 expression with a very sensitive dose-response, although the mechanism may not be direct (29)[D. D. Scripture-Adams, A. M. Arias, K. J. Elihu, M. Zarnegar, and E. V. R., unpublished results]. The details of mechanism explaining the timing, magnitude, and near-permanence of the repression need further study, but it is notable that GATA-3, Runx1/CBFβ, and TCF-1 all play direct roles in the positive regulation of T-cell genes as well.
It is revealing to compare these lineage choice mechanisms in early T cells with the gene regulatory networks that have been proposed to explain other cell fate choices, where each cell type requires PU.1 but at different levels (52,53). In myeloid vs. B lineage fate choice, access to the B-cell fate appears to depend on PU.1 restrained by Ikaros plus E2A (52). The switch function at the heart of this network comprises “myeloid” Id2 and Egr2 on one side, induced by high-level PU.1, with Gfi1 on the other, which is turned on by E2A and Ikaros and itself helps to limit PU.1 expression. Superficially, the early pro-T cell regulatory environment seems like that of the early B cell, with strong expression of Ikaros and Gfi1 and steady activity of E proteins, and with little C/EBPα, Egr2, or Id2. However, although both early B and early T cells need to restrain PU.1 using shared lymphoid factors, in one respect their two networks give quite different results as PU.1's activity is antagonized. In early T cells, the B cell option has already been closed off irreversibly, even before Runx1, TCF-1, and Gfi1 help to silence PU.1.
Candidate mechanisms in B lineage exclusion
How can the B cell fate be excluded so early? The B cell fate should be connected to the T cell program by an overabundance of “regulatory bridges”. B cells share with T cells the use of RAG-dependent gene rearrangement to assemble the immunoglobulin superfamily immune receptors, a common sequence of immune receptor-dependent developmental checkpoints, and and extensive analogies in morphology and signal processing, as well as sustained requirements for transcription factors including E2A and Ikaros, and IL-7Rα/γc signaling. Furthermore, whereas the thymic stroma does not produce myeloid growth factors normally, it provides a rich source of the IL-7 that B cell precursors need to grow (3). This makes it easy to understand why the thymus fills with B cells when Notch signaling is impaired (26,54). However, even after just a few days of contact with Notch signals, thymus-settling precursors lose the ability to make B cells even when switched to B-cell conditions, while their ability to make myeloid cells, under optimal in vitro conditions, remains intact.
The B lineage exclusion mechanism must inactivate some crucial B-cell function that T and B cells do not share. The B-cell specifically expressed factors EBF1 and Pax5, both essential for B cell development in a mutual positive feedback loop (55), are likely to be the crucial control points. Neither of these factors is detectably expressed in any early thymocyte or in vitro differentiated pro-T cell population from the ETP stage onward (56-58). Heavy histone H3K27me3 marks, deposited across these loci as early as the ETP stage, also imply that these genes are silenced epigenetically (J. Zhang, A. Mortazavi, B. Williams, B. J. Wold, & E.V.R., unpublished). This correlates well with the cells’ inability to switch to the B-cell fate from the later ETP stage onward.
Nevertheless, it is not yet fully clear how Notch signaling terminates access to the B-cell fate. While there appears to be a mechanism through which Notch signaling temporarily interferes with EBF1 function in EBF1-expressing cells (59), the permanent transcriptional silencing of the Ebf1 and Pax5 loci needs another explanation. This is not an immediate Notch response like Hes1 induction, as several days of Notch signaling are required for full loss of B-cell potential (11,60). It is more coordinated with upregulation of GATA-3 and TCF-1, the first T-cell specific transcription factors to be induced (60). Even low-dose GATA-3 appears to be highly antagonistic to the B-cell fate (29,61-63)(D. D. Scripture-Adams, A. M. Arias, & E.V.R., unpublished). However, GATA-3 may not antagonize the B-lineage program alone, as Notch signaling can also block B-cell development in vitro from GATA-3 deficient cells (64). This GATA-3-independent effect of Notch could indicate the presence of another T-lineage specific B-cell exclusion system.
Blocking the path to the NK fate: “work never done”
The loss of NK potential, as measured by the ability to generate NK1.1.+, DX-5+, Perforin+ cells lacking T cell markers when supportive cytokines are provided, occurs after the DN2a stage, similarly timed but probably slightly later than the loss of myeloid and dendritic potential (18-20). NK cell fate is now understood as a complex cluster of programs (65), but in general they all have a great deal in common with T-cell subset programs. Most of the transcription factors NK cells use are shared with T cells, and there is a thymically derived lineage of NK cells that makes use of GATA-3 and IL-7R very much like T cells (66). Although genes expressed by NK cells are often used as markers for NK cell fate choice, most are also expressed by invariant NK T cells and some TCRγδ cells after their TCR-dependent selection. Thus, the problem with defining how T-cell precursors lose NK potential is that it is uncertain whether access to the “NK” program is ever fully lost at all, or simply restrained until a later stage when it can be redeployed for cytolytic T cell functions.
For years, the only candidate “master regulator” for NK cells was the helix-loop-helix antagonist Id2. This was a frustrating candidate. Not only is it also used in later T-cell development, but also it is not a DNA-binding factor itself at all, rather just a decoy that blocks the ability of bHLH transcription factors (E2A, HEB) to bind to the DNA. Thus, Id2 itself has no direct regulatory targets that could explain the network of NK differentiation. However, a great advance has been the discovery of a true sequence-specific DNA-binding factor, Nfil3 (E4bp4), that has a crucial role in the programming of NK cells, including induction of Id2 itself (67,68)(reviewed by (61)). Furthermore, at least one transcription factor has been described recently, Zfp105, which may be the most specific NK cell regulator yet (69). These factors are not part of the normal adult T-cell precursor regulatory repertoire, and so their regulation can be considered a key to NK potential (61).
Surprisingly, it is the NK pathway for which a T-lineage-specific, dedicated negative regulator has emerged first. This is the highly T-lineage-specific transcription factor, Bcl11b (70-72). Bcl11b expression is induced immediately before the loss of access to the NK option. Acute deletion of Bcl11b not only channels early T-cell precursors into an NK fate under T-cell conditions, but also allows fully committed T-cell precursors to back differentiate to NK cells or NK-like effectors if deletion is postponed to a later stage (72). If cells delete Bcl11b just during β-selection, there is a severe decrease in their lifespans as CD4+ CD8+ thymocytes, associated with aberrant activation of a number of mature cytolytic effector genes (73) as well as other precociously activated maturation regulators suggesting an NKT-like fate (74). Bcl11b loss allows the innate-immune cell transcription factors Nfil3 and Zbtb16 (PLZF) to be upregulated, as well as the highly NK-specific regulatory gene Zfp105 (71,72). Bcl11b also may repress Id2 via direct binding (74). Within the T lineage, Bcl11b is expressed continuously from the DN2b stage onward. Interestingly, its levels of expression vary, and are in fact lowest in activated effector CD8 cells and NKT cells. NK cells also express some Bcl11b but at lower levels still (http://www.immgen.org) (75). Thus Bcl11b appears to be needed as a continuing restraint on the deployment of NK-like functions, not only during early T-lineage commitment but throughout all later T-cell development.
Commitment irreversibility: loss of self-renewal potential
Bcl11b's effects extend more broadly than to repress NK functions. One of the phenotypes that is most striking in Bcl11b knockout pro-T cells is that they can also continue apparent self-renewal in a precommitment DN2a-like state, growing better than wildtype cells in the presence of Notch signals in vitro, but retaining DN2-like properties rather than progressing in differentiation (70,71). Like normal DN2a cells, they retain access to myeloid fates (if myeloid growth factors are supplied), though not B lineage fates, even after weeks of exposure to strong Notch signals (70,71). They not only preserve expression of PU.1 under these circumstances (70,71), but also preserve expression of a cluster of stem/progenitor associated regulatory genes that are expressed in early thymocytes and normally downregulated during commitment (20). These genes are otherwise implicated in aspects of precursor self-renewal (71). and appear to define an initial subprogram for pro-T cell expansion (23). Thus, Bcl11b normally both restrains the NK option and helps to end a discrete, precommitment expansion program (Fig. 1, “phase 1”). Notably, the termination of this “phase 1” program (Fig. 1) is tightly correlated with an abrupt transition to extreme Notch-dependence for survival (76), coinciding with commitment at the DN2b stage (20). The shift to acute Notch dependence itself can contribute to the timing of commitment, as it effectively applies a death penalty to any cell, from DN2b through β-selection, that ventures to sample an environment free of the developmental constraints of Notch signals.
Controversial commitment trees: the Common Lymphoid Precursor and its rivals
The use of distinct repression mechanisms to block different alternative potentials makes the T-lineage program quite versatile in its ability to use input cells from different prethymic commitment states. If the suite of transcription factors activated by Notch signals can separately exclude the B-cell fate, whether the myeloid fate has been excluded before or not, and separately block the myeloid fate, whether the B-cell fate has been excluded or not, then there is no need to postulate only one true pipeline of prethymic cells feeding the thymus. This is important because there is evidence that a range of different precursors can in fact provide input into the T-cell program (rev. (77); (78-80)), though in vivo they may reach the thymus differently and undergo differentiation vs. self-renewal expansion with different kinetics.
While these different prethymic precursors share NK and dendritic-cell potential, and B-cell potential if they come from postnatal animals, they differ in their perceived ability to make myeloid cells. Thus, CCR9+ Lymphoid-primed Multipotent Precursors (LMPP) have strong myeloid activity as well as lymphoid activity, while Common Lymphoid Precursors (CLP) (81) and other lymphoid-biased early lymphoid precursor subsets (82-84) appear to have much stronger B, NK, and T lymphoid potential than granulocyte or macrophage potential, at least in vivo. But this seems to pose a logical contradiction: if many thymus-settling cells have already lost myeloid potential, then how can myeloid potential also persist after B-cell potential is lost?
Heterogeneity of thymus-immigrating cells can provide one answer: the myeloid potential may be persisting only in descendants of those immigrants that were not CLP. Another answer, however, is that the myeloid potential in CLP is not actually lost, but rather under constraint. In fact, CLPs are also highly efficient at generating myeloid cells, similar to intrathymic ETP and DN2a cells, provided that they are tested similarly in optimal in vitro conditions (5,25). The largest differences in myeloid potential between CLPs and LMPPs are seen when the cells are assayed in vivo by adoptive transfer, in which they are allowed to determine their own homing to distinct microenvironments. Thus a strong possibility is that “traffic control” mechanisms specific to CLPs help to make them lymphoid-restricted in vivo, by directing them to lymphopoietic environments that are not fully permissive for myeloid development.
This is more plausible than it might appear, because even the bone marrow itself now appears to contain distinct domains that are permissive or restrictive for B lymphoid vs. myeloid differentiation (85). In vivo, a G-protein coupled receptor sensitive to pertussis toxin appears to keep CLP within the lymphopoietic domain and keep them out of the myeloid domain. This localization is important for their behavior as lymphoid restricted precursors, since these cells generate myeloid progeny instead, if pertussis toxin is used in vivo to block their attraction to B lymphopoietic domains (85). The myeloid growth factor receptor-triggered positive feedback with PU.1 provides a strong candidate mechanism that may be needed for an environment to be myeloid-permissive. On the other hand, bone marrow lymphopoietic zones that promote B and NK development are likely to protect the cells from these myeloid growth factor receptor-like signals and Toll-like receptor signals, either of which can readily induce myeloid differentiation from CLP (86-88). Thus the nature of the mechanisms that provide the first restraint on myeloid differentiation of prethymic lymphoid precursors in vivo may be powerful in vivo, but qualitatively different from the cell-intrinsic, transcriptional regulatory circuits that later extinguish myeloid potential by silencing first EBF1 and Pax5, and then PU.1 and C/EBPα expression during T-lineage commitment.
Conclusions
Lineage commitment in the T cell system might have been a single event driven by a single factor, like erythroid commitment driven by GATA-1, which combines positive and negative regulatory activities in a single potent factor (24). But there has been no evidence to support this. Instead, the successive losses of different options for T-cell development depend on the operation of molecularly distinct exclusion mechanisms. The environmental effects of Notch signaling are applied only during the commitment process, to activate the T-cell program. Then multiple regulators that are likely contributors to commitment – GATA-3, Bcl11b, TCF-1, and members of the Runx family – not only are turned on but remain active in all T-cell subsets at various levels thereafter. The roles of these factors in commitment may each be more to exclude a particular alternative than to cause “T lineage commitment” in toto. For example, GATA-3 which helps to block B cell development can even enhance myeloid development from certain precursors (89). T-cell commitment overall appears to be a mosaic of separable though interlocking parts.
Much debate has been focused on the right ways to consider hierarchies of relatedness in the lymphoid lineages based on the order of lineage exclusion. But mechanisms of exclusion that cause loss of a given developmental potential simply attack the points of vulnerability of the alternative program. They work based on the minimum number of repression targets needed to block access to that fate, not on the number of shared regulatory strands that otherwise make two programs related. Therefore, commitment mechanisms need not operate in order of a hierarchical “relatedness index” between two fates. The large number of shared genes between B and T fates leave points of vulnerability in the B-cell specific need for EBF1 and Pax5, and the T-cell specific need for Notch1, and these genes become foci for mutual B/T lineage exclusion. Conversely, the persistence of myeloid and dendritic options in early thymocytes may reflect not a hierchical proximity of these fates to the T-cell program, but a side effect of the versatility of PU.1. Despite providing a regulatory bridge to myeloid fates, PU.1-driven Flt3 expression and other growth-promoting effects are crucial for development of LMPP cells and possibly all prethymic cells. Myeloid potentials, easily checked by Notch signals in vivo, may simply be a temporary price paid for some other useful role of PU.1 in the self-renewing “phase 1” regulatory state of the earliest thymocytes.
If different lineage exclusions are mediated by distinct components of the T-cell program, each with its own separate regulation, then the idea that different decisions can be made in nonhierarchical or even variable order, e.g. between fetal and adult precursor cohorts, is not so problematic. We have much to learn about what controls the activation and coordination of different parts of the T-cell program, including the mechanisms of action of factors that play a role in lineage exclusion. However, the identity of T cells emerges not from a simple command switch but from the network of these factors’ interactions.
Acknowledgments
The author apologizes to many colleagues whose work could not be adequately cited due to space limitations.
Footnotes
Support was from USPHS grants CA90233, HL089123, DK073658, CA98925, CA148278, and AI083514, and the Albert Billings Ruddock Professorship at Caltech.
Reference List
- 1.Radtke F, Fasnacht N, MacDonald HR. Notch signaling in the immune system. Immunity. 2010;32:14–27. doi: 10.1016/j.immuni.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 2.Yuan JS, Kousis PC, Suliman S, Visan I, Guidos CJ. Functions of notch signaling in the immune system: consensus and controversies. Annu.Rev.Immunol. 2010;28:343–365. doi: 10.1146/annurev.immunol.021908.132719. [DOI] [PubMed] [Google Scholar]
- 3.Petrie HT, Zuniga-Pflucker JC. Zoned out: functional mapping of stromal signaling microenvironments in the thymus. Annu.Rev.Immunol. 2007;25:649–679. doi: 10.1146/annurev.immunol.23.021704.115715. [DOI] [PubMed] [Google Scholar]
- 4.Schlenner SM, Rodewald HR. Early T cell development and the pitfalls of potential. Trends Immunol. 2010;31:303–310. doi: 10.1016/j.it.2010.06.002. [DOI] [PubMed] [Google Scholar]
- 5.Ehrlich LI, Serwold T, Weissman IL. In vitro assays misrepresent in vivo lineage potentials of murine lymphoid progenitors. Blood. 2010 doi: 10.1182/blood-2010-05-287102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Souabni A, Cobaleda C, Schebesta M, Busslinger M. Pax5 promotes B lymphopoiesis and blocks T cell development by repressing Notch1. Immunity. 2002;17:781–793. doi: 10.1016/s1074-7613(02)00472-7. [DOI] [PubMed] [Google Scholar]
- 7.Krueger A, Willenzon S, Lyszkiewicz M, Kremmer E, Förster R. CC chemokine receptor 7 and 9 double-deficient hematopoietic progenitors are severely impaired in seeding the adult thymus. Blood. 2010;115:1906–1912. doi: 10.1182/blood-2009-07-235721. [DOI] [PubMed] [Google Scholar]
- 8.Zlotoff DA, Sambandam A, Logan TD, Bell JJ, Schwarz BA, Bhandoola A. CCR7 and CCR9 together recruit hematopoietic progenitors to the adult thymus. Blood. 2010;115:1897–1905. doi: 10.1182/blood-2009-08-237784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu C, Saito F, Liu Z, Lei Y, Uehara S, Love P, Lipp M, Kondo S, Manley N, Takahama Y. Coordination between CCR7- and CCR9-mediated chemokine signals in prevascular fetal thymus colonization. Blood. 2006;108:2531–2539. doi: 10.1182/blood-2006-05-024190. [DOI] [PubMed] [Google Scholar]
- 10.Svensson M, Marsal J, Uronen-Hansson H, Cheng M, Jenkinson W, Cilio C, Jacobsen SE, Sitnicka E, Anderson G, Agace WW. Involvement of CCR9 at multiple stages of adult T lymphopoiesis. J.Leukoc.Biol. 2008;83:156–164. doi: 10.1189/jlb.0607423. [DOI] [PubMed] [Google Scholar]
- 11.Heinzel K, Benz C, Martins VC, Haidl ID, Bleul CC. Bone marrow-derived hemopoietic precursors commit to the T cell lineage only after arrival in the thymic microenvironment. J.Immunol. 2007;178:858–868. doi: 10.4049/jimmunol.178.2.858. [DOI] [PubMed] [Google Scholar]
- 12.Harman BC, Jenkinson WE, Parnell SM, Rossi SW, Jenkinson EJ, Anderson G. T/B lineage choice occurs prior to intrathymic Notch signalling. Blood. 2005;106:886–892. doi: 10.1182/blood-2004-12-4881. [DOI] [PubMed] [Google Scholar]
- 13.Masuda K, Itoi M, Amagai T, Minato N, Katsura Y, Kawamoto H. Thymic anlage is colonized by progenitors restricted to T, NK, and dendritic cell lineages. J.Immunol. 2005;174:2525–2532. doi: 10.4049/jimmunol.174.5.2525. [DOI] [PubMed] [Google Scholar]
- 14.Wu L, Li C-L, Shortman K. Thymic dendritic cell precursors: relationship to the T lymphocyte lineage and phenotype of the dendritic cell progeny. J.Exp.Med. 1996;184:903–911. doi: 10.1084/jem.184.3.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu M, Tayu R, Ikawa T, Masuda K, Matsumoto I, Mugishima H, Kawamoto H, Katsura Y. The earliest thymic progenitors in adults are restricted to T, NK, and dendritic cell lineage and have a potential to form more diverse TCRβ chains than fetal progenitors. J.Immunol. 2005;175:5848–5856. doi: 10.4049/jimmunol.175.9.5848. [DOI] [PubMed] [Google Scholar]
- 16.Shen HQ, Lu M, Ikawa T, Masuda K, Ohmura K, Minato N, Katsura Y, Kawamoto H. T/NK bipotent progenitors in the thymus retain the potential to generate dendritic cells. J.Immunol. 2003;171:3401–3406. doi: 10.4049/jimmunol.171.7.3401. [DOI] [PubMed] [Google Scholar]
- 17.Balciunaite G, Ceredig R, Rolink AG. The earliest subpopulation of mouse thymocytes contains potent T, significant macrophage, and natural killer cell but no B-lymphocyte potential. Blood. 2005;105:1930–1936. doi: 10.1182/blood-2004-08-3087. [DOI] [PubMed] [Google Scholar]
- 18.Schmitt TM, Ciofani M, Petrie HT, Zúñiga-Pflücker JC. Maintenance of T cell specification and differentiation requires recurrent Notch receptor-ligand interactions. J.Exp.Med. 2004;200:469–479. doi: 10.1084/jem.20040394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Masuda K, Kakugawa K, Nakayama T, Minato M, Katsura Y, Kawamoto H. T cell lineage determination precedes the initiation of TCRβ rearrangement. J.Immunol. 2007;179:3699–3706. doi: 10.4049/jimmunol.179.6.3699. [DOI] [PubMed] [Google Scholar]
- 20.Yui MA, Feng N, Rothenberg EV. Fine-scale staging of T cell lineage commitment in adult mouse thymus. J.Immunol. 2010;185:284–293. doi: 10.4049/jimmunol.1000679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bell JJ, Bhandoola A. The earliest thymic progenitors for T cells possess myeloid lineage potential. Nature. 2008;452:764–767. doi: 10.1038/nature06840. [DOI] [PubMed] [Google Scholar]
- 22.Wada H, Masuda K, Satoh R, Kakugawa K, Ikawa T, Katsura Y, Kawamoto H. Adult T-cell progenitors retain myeloid potential. Nature. 2008;452:768–772. doi: 10.1038/nature06839. [DOI] [PubMed] [Google Scholar]
- 23.Rothenberg EV, Zhang J, Li L. Multilayered specification of the T-cell lineage fate. Immunol.Rev. 2010;238:150–168. doi: 10.1111/j.1600-065X.2010.00964.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kerenyi MA, Orkin SH. Networking erythropoiesis. J.Exp.Med. 2010;207:2537–2541. doi: 10.1084/jem.20102260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schlenner SM, Madan V, Busch K, Tietz A, Laufle C, Costa C, Blum C, Fehling HJ, Rodewald HR. Fate mapping reveals separate origins of T cells and myeloid lineages in the thymus. Immunity. 2010;32:426–436. doi: 10.1016/j.immuni.2010.03.005. [DOI] [PubMed] [Google Scholar]
- 26.Feyerabend TB, Terszowski G, Tietz A, Blum C, Luche H, Gossler A, Gale NW, Radtke F, Fehling HJ, Rodewald HR. Deletion of Notch1 converts pro-T cells to dendritic cells and promotes thymic B cells by cell-extrinsic and cell-intrinsic mechanisms. Immunity. 2009;30:67–79. doi: 10.1016/j.immuni.2008.10.016. [DOI] [PubMed] [Google Scholar]
- 27.Franco CB, Scripture-Adams DD, Proekt I, Taghon T, Weiss AH, Yui MA, Adams SL, Diamond RA, Rothenberg EV. Notch/Delta signaling constrains re-engineering of pro-T cells by PU.1. Proc.Natl.Acad.Sci.U.S.A. 2006;103:11993–11998. doi: 10.1073/pnas.0601188103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laiosa CV, Stadtfeld M, Xie H, Andres-Aguayo L, Graf T. Reprogramming of committed T cell progenitors to macrophages and dendritic cells by C/EBPα and PU.1 transcription factors. Immunity. 2006;25:731–744. doi: 10.1016/j.immuni.2006.09.011. [DOI] [PubMed] [Google Scholar]
- 29.Taghon T, Yui MA, Rothenberg EV. Mast cell lineage diversion of T lineage precursors by the essential T-cell transcription factor GATA-3. Nat.Immunol. 2007;8:845–855. doi: 10.1038/ni1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Månsson R, Hultquist A, Luc S, Yang L, Anderson K, Kharazi S, Al Hashmi S, Liuba K, Thorén L, Adolfsson J, Buza-Vidas N, Qian H, Soneji S, Enver T, Sigvardsson M, Jacobsen SEW. Molecular evidence for hierarchical transcriptional lineage priming in fetal and adult stem cells and multipotent progenitors. Immunity. 2007;26:407–419. doi: 10.1016/j.immuni.2007.02.013. [DOI] [PubMed] [Google Scholar]
- 31.Miyamoto T, Iwasaki H, Reizis B, Ye M, Graf T, Weissman IL, Akashi K. Myeloid or lymphoid promiscuity as a critical step in hematopoietic lineage commitment. Dev.Cell. 2002;3:137–147. doi: 10.1016/s1534-5807(02)00201-0. [DOI] [PubMed] [Google Scholar]
- 32.Laiosa CV, Stadtfeld M, Graf T. Determinants of lymphoid-myeloid lineage diversification. Annu.Rev.Immunol. 2006;24:705–738. doi: 10.1146/annurev.immunol.24.021605.090742. [DOI] [PubMed] [Google Scholar]
- 33.Arinobu Y, Mizuno S, Chong Y, Shigematsu H, Iino T, Iwasaki H, Graf T, Mayfield R, Chan S, Kastner P, Akashi K. Reciprocal activation of GATA-1 and PU.1 marks initial specification of hematopoietic stem cells into myeloerythroid and myelolymphoid lineages. Cell Stem Cell. 2007;1:416–427. doi: 10.1016/j.stem.2007.07.004. [DOI] [PubMed] [Google Scholar]
- 34.Nutt SL, Metcalf D, D'Amico A, Polli M, Wu L. Dynamic regulation of PU.1 expression in multipotent hematopoietic progenitors. J.Exp.Med. 2005;201:221–231. doi: 10.1084/jem.20041535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yeamans C, Wang D, Paz-Priel I, Torbett BE, Tenen DG, Friedman AD. C/EBPα binds and activates the PU.1 distal enhancer to induce monocyte lineage commitment. Blood. 2007;110:3136–3142. doi: 10.1182/blood-2007-03-080291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Feng R, Desbordes SC, Xie H, Tillo ES, Pixley F, Stanley ER, Graf T. PU.1 and C/EBPα/β convert fibroblasts into macrophage-like cells. Proc.Natl.Acad.Sci.U.S.A. 2008;105:6057–6062. doi: 10.1073/pnas.0711961105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rieske P, Pongubala JM. AKT induces transcriptional activity of PU.1 through phosphorylation-mediated modifications within its transactivation domain. J Biol Chem. 2001;276:8460–8468. doi: 10.1074/jbc.M007482200. [DOI] [PubMed] [Google Scholar]
- 38.Wang J-M, Lai M-Z, Yang-Yen H-F. Interleukin-3 stimulation of mcl-1 gene transcription involves activation of the PU.1 transcription factor through a p38 mitogen-activated protein kinase-dependent pathway. Mol.Cell Biol. 2003;23:1896–1909. doi: 10.1128/MCB.23.6.1896-1909.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Himes SR, Sester DP, Ravasi T, Cronau SL, Sasmono T, Hume DA. The JNK are important for development and survival of macrophages. J.Immunol. 2006;176:2219–2228. doi: 10.4049/jimmunol.176.4.2219. [DOI] [PubMed] [Google Scholar]
- 40.Tagoh H, Himes R, Clarke D, Leenen PJ, Riggs AD, Hume D, Bonifer C. Transcription factor complex formation and chromatin fine structure alterations at the murine c-fms (CSF-1 receptor) locus during maturation of myeloid precursor cells. Genes Dev. 2002;16:1721–1737. doi: 10.1101/gad.222002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carotta S, Dakic A, D'Amico A, Pang SH, Greig KT, Nutt SL, Wu L. The transcription factor PU.1 controls dendritic cell development and Flt3 cytokine receptor expression in a dose-dependent manner. Immunity. 2010;32:628–641. doi: 10.1016/j.immuni.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 42.Wölfler A, Danen-van Oorschot AA, Haanstra JR, Valkhof M, Bodner C, Vroegindeweij E, van Strien P, Novak A, Cupedo T, Touw IP. Lineage-instructive function of C/EBPα in multipotent hematopoietic cells and early thymic progenitors. Blood. 2010;116:4116–4125. doi: 10.1182/blood-2010-03-275404. [DOI] [PubMed] [Google Scholar]
- 43.Lefebvre JM, Haks MC, Carleton MO, Rhodes M, Sinnathamby G, Simon MC, Eisenlohr LC, Garrett-Sinha LA, Wiest DL. Enforced expression of Spi-B reverses T lineage commitment and blocks β-selection. J.Immunol. 2005;174:6184–6194. doi: 10.4049/jimmunol.174.10.6184. [DOI] [PubMed] [Google Scholar]
- 44.Dionne CJ, Tse KY, Weiss AH, Franco CB, Wiest DL, Anderson MK, Rothenberg EV. Subversion of T lineage commitment by PU.1 in a clonal cell line system. Dev Biol. 2005;280:448–466. doi: 10.1016/j.ydbio.2005.01.027. [DOI] [PubMed] [Google Scholar]
- 45.Watowich SS, Liu YJ. Mechanisms regulating dendritic cell specification and development. Immunol.Rev. 2010;238:76–92. doi: 10.1111/j.1600-065X.2010.00949.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Georgescu C, Longabaugh WJ, Scripture-Adams DD, David-Fung ES, Yui MA, Zarnegar MA, Bolouri H, Rothenberg EV. A gene regulatory network armature for T lymphocyte specification. Proc.Natl.Acad.Sci.U.S.A. 2008;105:20100–20105. doi: 10.1073/pnas.0806501105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sakata-Yanagimoto M, Nakagami-Yamaguchi E, Saito T, Kumano K, Yasutomo K, Ogawa S, Kurokawa M, Chiba S. Coordinated regulation of transcription factors through Notch2 is an important mediator of mast cell fate. Proc.Natl.Acad.Sci.U.S.A. 2008;105:7839–7844. doi: 10.1073/pnas.0801074105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hoogenkamp M, Lichtinger M, Krysinska H, Lancrin C, Clarke D, Williamson A, Mazzarella L, Ingram R, Jorgensen H, Fisher A, Tenen DG, Kouskoff V, Lacaud G, Bonifer C. Early chromatin unfolding by RUNX1: a molecular explanation for differential requirements during specification versus maintenance of the hematopoietic gene expression program. Blood. 2009;114:299–309. doi: 10.1182/blood-2008-11-191890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang G, Zhang P, Hirai H, Elf S, Yan X, Chen Z, Koschmieder S, Okuno Y, Dayaram T, Growney JD, Shivdasani RA, Gilliland DG, Speck NA, Nimer SD, Tenen DG. PU.1 is a major downstream target of AML1 (RUNX1) in adult mouse hematopoiesis. Nat Genet. 2008;40:51–60. doi: 10.1038/ng.2007.7. [DOI] [PubMed] [Google Scholar]
- 50.Zarnegar MA, Chen J, Rothenberg EV. Cell type-specific activation and repression of PU.1 by a complex of discrete, functionally specialized cis-regulatory elements. Mol.Cell Biol. 2010;30:4922–4939. doi: 10.1128/MCB.00354-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rosenbauer F, Owens BM, Yu L, Tumang JR, Steidl U, Kutok JL, Clayton LK, Wagner K, Scheller M, Iwasaki H, Liu C, Hackanson B, Akashi K, Leutz A, Rothstein TL, Plass C, Tenen DG. Lymphoid cell growth and transformation are suppressed by a key regulatory element of the gene encoding PU.1. Nat Genet. 2006;38:27–37. doi: 10.1038/ng1679. [DOI] [PubMed] [Google Scholar]
- 52.Spooner CJ, Cheng JX, Pujadas E, Laslo P, Singh H. A recurrent network involving the transcription factors PU.1 and Gfi1 orchestrates innate and adaptive immune cell fates. Immunity. 2009;31:576–586. doi: 10.1016/j.immuni.2009.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Laslo P, Spooner CJ, Warmflash A, Lancki DW, Lee HJ, Sciammas R, Gantner BN, Dinner AR, Singh H. Multilineage transcriptional priming and determination of alternate hematopoietic cell fates. Cell. 2006;126:755–766. doi: 10.1016/j.cell.2006.06.052. [DOI] [PubMed] [Google Scholar]
- 54.Wilson A, MacDonald HR, Radtke F. Notch 1-deficient common lymphoid precursors adopt a B cell fate in the thymus. J.Exp.Med. 2001;194:1003–1012. doi: 10.1084/jem.194.7.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mandel EM, Grosschedl R. Transcription control of early B cell differentiation. Curr.Opin.Immunol. 2010;22:161–167. doi: 10.1016/j.coi.2010.01.010. [DOI] [PubMed] [Google Scholar]
- 56.Tabrizifard S, Olaru A, Plotkin J, Fallahi-Sichani M, Livak F, Petrie HT. Analysis of transcription factor expression during discrete stages of postnatal thymocyte differentiation. J.Immunol. 2004;173:1094–1102. doi: 10.4049/jimmunol.173.2.1094. [DOI] [PubMed] [Google Scholar]
- 57.Dik WA, Pike-Overzet K, Weerkamp F, de Ridder D, de Haas EF, Baert MR, van der SP, Koster EE, Reinders MJ, van Dongen JJ, Langerak AW, Staal FJT. New insights on human T cell development by quantitative T cell receptor gene rearrangement studies and gene expression profiling. J.Exp.Med. 2005;201:1715–1723. doi: 10.1084/jem.20042524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.David-Fung E-S, Butler R, Buzi G, Yui MA, Diamond RA, Anderson MK, Rowen L, Rothenberg EV. Transcription factor expression dynamics of early T-lymphocyte specification and commitment. Dev.Biol. 2009;325:444–467. doi: 10.1016/j.ydbio.2008.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Smith EM, Åkerblad P, Kadesch T, Axelson H, Sigvardsson M. Inhibition of EBF function by active Notch signaling reveals a novel regulatory pathway in early B-cell development. Blood. 2005;106:1995–2001. doi: 10.1182/blood-2004-12-4744. [DOI] [PubMed] [Google Scholar]
- 60.Taghon TN, David E-S, Zúñiga-Pflücker JC, Rothenberg EV. Delayed, asynchronous, and reversible T-lineage specification induced by Notch/Delta signaling. Genes Dev. 2005;19:965–978. doi: 10.1101/gad.1298305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Klein Wolterink RGJ, García-Ojeda ME, Vosshenrich CA, Hendriks RW, Di Santo JP. The intrathymic crossroads of T and NK cell differentiation. Immunol.Rev. 2010;238:126–137. doi: 10.1111/j.1600-065X.2010.00960.x. [DOI] [PubMed] [Google Scholar]
- 62.Anderson MK, Hernandez-Hoyos G, Dionne CJ, Arias A, Chen D, Rothenberg EV. Definition of regulatory network elements for T-cell development by perturbation analysis with PU.1 and GATA-3. Devel.Biol. 2002;246:103–121. doi: 10.1006/dbio.2002.0674. [DOI] [PubMed] [Google Scholar]
- 63.Heavey B, Charalambous C, Cobaleda C, Busslinger M. Myeloid lineage switch of Pax5 mutant but not wild-type B cell progenitors by C/EBPα and GATA factors. EMBO J. 2003;22:3887–3897. doi: 10.1093/emboj/cdg380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hozumi K, Negishi N, Tsuchiya I, Abe N, Hirano K, Suzuki D, Yamamoto M, Engel JD, Habu S. Notch signaling is necessary for GATA3 function in the initiation of T cell development. Eur.J.Immunol. 2008;38:977–985. doi: 10.1002/eji.200737688. [DOI] [PubMed] [Google Scholar]
- 65.Ramirez K, Kee BL. Multiple hats for natural killers. Curr.Opin.Immunol. 2010;22:193–198. doi: 10.1016/j.coi.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vosshenrich CA, Garcia-Ojeda ME, Samson-Villeger SI, Pasqualetto V, Enault L, Goff OR, Corcuff E, Guy-Grand D, Rocha B, Cumano A, Rogge L, Ezine S, Di Santo JP. A thymic pathway of mouse natural killer cell development characterized by expression of GATA-3 and CD127. Nat.Immunol. 2006;7:1217–1224. doi: 10.1038/ni1395. [DOI] [PubMed] [Google Scholar]
- 67.Gascoyne DM, Long E, Veiga-Fernandes H, de Boer J, Williams O, Seddon B, Coles M, Kioussis D, Brady HJM. The basic leucine zipper transcription factor E4BP4 is essential for natural killer cell development. Nat.Immunol. 2009;10:1118–1124. doi: 10.1038/ni.1787. [DOI] [PubMed] [Google Scholar]
- 68.Kamizono S, Duncan GS, Seidel MG, Morimoto A, Hamada K, Grosveld G, Akashi K, Lind EF, Haight JP, Ohashi PS, Look AT, Mak TW. Nfil3/E4bp4 is required for the development and maturation of NK cells in vivo. J.Exp.Med. 2009;206:2977–2986. doi: 10.1084/jem.20092176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chambers SM, Boles NC, Lin KY, Tierney MP, Bowman TV, Bradfute SB, Chen AJ, Merchant AA, Sirin O, Weksberg DC, Merchant MG, Fisk CJ, Shaw CA, Goodell MA. Hematopoietic fingerprints: an expression database of stem cells and their progeny. Cell Stem Cell. 2007;1:578–591. doi: 10.1016/j.stem.2007.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ikawa T, Hirose S, Masuda K, Kakugawa K, Satoh R, Shibano-Satoh A, Kominami R, Katsura Y, Kawamoto H. An essential developmental checkpoint for production of the T cell lineage. Science. 2010;329:93–96. doi: 10.1126/science.1188995. [DOI] [PubMed] [Google Scholar]
- 71.Li L, Leid M, Rothenberg EV. An early T cell lineage commitment checkpoint dependent on the transcription factor Bcl11b. Science. 2010;329:89–93. doi: 10.1126/science.1188989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li P, Burke S, Wang J, Chen X, Ortiz M, Lee SC, Lu D, Campos L, Goulding D, Ng BL, Dougan G, Huntly B, Gottgens B, Jenkins NA, Copeland NG, Colucci F, Liu P. Reprogramming of T cells to natural killer-like cells upon Bcl11b deletion. Science. 2010;329:85–89. doi: 10.1126/science.1188063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Albu DI, Feng D, Bhattacharya D, Jenkins NA, Copeland NG, Liu P, Avram D. BCL11B is required for positive selection and survival of double-positive thymocytes. J.Exp.Med. 2007;204:3003–3015. doi: 10.1084/jem.20070863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kastner P, Chan S, Vogel WK, Zhang LJ, Topark-Ngarm A, Golonzhka O, Jost B, Le Gras S, Gross MK, Leid M. Bcl11b represses a mature T-cell gene expression program in immature CD4+CD8+ thymocytes. Eur.J Immunol. 2010;40:2143–2154. doi: 10.1002/eji.200940258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Heng TSP, Painter MW, Consortium TIGP. The Immunological Genome Project: networks of gene expression in immune cells. Nat.Immunol. 2008;9:1091–1094. doi: 10.1038/ni1008-1091. [DOI] [PubMed] [Google Scholar]
- 76.Ciofani M, Zúñiga-Pflücker JC. A survival guide to early T cell development. Immunol Res. 2006;34:117–132. doi: 10.1385/IR:34:2:117. [DOI] [PubMed] [Google Scholar]
- 77.Bhandoola A, von Boehmer H, Petrie HT, Zuniga-Pflucker JC. Commitment and developmental potential of extrathymic and intrathymic T cell precursors: plenty to choose from. Immunity. 2007;26:678–689. doi: 10.1016/j.immuni.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 78.Karsunky H, Inlay MA, Serwold T, Bhattacharya D, Weissman IL. Flk2+ common lymphoid progenitors possess equivalent differentiation potential for the B and T lineages. Blood. 2008;111:5562–5570. doi: 10.1182/blood-2007-11-126219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Serwold T, Ehrlich LI, Weissman IL. Reductive isolation from bone marrow and blood implicates common lymphoid progenitors as the major source of thymopoiesis. Blood. 2009;113:807–815. doi: 10.1182/blood-2008-08-173682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Saran N, Lyszkiewicz M, Pommerencke J, Witzlau K, Vakilzadeh R, Ballmaier M, von Boehmer H, Krueger A. Multiple extrathymic precursors contribute to T-cell development with different kinetics. Blood. 2010;115:1137–1144. doi: 10.1182/blood-2009-07-230821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kondo M, Weissman IL, Akashi K. Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell. 1997;91:661–672. doi: 10.1016/s0092-8674(00)80453-5. [DOI] [PubMed] [Google Scholar]
- 82.Medina KL, Garrett KP, Thompson LF, Rossi MI, Payne KJ, Kincade PW. Identification of very early lymphoid precursors in bone marrow and their regulation by estrogen. Nat.Immunol. 2001;2:718–724. doi: 10.1038/90659. [DOI] [PubMed] [Google Scholar]
- 83.Lai AY, Kondo M. Identification of a bone marrow precursor of the earliest thymocytes in adult mouse. Proc.Natl.Acad.Sci.U.S.A. 2007;104:6311–6316. doi: 10.1073/pnas.0609608104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Perry SS, Welner RS, Kouro T, Kincade PW, Sun XH. Primitive lymphoid progenitors in bone marrow with T lineage reconstituting potential. J.Immunol. 2006;177:2880–2887. doi: 10.4049/jimmunol.177.5.2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lai AY, Watanabe A, O'Brien T, Kondo M. Pertussis toxin-sensitive G proteins regulate lymphoid lineage specification in multipotent hematopoietic progenitors. Blood. 2009;113:5757–5764. doi: 10.1182/blood-2009-01-201939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Iwasaki-Arai J, Iwasaki H, Miyamoto T, Watanabe S, Akashi K. Enforced Granulocyte/Macrophage Colony-Stimulating Factor signals do not support lymphopoiesis, but instruct lymphoid to myelomonocytic lineage conversion. J.Exp.Med. 2003;197:1311–1322. doi: 10.1084/jem.20021843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kondo M, Scherer DC, Miyamoto T, King AG, Akashi K, Sugamura K, Weissman IL. Cell-fate conversion of lymphoid-committed progenitors by instructive actions of cytokines. Nature. 2000;407:383–386. doi: 10.1038/35030112. [DOI] [PubMed] [Google Scholar]
- 88.Welner RS, Pelayo R, Nagai Y, Garrett KP, Wuest TR, Carr DJ, Borghesi LA, Farrar MA, Kincade PW. Lymphoid precursors are directed to produce dendritic cells as a result of TLR9 ligation during herpes infection. Blood. 2008;112:3753–3761. doi: 10.1182/blood-2008-04-151506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Höflinger S, Kesavan K, Fuxa M, Hutter C, Heavey B, Radtke F, Busslinger M. Analysis of Notch1 function by in vitro T cell differentiation of Pax5 mutant lymphoid progenitors. J.Immunol. 2004;173:3935–3944. doi: 10.4049/jimmunol.173.6.3935. [DOI] [PubMed] [Google Scholar]