Abstract
Background: While screening programs have reduced the risk of infectious disease transmission by donors in human and veterinary blood banking, bacterial contamination of blood products has emerged as a major complication in human medicine.
Objectives: To describe a Pseudomonas fluorescens (Pf)‐contaminated feline packed RBC (pRBC) unit and experimentally investigate Pf‐contaminated canine pRBCs.
Methods: Canine pRBCs were inoculated with Pf‐rich pRBCs from the sentinel feline unit and stored at 4°C or 20°C for 72 hours. Aliquots from the pRBCs were serially evaluated by microscopy, culture, and a eubacterial 16S rRNA real‐time PCR assay.
Results: One Pf‐contaminated feline unit turned black after 22 days of storage and was removed from the blood bank; a source was not found, and no other contaminated units were identified. Canine pRBCs spiked with 5 or 25 μL of the sentinel unit became culture‐ and/or 16S PCR‐positive at ≥8 hours at 20°C and 48 hours at 4°C and developed a color change at ≥24 hours. Sensitivity studies indicated that without incubation, inoculation of ≥100 μL Pf‐rich pRBCs was necessary for a positive 16S PCR test result.
Conclusions: P. fluorescens grows in stored pRBCs slowly at 4°C and rapidly at 20°C. Screening of blood products for color change, estimating bacterial concentration with microscopy, and 16S PCR testing are simple and fast ways to detect bacteria in stored blood. Aseptic collection, temperature‐controlled storage, and regular visual monitoring of stored units is recommended. Discolored units should not be transfused, but examined for bacterial contamination or other blood product quality problems.
Keywords: Blood contamination, cat, dog, PCR, Pseudomonas fluorescens, transfusion
Introduction
Ensuring the safety of blood component transfusions is of the utmost importance in human and veterinary transfusion medicine. In addition to appropriate blood compatibility testing by blood typing and cross matching, infectious disease screening of the donors and donated units is crucial to assure the quality of blood components. Although strict guidelines ensure that only healthy donors are used, there is still the potential that donors can have subclinical viremia, bacteremia, or parasitemia and that the organisms could be transmitted via transfusion to a patient. Patients receiving blood transfusions are often debilitated and/or immunocompromised; therefore, transmission of infectious organisms can be a particularly devastating complication of transfusion therapy. Because emerging infectious diseases can potentially be transmitted through blood component transfusion, new screening technologies and schemes are being introduced in human and animal blood banks.
In contrast to human medicine, where the main focus has been on prevention of the transmission of viral agents, screening for donor bacteremia and parasitemia has been the major concern in dogs, as babesiosis and leishmaniasis following blood transfusion have been reported. 1 , 2 In cats, not only is there similar concern about transmission of blood‐borne bacteria and parasites, but also the transmission of viral agents. 3 , 4 , 5 Direct blood smear examination, serology, and antigen‐based assays (including PCR) are currently used techniques to screen canine and feline donors for infections. 4 , 5 , 6 As many of these organisms are vector‐transmitted, flea and tick prevention and treatment also play a pivotal role in the health of the donor population.
Aside from the risk of transmitting an infectious disease via blood components from an infected but clinically healthy blood donor to a patient, bacterial contamination of blood products during collection, processing, storage, and administration may occur and has emerged as a cause of morbidity and mortality in human transfusion medicine. 7 , 8 , 9 , 10 In particular, human platelet concentrates (stored at 20°C for up to 5 days and constantly agitated to remain functional) and packed RBC (pRBC) units (stored at 4°C for up to 40 days) have been found to be contaminated with bacteria at rates of 0.09–0.43% and ≤0.1%, respectively. 7 , 8 , 11 To minimize the risk of bacterial contamination, major efforts are ongoing to improve the techniques used to detect bacterial contamination of blood components in human blood banks. 7 , 8 , 10 , 12
While aseptic techniques are practiced in veterinary transfusion medicine, to our knowledge bacterial surveillance programs are not used. In the only published report of bacterial contamination of veterinary blood products, units of feline whole blood at 1 institution were found to be contaminated during collection by Serratia marcescens via the saline flush used for sedation of feline donors, which resulted in recipient morbidity and mortality. 13 We describe here the investigations surrounding bacterial contamination of a single feline pRBC unit (sentinel unit) and experimental bacterial contamination of canine pRBCs stored at 4°C and 20°C in which standard gross and microscopic examinations, bacterial culture, and a novel semiquantitative real‐time eubacterial 16S rRNA gene PCR test were used for blood product screening.
Materials and Methods
Blood donors, blood collection, and storage of pRBCs
The Penn Animal Blood Bank follows established standard operating procedures in accordance with guidelines set by the American Association of Blood Banks and the American Association of Veterinary Blood Banks. 7 , 14 Dogs donate blood through a community volunteer canine donor program. Hemoglobin concentration is measured at each donation and an annual screening includes a CBC, serum chemistry panel, and infectious disease testing for the following: Dirofilaria immitis antigen; antibodies against Borrelia burgdorferi, Ehrlichia canis, and Anaplasma phagocytophilum (Snap 4Dx, IDEXX Laboratories Inc., Westbrook, ME, USA), Bartonella sp. (FeBart, National Veterinary Laboratory Inc., Franklin Lakes, NJ, USA), and Babesia sp. (Protatek Reference Laboratory, Chandler, AZ, USA); and Bartonella sp., Babesia sp., Mycoplasma sp., Ehrlichia sp., Anaplasma sp., Leishmania sp., and Rickettsia rickettsii antigen using a real‐time PCR‐based assay (IDEXX Laboratories Inc.). In addition, all donors are required to have annual physical examinations and vaccinations by their regular veterinarian, and regular flea and tick control is strongly recommended to owners. Dogs are excluded as donors if they have hematologic or biochemical abnormalities, evidence of systemic abnormalities on physical examination, positive serologic results for E. canis or Bartonella sp., a titer ≥1:80 for Babesia sp., a positive test result for D. immitis antigen, or are PCR‐positive for any organism tested.
Feline donors are either part of a closed colony at the Veterinary Hospital of the University of Pennsylvania (originating from a specific‐pathogen‐free colony) or owned by hospital staff and veterinary students. All donor cats are kept indoors and must be free of fleas and ticks. Similar to the canine donors, hemoglobin concentration is measured at each donation, and donors undergo annual screening with a CBC, chemistry panel, and infectious disease testing. Infectious disease screening for cats includes tests for feline leukemia virus antigen and feline immunodeficiency virus antibody (Snap FIV/FeLV Combo, IDEXX Laboratories Inc.), coronavirus serology (Animal Health Diagnostic Center, Cornell University, Ithaca, NY, USA), and PCR testing for Mycoplasma haemofelis and ‘Candidatus M. haemominutum’ (Diagnostic Center for Population and Animal Health, Michigan State University, Lansing, MI, USA). Cats are excluded if any retroviral or PCR tests are positive or if they have hematologic or biochemical abnormalities.
For blood collection, feline donors are sedated with an intravenous injection into the medial saphenous or cephalic vein, while dogs donate without sedation. The sedative used for feline patients is taken from a multidose vial used exclusively by the blood bank for ∼8 weeks and swabbed with alcohol before use; a sterile needle is used and repeated entry into the vial with the same needle is not permitted. The area over 1 jugular vein is carefully shaved and cleaned with chlorhexidine 3 times followed by 70% alcohol. Scrub and alcohol‐soaked cotton balls are prepared individually for each donation in disposable plastic containers.
Standard, closed, 450 mL blood collection systems (Baxter Healthcare Corp., Fenwal Division, Deerfield, IL, USA) are used for canine blood collection. 15 To create an aseptic collection system for the small blood volumes of feline units (35–50 mL), we designed a closed system using a commercially available pediatric blood bag and apheresis products. 16 Single pediatric transfer bags (Pedi‐Pak Single, National Hospital Specialities, Hackensack, NJ, USA) are aseptically welded (Terumo SCD312 tube welder, Terumo Medical Corp., Elkton, MD, USA) to a 450 mL single blood unit containing citrate–phosphate–dextrose–adenine solution (CPDA‐1; Baxter Healthcare Corp.) in a laminar flow cell culture hood. Five milliliters of CPDA‐1 is transferred to each pediatric transfer bag in the standard ratio of 1 mL anticoagulant to 9 mL blood, after which the bag is welded to a single apheresis needle (SysLoc Safety A.V. Fistula Needle Set, JMS Singapore Pte Ltd., Singapore). The final product is heat‐sealed into a foil bag for storage (Kapak Pouches, Kapak Corp., Minneapolis, MN, USA) until immediately before use. Each batch of feline blood collection systems is assigned a lot number for tracking purposes.
From canine and feline donors, ∼450 and ∼40 mL of whole blood, respectively, are collected in citrate–phosphate–dextrose (CPD; Baxter Healthcare Corp.) and CPDA‐1, respectively. Canine and feline units are processed into pRBCs and fresh frozen plasma (FFP) within 8 and 4 hours of collection, respectively, by refrigerated centrifugation (5000g at 4°C) for 15 minutes with a slow break. Plasma is extracted into a satellite bag and frozen as FFP for up to 1 year at −30°C. Canine pRBCs are suspended in a preservative solution (Adsol, Baxter Healthcare Corp.) and feline units are suspended in approximately 10 mL of autologous plasma and immediately refrigerated in a blood‐storage refrigerator (Forma Scientific, Marietta, OH, USA) at 4°C for up to 33 and 26 days, respectively. Storage temperature deviations of the refrigerator are monitored with an alarm that is triggered by deviations of ±2°C, and a recording chart; temperature also is recorded manually in a logbook twice daily. Each pRBC unit is visually inspected daily for gross color changes, and rotated every‐other‐day by the nursing staff. If any color or other visible changes are noted in a unit at any time, the unit is removed from circulation for further evaluation and then is discarded. Every transfusion reaction at our hospital is investigated.
Inoculation of naïve units
Blood from the contaminated feline pRBC unit (sentinel, Pseudomonas fluorescens [Pf]‐rich pRBCs) was used to experimentally inoculate naïve canine pRBC units. Canine units were used because of their larger size and the limited supply of feline RBC products. One canine pRBC unit (∼250 mL) stored for 32 days was divided with a sterile tube welder into 8 smaller units of 25 mL each. A 1 mL sample of the original unit was aseptically removed before dividing, stored in a blood collection tube without additives, and frozen at −20°C for up to 7 days until testing. The 8 small units were divided into 2 groups for storage at 4°C or room temperature (∼20°C), similar temperatures to those used in a similar study with human blood. 17 In each group, 2 units served as controls (1 was not inoculated and the other was inoculated with 1 mL of sterile saline) and 2 units were inoculated with either 5 or 25 μL of Pf‐rich pRBCs from the sentinel unit diluted in 1 mL of sterile saline. All inoculation and sampling was performed using aseptic technique.
Units were thoroughly mixed before sampling and 1 mL aliquots were collected immediately after inoculation (0 hour) and at 4, 8, 24, 48, and 72 hours postinoculation. After 48 hours, the units being stored at 4°C were removed from refrigeration and placed at room temperature until the end of the study; the Pf‐inoculated units were also sampled at 52 hours. All aliquots were frozen at −20°C until testing, within 7 days. Digital photographs were taken at each time point to document gross color changes.
16S PCR test and bacterial identification by sequencing
DNA was extracted from a 200 μL aliquot of blood collected in EDTA from the collection system at the time of donation (QIAamp DNA Blood Mini Kit, Qiagen, Hilden, Germany). Real‐time PCR amplification was performed (SmartCycler, Cepheid, Sunnyvale, CA, USA) on a 5 μL sample using the primers 5′‐TCC TACGGGAGGCAGCAGT‐3′ and 5′‐GGACTACCAGGG TATCTAATCCTGTT‐3′ and a fluorescent‐labeled probe (6‐carboxyfluorescein‐aminohexyl amidite [FAM])‐5′CGTATTACCGCGGCTGCTGGCAC‐3′‐(Black Hole Quencher‐1) (Integrated DNA Technologies, Coralville, IA, USA) as described previously. 18 A total of 35 cycles of amplification were performed as follows: 94°C for 30 seconds, 61°C for 20 seconds, and 72°C for 5 seconds. The FAM fluorophore threshold is the level of background fluorescence and was set to 15 as the cutoff for a positive result. Negative controls consisted of 20 μL of PCR master mix (Omnimix, Cepheid) and 5 μL of water. All samples were run in duplicate with the mean of the cycle threshold values recorded.
Bacteria detected by the 16S PCR test were identified by sequencing the PCR product using a bacterial identification kit (Fast MicroSeq 500, ABI, Foster City, CA, USA), which ensured that a positive 16S PCR test was confirmed with a different set of PCR primers. The DNA sequence was compared with validated DNA sequences (MicroSeq Microbial Identification System Database, ABI).
To determine the detection limit of the 16S PCR assay and estimate the volume of contaminated blood needed for the 16S PCR assay to test positive, aliquots of an additional canine pRBC unit from a different donor were inoculated in duplicate with different volumes of Pf‐rich pRBCs from the sentinel unit and tested without incubation. Aliquots (25 mL) of canine pRBCs stored at 4°C for 32 days were aseptically transferred to sterile 50 mL plastic tubes and 2 aliquots of 0.5 mL each were placed into separate sterile tubes and frozen at −20°C until testing to serve as negative controls. Increasing volumes (5–750 μL) of Pf‐rich feline pRBCs from the sentinel unit were then added to the canine pRBCs. After each inoculation, the tubes were vigorously mixed with a vortexer for 30 seconds; two 0.5 mL aliquots were collected and frozen at −20°C until testing by 16S PCR. Final test concentrations were 0.2, 1, 2, 4, 10, 25, and 30 μL Pf‐rich pRBCs/mL of canine pRBCs.
Bacterial culture
Aerobic cultures of the sentinel unit (pRBCs and FFP), and the 3 additional canine units used in the inoculation study (Table 1) were incubated at 37°C and/or 20°C for up to 5 days using sheep blood and MacConkey agar plates (Remel, Lenexa, KS, USA). Isolates were identified as P. fluorescens using the Sensititre ARIS 2X automated bacterial identification system (Trek Diagnostic Systems, Cleveland, OH, USA).
Table 1.
Results of 16S PCR testing of canine pRBC units inoculated with 5 or 25 μL of the Pseudomonas fluorescens (Pf)–contaminated sentinel unit and stored at 4°C and 20°C for up to 72 hours.
| Storage Temperature | Inoculum (μL of Pf‐rich pRBCs) | 16S PCR Results | ||||||
|---|---|---|---|---|---|---|---|---|
| Incubation Period (Hours) | ||||||||
| 0 | 4 | 8 | 24 | 48 | 52 | 72 | ||
| 4°C (moved to 20°C after 48 hours) | 0 | − | − | − | − | − | ND | − |
| 0 (1 mL saline) | − | − | − | + (c−) | − | ND | − | |
| 5 | − | − | − | − | − | + | + | |
| 25 | − | − | − | − (c+) | + | + | + | |
| 20°C | 0 | − | − | − | − | − | ND | − |
| 0 (1 mL saline) | − | − | − | − | − | ND | − | |
| 5 | − | − | − | − | + | ND | + | |
| 25 | − | − | + | + (c+) | + | ND | + | |
Results for samples that were cultured are indicated as (c+), culture‐positive for P. fluorescens and (c−), culture negative. Shaded results indicate a color change was also observed.
+, positive; −, negative; ND, not done.
Estimate of bacterial load
To estimate the number of bacteria in each microliter of pRBCs, a blood smear was made from the sentinel unit at the time of color change and stained with a modified Wright–Giemsa stain (EM Diagnostics, Gibbstown, NY, USA) using an automatic stainer (MIDAS III, EMD Chemicals Inc., Darmstadt, Germany). The stain is changed and filtered daily and the boats are washed daily with methanol to ensure a clean staining process. A manual count of bacteria was performed by light microscopic examination: the number of bacterial rods observed was counted in 10–20 oil‐immersion × 100 objective fields (0.22 mm field diameter) and averaged. For the purpose of this study, the average bacterial count per oil‐immersion field was then multiplied by 15,000 to give an estimate of the number of colony‐forming units (CFU)/μL, and each bacterial rod was considered a CFU.
Results
Sentinel feline pRBC unit
A feline pRBC unit, which was collected from a young adult, male, healthy colony cat, processed from whole blood, and stored according to standard protocols (at 4°C) turned from a normal red color on day 21 of storage to black on day 22 of storage (Figure 1A and B). This was the donor's 21st donation, which was uneventful. The unit was immediately removed from the blood bank refrigerator for further testing. Although there was no evidence of a physical breach in the unit, the unit appeared hemolyzed, with a red‐colored supernatant, and on examination of a blood smear many extracellular rod‐shaped bacteria were observed. Bacteria were estimated at a concentration of ∼3.45 × 105 CFU/μL (∼3.45 × 108 CFU/mL) (Figure 1C). Aerobic bacterial culture of an aliquot of blood from this unit was negative following 5 days incubation at 37°C. A subsequent culture incubated at 20°C was positive for P. fluorescens after 72 hours. The 16S PCR test of an aliquot from this sentinel unit was positive for bacterial DNA, and subsequent DNA sequencing identified the organism as P. fluorescens.
Figure 1.
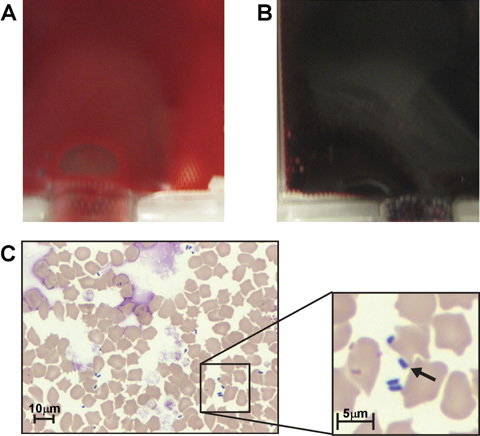
Gross visual and microscopic inspection results of a contaminated pRBC unit. (A) A regular feline pRBC unit compared with (B) the sentinel feline pRBC unit contaminated with Pseudomonas fluorescens, both at 22 days of storage. (C) Poikilocytosis and free bacterial rods in a smear of the sentinel unit of pRBCs. The arrow indicates 1 of 4 P. fluorescens organisms in the field. Modified Wright–Giemsa.
The FFP unit prepared from the same whole blood as the contaminated pRBCs and a 2nd feline pRBC unit that had been collected on the same day with a collection system having the same lot number were also withdrawn from the blood supply as a precautionary measure and tested. Neither yielded positive 16S PCR or culture results (at both 37°C and 20°C); a blood smear of this 2nd pRBC unit did not show any evidence of bacterial contamination. Other unused collection systems from the same lot were not tested, nor were the sedation vials.
The donor remained clinically healthy with no evidence of infection or trauma at the venipuncture site. 16S PCR tests of the donor's skin (both shaved and unshaved areas) and random items in the donor's environment and collection room were negative. Two nurses involved in the collection of all the feline blood products collected the day the sentinel unit was collected were not tested for contamination, although hand‐washing was performed before and after each donation. No other bacterial contamination or color change was observed in any of the other stored pRBCs units, which were used for >900 canine and feline transfusions in the 8 months before, and 10 months following the detection of the sentinel unit in August 2008. No cats receiving pRBC transfusions during this time period developed signs of acute hemolytic, anaphylactic, or septic transfusion reactions referable to bacterial contamination of blood products.
Bacterial contamination of naïve canine pRBC units
Based upon the estimated bacterial concentration in the smear from the sentinel unit, ∼1.73 × 106 CFU of P. fluorescens was present in the 5 mL inoculum (final concentration, 69 CFU/μL canine pRBCs) and ∼8.65 × 106 CFU was present in the 25 mL inoculum (final concentration, 345 CFU/μL canine pRBCs). Following inoculation of canine pRBCs units with the 5 or 25 μL aliquots of Pf‐rich pRBCs, those incubated at 20°C became positive by 16S PCR at 48 and 8 hours, respectively (Table 1). Of the units stored at 4°C, only the unit inoculated with 25 μL Pf‐rich pRBCs tested positive before changing incubation temperature at 48 hours. When, after 48 hours, the units stored at 4°C were incubated at 20°C, only those inoculated with aliquots from the sentinel unit tested 16S PCR positive. The only Pf‐inoculated unit stored at 4°C that was still negative at 48 hours tested 16S PCR positive within 4 hours of being moved to room temperature. All control units (no inoculation or inoculation with sterile saline) remained negative by 16S PCR testing regardless of incubation temperature, with 1 exception: an aliquot from the unit stored at 4°C and inoculated with sterile saline was 16S PCR positive at 24 hours; however, a culture of this aliquot at 20°C was negative, and 3 additional samples from this unit were 16S PCR negative after continued incubation (including incubation at 20°C) for 72 hours, suggesting the initial positive PCR result was spurious.
All pRBC units remained a normal red color until 24 hours when a color change to black was noted in the Pf‐inoculated units stored at 20°C. The color change was more pronounced in the unit inoculated with 25 μL than with 5 μL Pf‐rich pRBCs (Figure 2). When the refrigerated units were moved to 20°C, the first color change was noted at 24 hours. All units changed color after testing 16S PCR positive, except for 1 unit that changed color at 24 hours, but was 16S PCR negative at that time point (this sample was not cultured). At 72 hours, all control units were the same bright red color as when the study began, independent of storage temperature, and were 16S PCR negative; all inoculated units were dark red to black.
Figure 2.

Color changes in canine pRBC units spiked with Pseudomonas fluorescens (Pf) after 24 hours of incubation at (A) 4°C and (B) 20°C. The 25 mL units were either not inoculated (left), or inoculated with 1 mL sterile saline (left middle), 5 μL Pf‐rich feline pRBCs in 1 mL sterile saline (right middle), or 25 μL Pf‐rich feline pRBCs in 1 mL sterile saline (right).
When canine pRBCs were inoculated with increasing amounts of Pf‐rich pRBCs and immediately frozen for testing without incubation to determine the sensitivity of the 16S PCR test for detecting bacteria in canine pRBCs, only the 25 mL units inoculated with ≥100 μL of Pf‐rich pRBCs tested positive by 16S PCR, indicating an estimated final concentration of ∼1.38 × 103 CFU/μL of pRBCs.
Discussion
Despite the use of aseptic techniques in human and veterinary medicine, there are many iatrogenic sources of bacterial contamination, which may result in local or systemic life‐threatening infections in a patient. In particular, blood transfusion therapy has emerged as a potential source of iatrogenic septicemia in human patients, 7 , 9 , 10 , 12 , 19 , 20 , 21 while in veterinary medicine, only 1 incident has been reported. 13 In the present study, we describe and characterize the contamination and growth of P. fluorescens, and show that it is capable of growing in canine and feline pRBC units stored at both refrigerated and room temperatures.
In human transfusion medicine, estimates of pRBC contamination rates range from <0.001% to 0.1%. A ≥60% mortality rate has been estimated when contaminated units are transfused into patients, yet it is likely that some nonfatal reactions are not reported or are attributed to other sources (eg, surgery, trauma, or underlying illness). 7 , 11 Between 1976 and 1998, 26 fatalities were reported to be secondary to RBC transfusion‐related septicemia in humans, of which 53.8% were due to Yersinia entercolotica, 11.5% to Pseudomonas sp., and the rest to a variety of other, mostly Gram‐negative bacteria. 10 In another study, 2 of 21 Gram‐negative transfusion‐associated infections were due to P. fluorescens. 21 In a 3rd survey, P. fluorescens was implicated in 26.5% of cases of transfusion‐related septicemia and was cultured from 6 of 12 recipients of Pf‐contaminated blood. 19 As P. fluorescens is ubiquitous in soil and water, it is assumed that most cases of Pf‐associated septicemia are the result of contamination of donor skin or of seemingly sterile equipment and supplies. 22 P. fluorescens contamination of human blood products has been traced back to skin, 23 heparin flushes, 24 and, in a preliminary report, cold cloths, 25 although in many cases the source remains unknown. 26 Permanent skin damage (ie, scarring) from multiple donations can be associated with increased risk of infection, especially with platelet donations, as the advent of collecting platelet concentrates via pheresis have allowed human donors to donate platelets more frequently and therefore develop scarring of the venipuncture site. 27 , 28 The donor of the sentinel unit in this study had donated 20 previous times such that scarring and subsequent skin infection may have played a role despite the venipuncture site looking clean and healthy. Subsequent donations from this donor have not shown signs of contamination. The source of contamination of the isolated feline unit with P. fluorescens in this report could not be determined, despite considerable investigation of donor and blood bank environment, tools, and reagents.
Excluding cases of infections transmitted via the donor, 1 , 2 the only report of blood product contamination in veterinary medicine is that caused by S. marcescens contamination of saline flush and alcohol‐soaked cotton balls used during feline donor sedation for blood collection over a 7‐month period. 13 In that report, 29 of 174 units of feline whole blood were contaminated with S. marcescens. Unknowingly, 15 of the contaminated units were transfused to 14 patients, resulting in illness in 10 cats and death in 4 cats. Three of 5 units examined were discolored at the time of transfusion. Signs of a septic transfusion reaction in these cats included vomiting, pyrexia, collapse, panting, and acute death, consistent with septic shock. 13 , 29 In the present report, the Pf‐contaminated unit was removed from circulation as soon as the color change was noted, and, therefore, it was not transfused and did not cause harm to any patients.
P. fluorescens is a Gram‐negative rod that is ubiquitous in the environment, grows optimally at or below 30°C and as low as 2°C, and does not grow well at temperatures ≥37°C. These growth characteristics differentiate P. fluorescens from other pseudomonads such as P. aeruginosa, 22 , 30 , 31 , 32 and may explain why P. fluorescens was not isolated in the initial aerobic and anaerobic cultures at 37°C of the sentinel unit, although technical error cannot be excluded. Whole blood and pRBCs stored at 4°C provide an ideal growth medium for P. fluorescens and the potential for transfusing large numbers of this organism to patients must be seriously considered. 17 , 22 , 23 , 25 , 26 , 31 Clinical infection with P. fluorescens in cats and dogs is mostly limited to otitis externa 33 ; P. fluorescens has yet to be reported as a cause of transfusion‐related septicemia in animals. Despite faster growth at 30°C than at 5°C on glucose–salt–agar, growth at the latter temperature is possible. 34
When human blood contaminated with P. fluorescens was incubated at 20°C, the doubling time was 2 hours, whereas at 4°C it was 14.4 hours. 17 These doubling times correspond well to the estimates of the growth in experimentally contaminated canine pRBC units in this study. Approximately 2 doubling times would be needed for the 25 μL inoculum (∼345 CFU/μL) to reach a concentration of ≥1.38 × 103 CFU/μL and, therefore, test positive with the 16S PCR assay. At 20°C this would take ≥4 hours; the sample evaluated in this study tested positive between 4 and 8 hours. At 4°C this would take ≥28.8 hours, and the samples in this study tested positive between 24 and 48 hours. Similarly, it would take ∼5 doubling times for a 5 μL inoculum (∼69 CFU/μL) to reach a concentration of ≥1.38 × 103 CFU/μL; at 20°C this would take ≥10 hours (sample tested positive between 8 and 24 hours) and at 4°C this would take ∼72 hours. After 48 hours of incubation (∼3.3 doubling times and an estimated concentration of ∼700 CFU/μL), the refrigerated samples were moved to room temperature, at which point they still were negative by 16S PCR tests. At room temperature, only 1 additional doubling time (2 hours) was predicted to allow this sample to test positive, which it did 2–4 hours after the incubation temperature was changed.
In the present study, the sentinel unit changed color after 22 days of incubation at 4°C, which is estimated to be ∼36.7 doubling times at this temperature in human blood. 17 If the unit was contaminated at the time of collection then the initial inoculum must have been extremely minute (∼5 × 10−3 CFU/mL or ≤1 organism/pRBC unit) in order to reach a CFU count of 3.45 × 108 CFU/mL in 22 days of storage at 4°C. Gibb et al 17 showed that P. fluorescens had a growth plateau at a concentration of ∼3 × 108 CFU/mL human blood, which is similar to the number of organisms estimated in the sentinel unit when a color change was observed (∼3.45 × 108 CFU/mL).
In human transfusion medicine, different methodologies to detect bacterial contamination have been examined in an attempt to minimize transfusion‐associated sepsis. Screening tools include Gram stain, visual examination of color changes, swirling the units to identify changes in consistency, glucose measurement, and reagent strips to identify oxygen depletion by microbial organisms or to detect pH changes. 7 , 10 , 20 , 21 , 35 , 36 , 37 It has been postulated that the cause of the color change in contaminated units is a combination of decreased pO2 and hemolysis. 37 Methemoglobin may also contribute to the color change but was not measured in the present study because the reference laboratory would not accept contaminated blood for testing. To date, culture has been the gold standard for bacterial isolation in human blood products, however, some organisms such as P. fluorescens require nonstandard culture conditions for growth, and culture is an insensitive method if performed on the first day of collection when bacterial counts are low. 21
Recently, use of the eubacterial 16S PCR test to identify bacteria in the blood of bacteremic patients has been examined for screening blood products in human blood banks. 38 This method has been shown to detect 102–103 CFU/mL for some bacterial species. 21 The advantage of a real‐time PCR assay are its sensitivity and rapid detection of fastidious bacteria with reproducible results; however, the 16S PCR test also has limitations, including laboratory contamination and the detection of dead or degraded bacterial DNA, leading to false‐positive results. 39 , 40 In the present study, 3 of 4 Pf‐inoculated units tested positive with 16S PCR shortly before a gross color change was observed, while 1 control unit had 1 false‐positive 16S PCR test result (culture negative and subsequently 16S PCR negative) from among the 24 control samples tested across all time points. Furthermore, in 1 study that evaluated color changes in 15 stored pRBC units deliberately contaminated with 5 different concentrations of Y. enterocolitica, every contaminated unit showed a color change with prolonged storage that was preceded by positive bacterial culture, similar to our observations. 37
A review of accuracy studies of eubacterial 16S PCR screening in conjunction with blood cultures in human patients showed sensitivity and specificity ranged from 54% to 100% and 58% to 99%, respectively. 40 The sensitivity of eubacterial 16S PCR testing was 100% when the potential for septicemia was high (eg, newborn infants at risk for early‐onset sepsis and surgical patients with multiple trauma, major operations, or organ transplantation), although the total number of patients with positive cultures was low. 40 With the few experimentally inoculated canine units described here, it was not possible to determine sensitivity and specificity of the assay for bacterial detection in canine blood. Although bacterial colonies were not actually grown for quantification, CFU estimates based on microscopic review of a smear prepared from the sentinel unit suggested that ≥1.38 × 103 CFU/μL (1.38 × 106 CFU/mL) was needed for a positive result, which is 3 orders of magnitude higher than the 102–103 CFU/mL described for human blood. 21 Beside being cost prohibitive to screen all collected units, it would be challenging to use PCR testing as a screening tool for bacterial contamination of all animal blood products because they are often used immediately after collection, on‐site testing of units shortly before transfusion would not be generally available, and samples would need to be shipped to specific reference laboratories for 16S PCR testing, thus delaying results by 1–2 days. The assay may prove useful, however, for testing units that are approaching expiration or for following up units in which a color change has occurred or which resulted in transfusion reactions. Moreover, the PCR product can also be sequenced, permitting precise bacterial species identification. If 16S PCR testing is unavailable, screening tools such as evaluation of color change, Gram stain, and monitoring of pH and glucose concentration, as well as bacterial culture at 37°C and 20°C may prove useful. 7 , 20 , 35
Other guidelines in human blood banking are directed at bacterial avoidance at the time of collection and bacterial elimination before transfusion. Methods of bacterial avoidance include appropriate disinfection of the venipuncture site and diversion of the initial collection into a separate receptacle to avoid collection of a skin core at venipuncture that may be a nidus for contamination in the unit of blood. 7 , 10 , 21 Methods of bacterial elimination before transfusion include filtration and leukodepletion (to remove phagocytized bacteria), addition of antibiotics to blood products (a controversial approach), and inactivation of bacteria by irradiation and/or chemicals. 7 , 10 , 21 In veterinary blood banking, appropriate aseptic techniques are of the utmost importance, as these other methods of avoidance and elimination have not yet been verified clinically and may be impractical. 29
Based upon this study, P. fluorescens has the capacity to grow slowly in feline and canine pRBCs stored at cold temperatures and rapidly at room temperature. Because slow growth at refrigerated temperatures can lead to rapid growth at room temperature within 4 hours, transfusion of any blood unit should be administered as quickly as the patient will tolerate and in no more than 4 hours, consistent with guidelines set forth by the American Association of Blood Banks and the American Association of Veterinary Blood Banks. 7 , 14 , 29 , 41 Aseptic collection and processing methods and temperature‐controlled storage, along with regular visual evaluation of all blood products is also recommended. If gross abnormalities are noted, the products in question should be immediately removed from circulation and a blood smear, Gram stain, and additional testing such as 16S PCR should be considered. Although in the present case the contaminated unit was not transfused to a patient and a source of contamination or storage temperature error was not determined, bacterial contamination of blood products could be the first indication of a systemic error in the blood banking process and therefore should be thoroughly investigated.
Acknowledgment
The authors would like to extend their gratitude to Dr. Karen Jackson for the cytology review.
References
- 1. Stageman JR, Birkenheuer AH, Kruger JM, Breitschwerdt EB. Transfusion‐associated Babesia gibsoni infection in a dog. J Am Vet Med Assoc. 2003;222:959–963. [DOI] [PubMed] [Google Scholar]
- 2. Owens SD, Oakley DA, Marryott K, et al Transmission of visceral leishmaniasis through blood transfusions from infected English foxhounds to anemic dogs. J Am Vet Med Assoc. 2001;219:1076–1083. [DOI] [PubMed] [Google Scholar]
- 3. Gary AT, Richmond HL, Tasker S, Hackett TB, Lappin MR. Survival of Mycoplasma haemofelis and ‘Candidatus Mycoplasma haemominutum’ in blood of cats used for transfusions. J Fel Med Surg. 2006;8:321–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Reine NJ. Infection and blood transfusion: a guide to donor screening. Clin Tech Small Anim Pract. 2004;16:68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wardrop KJ, Reine N, Birkenheuer A, et al Canine and feline blood donor screening for infectious disease. J Vet Intern Med. 2005;19:135–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hackett TB, Jensen WA, Lehman TL, et al Prevalence of DNA of Mycoplasma haemofelis, ‘Candidatus Mycoplasma haemominutum’, Anaplasma phagocytophilu, and species of Bartonella, Neorickettsia, and Ehrlichia in cats used as blood donors in the United States. J Am Vet Med Assoc. 2006;229:700–705. [DOI] [PubMed] [Google Scholar]
- 7. Vengelen‐Tyler V. Blood Collection, Storage, and Component Preparation Methods. Technical Manual. 14th ed Bethesda, MD: American Association of Blood Banks; 2002. [Google Scholar]
- 8. Kuehnert MJ, Roth VR, Haley NR, et al Transfusion‐transmitted bacterial infection in the United States, 1998 through 2000. Transfusion. 2001;41:1493–1499. [DOI] [PubMed] [Google Scholar]
- 9. Hogman CF, Engstrand L. Serious bacterial complications from blood components – how do they occur? Transfus Med. 1998;8:1–3. [DOI] [PubMed] [Google Scholar]
- 10. Reading FC, Brecher ME. Transfusion‐related bacterial sepsis. Curr Opin Hematol. 2001;8:380–386. [DOI] [PubMed] [Google Scholar]
- 11. Vamvakas EC, Blajchman MA. Transfusion‐related mortality: the ongoing risks of allogeneic blood transfusion and the available strategies for their prevention. Blood. 2009;113:3406–3417. [DOI] [PubMed] [Google Scholar]
- 12. Perrotta PL, Snyder EL. Non‐infectious complications of transfusion therapy. Blood Rev. 2001;15:69–83. [DOI] [PubMed] [Google Scholar]
- 13. Hohenhaus AE, Drusin LM, Garvey MS. Serratia marcescens contamination of feline whole blood in a hospital blood bank. J Am Vet Med Assoc. 1997;210:794–798. [PubMed] [Google Scholar]
- 14. Hale AS, Kaufman P, Ziller M. Standards for Blood Banks and Transfusion Services. Orland, CA: American Association of Veterinary Blood Banks; 2005. [Google Scholar]
- 15. Schneider A. Blood components. Collection, processing, and storage. Vet Clin North Am Small Anim Pract. 1995;25:1245–1261. [DOI] [PubMed] [Google Scholar]
- 16. Springer T, Hatchett WL, Oakley DA, Niggemeier A, Giger U. Feline blood storage and component therapy using a closed collection system [abstract]. J Vet Intern Med. 1998;12:248. [Google Scholar]
- 17. Gibb AP, Martin KM, Davidson GA, Walker B, Murphy WG. Rate of growth of Pseudomonas fluorescens in donated blood. J Clin Pathol. 1995;48:717–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nadkarni MA, Martin FE, Jacques NA, Hunter N. Determination of bacterial load by real‐time PCR using a broad‐range (universal) probe and primers set. Microbiology. 2002;148:257–266. [DOI] [PubMed] [Google Scholar]
- 19. Wagner SJ, Friedman LI, Dodd RY. Transfusion‐associated bacterial sepsis. Clin Microbiol Rev. 1994;7:290–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Murphy WG, Foley M, Doherty C, et al Screening platelet concentrates for bacterial contamination: low numbers of bacteria and slow growth in contaminated units mandate an alternative approach to product safety. Vox Sang. 2008;95:13–19. [DOI] [PubMed] [Google Scholar]
- 21. Brecher ME, Hay SN. Bacterial contamination of blood components. Clin Microbiol Rev. 2005;18:195–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Forbes BA, Sahm DF, Weissfeld AS. Bailey & Scott's Diagnostic Microbiology. 12th ed St Louis, MO: Mosby; 2007:340–350. [Google Scholar]
- 23. Puckett A, Davison G, Entwistle CC, Barbara JAJ. Post‐transfusion septicaemia 1980–1989: importance of donor arm cleansing. J Clin Pathol. 1992;45:155–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gershman MD, Kennedy DJ, Noble‐Wang J, et al Multistate outbreak of Pseudomonas fluorescens bloodstream infection after exposure to contaminated heparinized saline flush prepared by a compounding pharmacy. Clin Infect Dis. 2008;47:1372–1379. [DOI] [PubMed] [Google Scholar]
- 25. Chaffin DJ, Kuehnert MJ. Pseudomonas fluorescens‐related septic transfusion reaction resulting from contaminated cold cloths [abstract]. Transfusion. 2002;42 (Suppl):41S. [Google Scholar]
- 26. Khabbaz RF, Arnow PM, Highsmith AK, et al Pseudomonas fluorescens bacteremia from blood transfusion. Am J Med. 1984;76:62–68. [DOI] [PubMed] [Google Scholar]
- 27. Gibson T, Norris W. Skin fragments removed by injection needles. Lancet. 1958;2:983–985. [DOI] [PubMed] [Google Scholar]
- 28. Anderson KC, Lew MA, Gorgone BC, Martel J, Leamy CB, Sullivan B. Transfusion‐related sepsis after prolonged platelet storage. Am J Med. 1986;81:405–411. [DOI] [PubMed] [Google Scholar]
- 29. Harrel KA, Kristensen AT. Canine transfusion reactions and their management. Vet Clin North Am Small Anim Pract. 1995;25:1333–1364. [DOI] [PubMed] [Google Scholar]
- 30. Redy CA. Methods for General and Molecular Microbiology. 3rd ed Washington, DC: ASM Press; 2007:232. [Google Scholar]
- 31. Ingraham JL. Growth of psychrophilic bacteria. J Bacteriol. 1958;76:75–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Madigan MT, Martinko JM, Parker J. Brock Biology of Microorganisms. 9th ed Upper Saddle River, NJ: Prentice Hall; 2000:470–474. [Google Scholar]
- 33. Barrasa MJL, Gomez LP, Lama GZ, Junco TMT. Antibacterial susceptibility patterns of Pseudomonas strains isolated from chronic canine otitis externa. J Vet Med B Infect Dis Vet Public Health. 2000;47:191–196. [DOI] [PubMed] [Google Scholar]
- 34. Johnson MG, Palumbo SA, Rieck VT, Witter LD. Influence of temperature on steady‐state growth of colonies of Pseudomonas fluorescens . J Bacteriol. 1970;103:267–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Burstain JM, Brecher ME, Workman K, Foster M, Faber GH, Mair D. Rapid identification of bacterially contaminated platelets using reagent strips: glucose and pH analysis as markers of bacterial metabolism. Transfusion. 1997;37:255–258. [DOI] [PubMed] [Google Scholar]
- 36. Goldman MR. Should we attempt to detect bacteria in red blood cells? Transfusion. 2008;48:1538–1540. [DOI] [PubMed] [Google Scholar]
- 37. Kim DM, Brecher ME, Bland LA, Estes TJ, Carmen RA, Nelson EJ. Visual identification of bacterially contaminated red cells. Transfusion. 1992;32:221–225. [DOI] [PubMed] [Google Scholar]
- 38. Dreier J, Stormer M, Kleesiek K. Real‐time polymerase chain reaction in transfusion medicine: applications for detection of bacterial contamination in blood products. Transfus Med Rev. 2007;21:237–254. [DOI] [PubMed] [Google Scholar]
- 39. Klouche M, Schroder U. Rapid methods for diagnosis of bloodstream infections. Clin Chem Lab Med. 2008;46:888–908. [DOI] [PubMed] [Google Scholar]
- 40. Peters RPH, Van Agtmael MA, Danner SA, Savelkoul PHM, Vandenbroucke‐Grauls CMJE. New developments in the diagnosis of bloodstream infections. Lancet Inf Dis. 2004;4:751–760. [DOI] [PubMed] [Google Scholar]
- 41. Kristensen AT, Feldman BF. General principles of small animal blood component administration. Vet Clin North Am Small Anim Pract. 1995;25:1277–1290. [DOI] [PubMed] [Google Scholar]


