Abstract
The thiamine pyrophosphate (TPP) riboswitch employs modular domains for binding TPP to form a platform for gene expression regulation. Specifically, TPP binding triggers a conformational switch in the RNA from a transcriptionally active “on” state to an inactive “off” state that concomitantly causes the formation of a terminator hairpin and halting of transcription. Here, clustering analysis of energy landscapes at different nucleotide lengths suggests a novel computational tool for analysis of the mechanics of transcription elongation in the presence or absence of the ligand. Namely, we suggest that the riboswitch’s kinetics are tightly governed by a length-dependent switch, whereby the energy landscape has two clusters available during transcription elongation, and where TPP’s binding shifts the preference to one form. Significantly, the biologically active and inactive structures determined experimentally matched well the structures predominant in each computational set. These clustering/structural analyses combined with modular computational design suggest design principles that exploit the above features to analyze as well as create new functions and structures of RNA systems.
Keywords: thiamine pyrophosphate (TPP) riboswitch, aptamer, energy landscape, cluster analysis, gene regulation, non-coding RNA
Introduction
Riboswitches regulate gene expression by exploiting the effects of selective binding of small molecules on secondary and tertiary structural changes.1,2 In prokaryotes, these modular RNA elements are normally found in the 5′ untranslated regions (5′ UTR) of genes and affect expression levels through various mechanisms: formation or destruction of transcription terminator hairpins,3–5 sequestration of ribosome binding-sites,6 or emergence of alternative cleavage sites.7–9
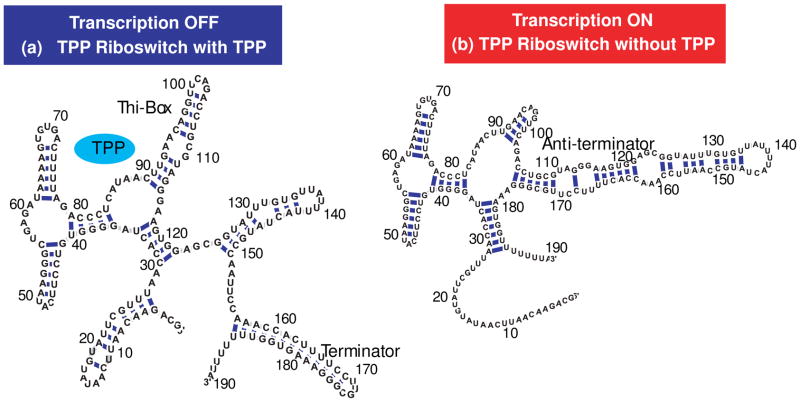
The thiamine pyrophosphate (TPP) riboswitch is a prominent example. This riboswitch can adapt two structures, depending on whether it is bound or unbound to TPP.10,11 TPP can bind only when the secondary structure that links the aptamer domain to the expression platform, termed thi-box, forms, as shown in Figure 1a. However, it is currently not known how this riboswitch achieves such specific structural changes without the aid of proteins. Ligand binding triggers a conformational switch in the entire RNA from a transcriptionally active “on” state to an inactive “off” state that causes the formation of a terminator hairpin. Due to a high activation barrier between these states, we suggest that this “conformational switch” is actually regulated by a thermodynamic/kinetic mechanism whereby the presence or absence of the ligand favors one particular folding pathway and shifts the folded state from one structural conformational to the other. That is, if the ligand is absent, the anti-terminator conformation is favored and transcription of the downstream gene is turned on (Figure 1b); when TPP is present, that cascade of events is hampered due to the terminator hairpin configuration. Previous studies on the function of FMN and TPP terminator hairpin riboswitches in Bacillus subtilis11 suggested a model for small molecule binding to nascent RNA riboswitches, whereby a decision between two alternative structures is likely to be made during transcription. Fluorescent experiments of the full-length thiM TPP riboswitch led to the conclusion that little to no structural changes occur upon TPP binding,12 while shorter riboswitch intermediates displayed competing alternative folds.
Figure 1.
Two alternative structures of thiamine pyrophosphate (TPP) riboswitch in Bacillus subtilis (B. subtilis). (a) Full-length riboswitch when TPP present. Thi-box and terminator are formed (lowest energy structure predicted by mfold). (b) TPP riboswitch when TPP is absent. Anti-terminator is paired with terminator (lowest energy structure with the constraint that prohibits base pairing in terminator stem-loop in (a)).
In this work, we examine this structural-equilibrium regulation in the TPP riboswitch as well as related systems using various computational tools. Our investigations show that the predominant structure in the nascent RNA is conformationally sensitive to the binding of TPP, which is present in the RNA up to a specific length (170 nt), after which the more stable structure is the anti-terminator structure. This mechanism points to a “switching threshold” for the riboswitch folding pathway, where one functional state is favored over the other at a critical length of the RNA. Our structural and thermodynamic analyses during transcription elongation help interpret how the presence or absence of the ligand regulates this conformation-dependent gene expression process and thus suggest an application of the tools offered here to RNA design.
Results
In the first subsection, we describe simulation results of the transcription folding process by structural and clustering analysis. These analyses help interpret key structural changes occurring when TPP binds and transcription terminates. For structural analysis, intermediate sequences are folded into their native secondary structures by free energy minimization. For thermodynamic analysis, we sample alternative secondary structures and plot the distance between the minimum free energy structure and all other structures against the free energy of folding to illustrate the folding pathways, which we call the energy landscape plot. Using a clustering algorithm, we partition the entire set of structures into their respective clusters and analyze the most dominant secondary structural features in the clusters (see details in Methods). These results are summarized in Table 1.
Table 1.
Riboswitches and energy landscape clustering. Clusters in each energy landscape are determined using the k-means algorithm (see Methods). Proportions of structures are based off of the set containing the transcriptionally inactive, ligand-binding structures.
| Riboswitch Name (reference) | Ligand | Number of Clusters in Structure Set | Proportion of structures in termination set |
|---|---|---|---|
| tenA, B. subtilis11 | Thiamine Pyrophosphate (TPP, Vit. B1) | 2 | 85% |
| 118 mutant | TPP | 1 | ~100% |
| 30 mutant | TPP | 2 | 39.8% |
| 80 mutant | TPP | 2 | 87% |
| 97 mutant | TPP | 2 | 8.1% |
| ribD, B. subtilis11 | Flavin Mononucleotide (FMN) | 2 | 85% |
| ypaA, B.subtilis18,19 | FMN | 2 | 77% |
| gcvT, B. subtilis22 | Glycine | 1 | ~100% |
| VCI-II, V. cholera 22 | Glycine | 1 | 51% |
| btuB, E. coli20 | Coenzyme B12 (AdoCbl) | 2 | 92.3% |
| thiM, E. coli4 | TPP | 2 | 35.1% |
Switches of TPP riboswitch at 170 nt (State 1) and at 175 nt (State 2)
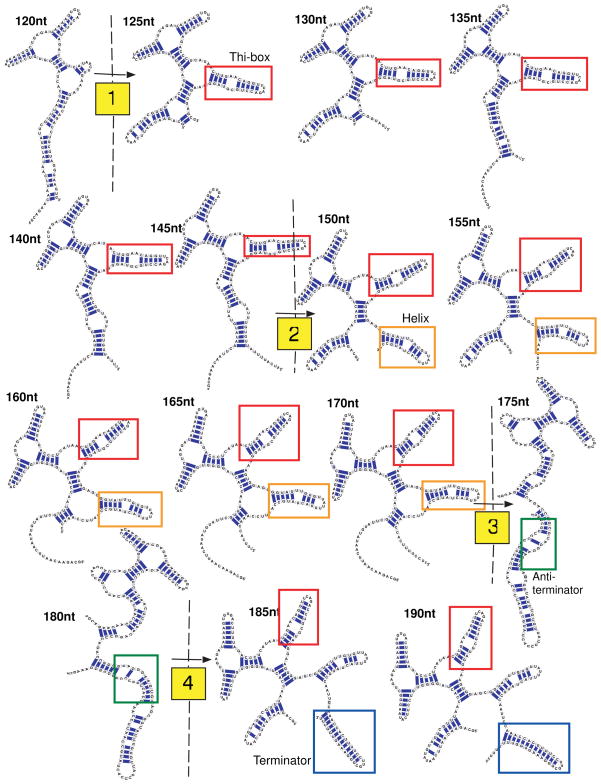
Structural predictions for the TPP riboswitch from 50 nt to the full-length 190 nt sequence were performed in 5 nt increments using secondary structure prediction tools such as mfold 43, as described in Methods. The thi-box domain 5′ start site begins at +77 nt relative to the transcription start site and ends at +122 nt.13 At each length, we “folded” the subsequence into its minimum free energy structure for comparison. As Figure 2 shows, from 125 until 170 nt the RNA riboswitch forms a structure that would permit ligand-binding, since the thi-box is fully formed. Based on the availability and energetic favorability of this ligand-binding domain, we propose a model where complete transcription of the TPP binding region (+121), remains intact up until 170 nt. During this length interval, the riboswitch can sense or detect the presence of intracellular ligand. Preorganization of the ligand-binding-domain has been confirmed through force-based spectroscopy14 and crystallographic evidence.2
Figure 2.
TPP riboswitch folding pathway snapshots from 120 nt to 190nt (full length) with the 5nt incremental step. Note four large-scale structural changes (120 to 125 nt, 145 to 150 nt, 170 to 175 nt, and 180 to 185 nt).
If the ligand is present at sufficiently high concentrations for binding, the ligand-bound configuration of the RNA (Figure 1a) is stabilized, and thus formation of alternative structures is prevented. Support for this theory comes from the high specificity and strength of binding between the thi-box domain and ligand4 as well as a similar model for an FMN-sensitive riboswitch.11
At a particular transcriptional length, the non-ligand bound state can become energetically favored, and a concomitant misfolding of the thi-box results. Past 175 nt, the favored RNA structure has different topology (Figure 2) and corresponds to that which forms the anti-terminator hairpin. This configuration may arise in TPP-poor environments due to the lack of stability provided by ligand-binding.
Clustering of energy landscape of TPP riboswitches: 2 groups or 1 group
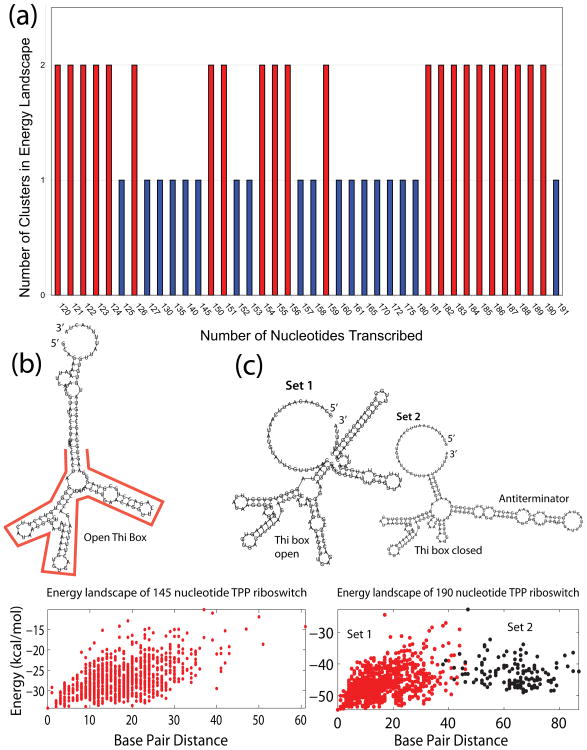
We evaluate the feasibility of the above model by performing a series of energy landscape plots at varying transcriptional lengths. We generate the energy landscape plots for the TPP riboswitch from 50 to 190 nt in 5 or 1 nucleotide increments. Using clustering techniques (see Methods), we found that the entire space of the sequence displays two distinct clusters in the energy plot (Figure 3). As shown in Figure 3a, the RNA forms two dominant clusters at the two end sections of the elongation process (120–124 nt and 181–190 nt), and these represent the two riboswitch conformations. The energy plot of 145 nt shows one cluster (Figure 3b), the majority of which folds into a helical structure with an open thi-box (see Figure 3b representative structure). On the other hand, the energy landscape at 190 nt shows two distinct clusters with two different structures (Figure 3c), corresponding to the transcriptionally active and inactive structures. The structures in the smaller-distance cluster closely resemble the transcriptionally inactive structure, where the thi-box domain has formed along with the downstream transcription terminator hairpin. Conversely, the structures in the larger-distance cluster resemble the riboswitch in the anti-terminator form which permits transcription to proceed. Thus, the presence or absence of TPP shifts the balance from one configuration over another.
Figure 3.
Energy landscape analysis of TPP riboswitch from 120 nt to 190 nt (full length) with 1–5 incremental step. (a) Clustering results of TPP riboswitch energy landscape along the length of the sequence. Note that two clusters are present after 180 nt. Two representatives of energy landscapes and structures display (b) one cluster at 145nt or (c) two distinct clusters at 190 nt. The Set 1 (left) and Set 2 (right) structures approximate the average structures represented by set 1 (red) and set 2 (black) on the energy landscape, respectively.
The average structures in each cluster agree well with their respective RNA secondary structures found in-vitro in the TPP-bound and TPP-free state.10 Each cluster is structurally homogeneous (88% and 84% respectively), but the difference between the two clusters is large (base pair distance between cluster centers: 48.3). A significant energetic barrier is evident by a low density area of structures between the two peaks (Figure 3c energy landscape plot). This suggests that only two global topologies are permissible for the folded riboswitch, each of which is represented by a cluster on the landscape.
Cluster analysis of mutants
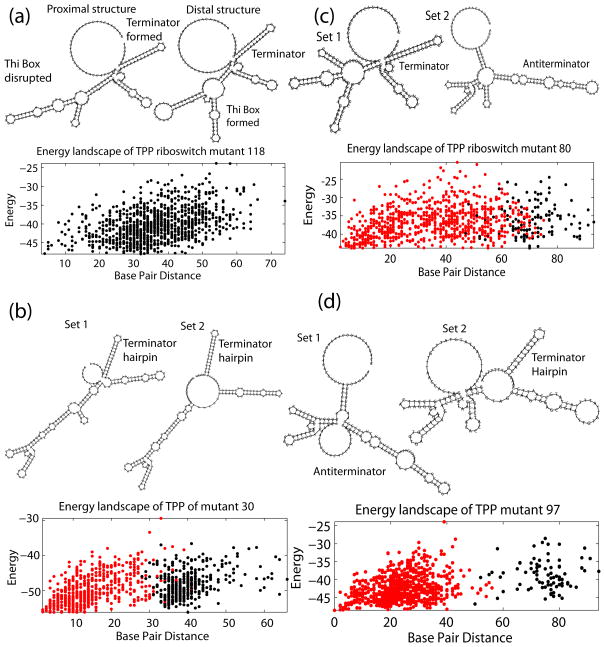
To test this hypothesis further, we analyzed a set of mutants constructed by the Nudler group11 with noted varying effects on the efficiency of transcription termination in vitro. A series of consecutive mutations inserted into the putative anti-terminator region of mutant sequence 118 (G118C, T119A, G120C, and G121C) increases termination efficiency up to ~100% (Table 1).
Thermodynamic analysis of mutant 118 showed a continuous energy funnel with no energy barrier, but high structural similarity (81.1%) when treated as a single cluster. This contrasts the wild-type sequence that displays two clusters (Figure 3c). As predicted, the terminator hairpin was present in all structures sampled in the set (Figure 4a).
Figure 4.
Energy landscape and representative structures of mutant riboswitches 118 (a), mutant 30 (b), mutant 80 (c), mutant 97 (d). Note that the two structures shown for mutant 118 do not represent different sets of structures but are only two sample points along the energy landscape. For mutants 30, 80, and 97 Set 1 (left) and Set 2 (right) structures approximate the average structures represented by set 1 (red) and set 2 (black) on the energy landscape, respectively.
Mutant sequence 30 (C30G, C31G, A32T, C33G), was designed to allow extensive base-pairing between the thi-box (+77–122) and mutated upstream bases, thus disrupting the TPP binding domain. The energy landscape of mutant 30 displays two clusters with little separation, both representing the terminator hairpin state; as predicted, none of the structures displayed the open thi-box domain (Figure 4b).
The 80 (C80A, C81A, C82A) and the 97 (G97C, G98C, T99A) mutants have point mutations of the thi-box domain that disable the riboswitch’s ability to bind to a ligand. Clustering confirms the presence of two structural clusters, extensively overlapping (Figure 4c, d), yet with high intra-cluster structural similarity (76%, 82% respectively). Similarly, the effect of TPP on transcription termination was completely abolished in the 97 mutant. The 97 mutant displays a two-cluster landscape predominantly composed (91.9%) of the low-energy family of anti-terminator structures (Figure 4d).
Crystallographic studies show that binding between the thi-box and TPP is dependent on an induced fit mechanism between highly conserved residues, divalent cations, and the moieties of TPP.15–17 These interactions were disrupted in the 80 and 97 mutations, whose substitutions had deleterious effects on the ability of TPP to favor one of the configurations (in vitro, both mutants showed a +2% termination efficiency increase in the presence of TPP; wild-type showed +71%). However, 2D folding analyses for these mutants display two-cluster energy landscapes which suggest preservation of function to some degree. Thus, this mutation has a deleterious effect on binding but not on folding.
Clustered landscapes are present in diverse riboswitch systems
We further applied our method to a number of riboswitch systems that utilize other ligands and alternative mechanisms of genetic control (see Table 1). The thiM (TPP)4 and ypaA (FMN)18 mRNA 5′ UTR sequences are translation terminator riboswitches that function by base-pairing of the Shine-Dalgarno (SD) sequence. Both sequences were found to display clustered energy landscape properties similar to those of the tenA riboswitch (see Figure 5a, b). However, the ypaA riboswitch is unique in that the higher-energy cluster (Set 2) corresponds to the FMN-bound structure, opposite of what was found for the wild-type tenA TPP riboswitch. This suggests that the lowest energy state is the non-bound form. Microarray analysis indicates that the ypaA sequence shows no change in the quantitative level of transcripts when B. subtilis strains are grown in the presence or absence of riboflavin, alluding to the function of ypaA as a translation inhibitor.19 In addition, the coenzyme B12-sensing btuB leader sequence in Escherchia coli shows a two-state energy landscape (see Figure 5c). This riboswitch utilizes a variety of tertiary interactions and pseudoknots to perform silencing of downstream genes in the presence of a ligand.20 These studies thus suggest that the energy landscape properties developed here are generalizable to different classes of riboswitches.
Figure 5.
Energy landscape plot and structures of the ypaA (a), ribD (b), and btuB (c) riboswitches. The Set 1 (left) and Set 2 (right) structures approximate the average structures represented by set 1 (red) and set 2 (black) on the energy landscape, respectively.
The gcvT and VCI-II elements are dual, tandem-aptamer RNA riboswitches that display cooperative binding of glycine and genetic control through transcription termination.21,22 Each aptamer domain can bind one separate molecule of glycine. Noteworthy is the fact that the presence of glycine decreases the level of gcvT termination, representing a case where the ligand is required to activate gene transcription. Computational analysis of both glycine riboswitches, however, reveals no clustering in the energy landscape (data not shown). This may be attributable to the unique function of the riboswitch as a subtle sensor that significantly modifies its structure with only moderate changes in ligand concentration.23 Thus, more global changes in secondary structure may not be required to affect riboswitch function. It is this cooperative nature of ligand-binding that may account for this difference in energy landscape properties.
Discussion
The total number of conformations that a linear macromolecule like RNA can adopt is astronomical. RNA’s ability to navigate this huge folding space quickly has been continuously subject to selective pressure. Many factors in the folding process, including thermally-driven chain fluctuations, ion-mediated electrostatics, base-pairing and stacking, and other non-canonical interactions guide the folding process.24 The advantage of the energy landscape approach presented here for analyzing the structural properties of functional RNAs is a simplified representation that approximates the feasible secondary structures at the low energy portion of the energy funnel. Funnel energy landscapes have proven invaluable in protein folding studies.25–27 The minimum free energy (mfe) approach, while theoretically most stable, represents a single point on the energy landscape and is subject to small changes in energy parameters and kinetic barriers to folding.28 A growing RNA molecule may not fold immediately into the most stable structure but explore multiple folding routes or fluctuate between nearby “suboptimal” structures. Additionally, significant kinetic traps have been found to hold long RNAs in suboptimal folds.29
Thus, it remains unknown whether RNA folding is a direct pathway to the native fold, singularly controlled by the more stabilizing interactions of the native fold, or a process complicated by a series of frustrated intermediate states, in which the RNA inhabits suboptimal structures. The former notion suggests that the folding landscape is a smoothly sloping energy funnel with a unique, minimum free energy structure. The latter theory supports an exhaustive search between all possible sub-interactions, in which the energy landscape is decorated with local minima that may trap the macromolecule. Our investigations suggest that the folding landscape of certain classes of riboswitches is length-dependent: during the process of transcription certain lengths utilize a smooth transition energy funnel to the mfe, while at other lengths, the energy landscape forms a frustrated equilibrium between two major topologies. This suggests that a complex interplay between kinetics and thermodynamics is required for the structure to attain the proper conformation. For all the riboswitches tested, we suggest that ligand-binding occurs on top of a preformed macromolecular backbone and that this step determines the future outcome of gene regulation. This has been confirmed recently in thermodynamic simulations of the S-Adenosylmethione riboswitch aptamer domain, in which a helical platform for SAM binding is pre-formed.30 Ligand-binding then stabilizes the rate-limiting step to folding.
Similarly, protein folding is hypothesized to be co-translational, which means that smaller, compactly folded intermediates and completed native-like structures are attached to the ribosome during the process of translation.31 This hypothesis is strongly supported by experimental results, such as enzymatic activities immediately after the protein’s release from the ribosome.31–33 The long history of research into protein folding has shown that the free energy surface is also frustrated,34 decorated with many minima asides from the native state, separated by varying energy barriers. Like in vivo protein folding, naturally occurring RNAs have been found to fold co-transcriptionally. Experimental studies done by the Pan and Sosnick groups have shown that RNA co-transcriptional folding in the cell is facilitated by pausing-induced non-native structures.35,36 Our computational approach identified such possible switches along the transcription elongation through a structural and thermodynamic analysis of the B. subtilis tenA TPP binding riboswitch. Though various folding algorithms analyzing RNAs during transcription have been developed – using genetic algorithms,29,37 Monte carlo simulations,38–40 and stochastic methods,41 – this is the first attempt to study minimum free energy folding as a function of nucleotide length, to the best of our knowledge.
Two major classes have emerged from crystal structures studies of riboswitches, functioning through differing allosteric mechanisms.2 Type 1 riboswitches, characterized by the TPP riboswitch, display global conformational changes upon ligand-binding. Type 2 riboswitches, exemplified by the purine and S-Adenosylmethionine riboswitches, function through subtle tertiary interactions stemming from changes in the binding pocket.2 In agreement with our results of the TPP riboswitch, those riboswitches that display global conformational clustering may indeed fall into the Type 1 classification. In contrast, it may be possible that the gcvT and VCI-II glycine riboswitches, characterized by their cooperative nature of ligand-binding, may act as Type 2 riboswitches and as a result do not display the same energy landscape properties in our analysis.
Interestingly, we also found that the experimentally-reported efficiency of functional gene regulation is somewhat correlated to the density of the ligand-bound structure set in our corresponding computed energy landscapes. Because the experimental values reflect nonuniform conditions in each experiment (see Supplementary Table), exact comparisons to our energy-landscape derived values are not prudent, but overall we note a general correspondence for most cases examined. This general correspondence suggests a structure-function connection of our energy landscape views to transcription activity.
Riboswitches can be exploited to design new functional RNAs for biotechnology or biomedical applications, for example, using rational modular design to engineer assemblies of riboswitches and other aptamer modules. Such design efforts can be made more productive by selecting RNAs with desired structural and thermodynamic properties, as examined in this work. Recently, Wieland et. al. designed a TPP riboswitch-hammerhead ribozyme fusion that controls ribosome-binding through the presence or absence of TPP.42 This process required exhaustive search of approximately 4,000 sequences for activity. We anticipate that a computational framework, such as the one we presented, could streamline the search for functional transcripts. Applications of such computational tools to novel RNA design are currently underway.
Materials and Methods
Simulation of transcription elongation
Various programs43,44 have been used to predict the “optimal” secondary structure of a short RNA sequences by minimizing the free energy of folding45 from a standard set of energetic parameters.46,47 These methods are based on the principles of hierarchal folding of RNA so that secondary structural elements form a scaffold upon which tertiary interactions are then achieved.48–50 It is also assumed that, at equilibrium, the molecule will thermodynamically favor its lowest energy state. Such RNA prediction of secondary structures have been shown to be around 70% accurate on average for short RNAs of less than 200 nucleotides,46 despite the approximations of thermodynamic parameters. Additionally, the kinetics of folding, tertiary interactions, and pseudoknot formation are not fully taken into account. Thus, the structures predicted by free energy minimization provide a valuable but incomplete view of structures. For recent approaches to RNA folding see 51.
In both mfold and Vienna RNAfold, the minimum free energy (mfe) and suboptimal structures of a single RNA sequence are predicted by the algorithm of Zuker and Stiegler.52 Essentially, the overall free energy is approximated by sum of the loop and base pair energies.53 These energy parameters are estimated based on melting temperature studies of synthetically constructed oligoribonucleotides at arbitrary temperatures.46
In our applications, we simulate the elongation of the TPP-binding riboswitch from the 5′ UTR of the tenA gene from B. subtilis at 1–5 nt incremental steps up to the transcription start site (191 nt) to investigate the structural and thermodynamic switches along the transcription elongation pathway. For structural analysis, sub-sequences were folded into their native (“optimal”) secondary structures using mfold 43 by free energy minimization.
Clustering analysis of energy landscape
For thermodynamic analysis, we sample 1,000 possible (“sub-optimal”) secondary structures using the RNAsubopt module of the Vienna RNA package.54 Sampling was performed at 55 Celsius from the Boltzmann-weighted distribution of secondary structures because we found that the sampling at elevated temperatures produces greater diversity and was more representative of in vitro experimental results. Our simulation shows that temperatures below 55°C generate structures that are mostly similar to the optimal secondary structures, while higher temperatures generate unfolded structures. Obtaining the partition function for folding at elevated temperatures requires extrapolation of free energy parameters from their reference at 37° C. However, this has been shown to accurately predict folding landscape properties and more importantly effectively enhance the range of conformational states sampled.55 Free energies of sampled structures were recalculated at 37° C using RNAeval to be physiologically relevant.
We compute the base pair distance matrix between all sampled structures using RNAdistance.56 The base pair distance measures the number of base pairings that require breaking or forming in order to convert one structure into another. A plot of the distance between the minimum free energy structure and all other structures (i.e., the first column of the distance matrix) against the free energy of folding produces an illustrative representation of the folding pathways, which we call the energy landscape plot. Such a plot indicates the range of possible RNA secondary structures for a given sequence with relationship to the mfe (minimum free energy) represented as the lowest point on the energy axis.
To assign individual secondary structures of the energy landscape to clusters of structures with similar topological characteristics, we use the k-means algorithm (with k=2) for partition clustering in the R statistical software package.57 The algorithm aims to partition the points into k groups such that the sum of squares between the assigned cluster centers and each point is minimized.58 For cluster validation, the average silhouette width was used, quantifying a measure of the clustering (for full description see: 58). We define a threshold value of ≥0.4 as a well-clustered result (note that the highest silhouette coefficient with k = 3, 4 and 5 for the B. Subtilis TPP riboswitch is 0.35, while the value with k=2 is 0.62).
We partition the entire set of structures into their respective clusters. We then developed an automated procedure to analyze the most dominant secondary structural features in the clusters. In abstract form, a given RNA secondary structure can be represented as a set of balanced parentheses and points associated with each nucleotide to indicate base pairing patterns. For example, the string: (((((.....))))) represents the secondary structure of a simple 5 base pair helix. In each cluster, we compute the frequency of finding one of the string characters at each position in the string. The most dominant characters (i.e., signature corresponding to the greatest number of structures) were assembled together into a string of characters. If no single element was present in the majority of the structures, an underscore character was assigned. The complete string of characters approximates the dominant structural elements in the cluster, which we term the average structure of the cluster. As an example, a set of structures was generated for the helix presented above (see Table 2).
Table 2.
Base pair distance between a minimum free energy structure and suboptimal structure. The average structure is composed of the most common structural elements at each position in the set of structures. The second structure requires unzipping of a short-helix (4 moves), followed by matching the distal 5′ and 3′ ends (5 moves) to reproduce the minimum free energy structure. In the last entry, at position 5, the average structure is marked with an underscore since no character is present in >50% of the set.
| Structure Set | Base Pair Distance |
|---|---|
| (((((.....))))) | Minimum free energy structure |
| ((((......)))). | 9 |
| ((((.......)))) | 1 |
| (((.(.......).))) | 1 |
| (.(((.....))).) | 1 |
| (((.........))) | 2 |
| ((((_.....))))) | Average structure |
To determine the accuracy of our method, we calculate the number of structural elements that matched well our average structure to structures derived experimentally or through sequence alignment analysis (see Table 3). The aptamer domains of many riboswitches are known to be highly-conserved across species3,6 and thus we limited this comparison to the aptamer region only. In each riboswitch data set analyzed, we found high structural similarity, indicating the validity of our computational approach.
Table 3.
Percent of matching base pairs between the ligand-binding domains of the computationally predicted average structure (see Methods) and experimentally derived structure. Asterisks (*) denote experimental structures published using mfold.43
Supplementary Material
Acknowledgments
We are deeply grateful to Dr. Hin Hark Gan for insightful comments and recommendations regarding various aspects of this work. Funding support from the NSF, NIH, and HFSP are gratefully acknowledged.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- 1.Serganov A, Yuan YR, Pikovskaya O, Polonskaia A, Malinina L, Phan AT, Hobartner C, Micura R, Breaker RR, Patel DJ. Structural basis for discriminative regulation of gene expression by adenine- and guanine-sensing mRNAs. Chemistry & Biology. 2004;11:1729–1741. doi: 10.1016/j.chembiol.2004.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Montange RK, Batey RT. Riboswitches: emerging themes in RNA structure and function. Annu Rev Biophys. 2008;37:117–133. doi: 10.1146/annurev.biophys.37.032807.130000. [DOI] [PubMed] [Google Scholar]
- 3.Nudler E, Mironov AS. The riboswitch control of bacterial metabolism. Trends Biochem Sci. 2004;29:11–17. doi: 10.1016/j.tibs.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 4.Winkler W, Nahvi A, Breaker RR. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature. 2002;419:952–956. doi: 10.1038/nature01145. [DOI] [PubMed] [Google Scholar]
- 5.Winkler WC, Breaker RR. Genetic control by metabolite-binding riboswitches. Chembiochem. 2003;4:1024–1032. doi: 10.1002/cbic.200300685. [DOI] [PubMed] [Google Scholar]
- 6.Winkler WC, Breaker RR. Regulation of bacterial gene expression by riboswitches. Annu Rev Microbiol. 2005;59:487–517. doi: 10.1146/annurev.micro.59.030804.121336. [DOI] [PubMed] [Google Scholar]
- 7.Roth A, Nahvi A, Lee M, Jona I, Breaker RR. Characteristics of the glmS ribozyme suggest only structural roles for divalent metal ions. RNA. 2006;12:607–619. doi: 10.1261/rna.2266506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheah MT, Wachter A, Sudarsan N, Breaker RR. Control of alternative RNA splicing and gene expression by eukaryotic riboswitches. Nature. 2007;447:497–500. doi: 10.1038/nature05769. [DOI] [PubMed] [Google Scholar]
- 9.Kubodera T, Watanabe M, Yoshiuchi K, Yamashita N, Nishimura A, Nakai S, Gomi K, Hanamoto H. Thiamine-regulated gene expression of Aspergillus oryzae thiA requires splicing of the intron containing a riboswitch-like domain in the 5′-UTR. FEBS Lett. 2003;555:516–520. doi: 10.1016/s0014-5793(03)01335-8. [DOI] [PubMed] [Google Scholar]
- 10.Rentmeister A, Mayer G, Kuhn N, Famulok M. Conformational changes in the expression domain of the Escherichia coli thiM riboswitch. Nucleic Acids Res. 2007;35:3713–3722. doi: 10.1093/nar/gkm300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mironov AS, Gusarov I, Rafikov R, Lopez LE, Shatalin K, Kreneva RA, Perumov DA, Nudler E. Sensing small molecules by nascent RNA: a mechanism to control transcription in bacteria. Cell. 2002;111:747–756. doi: 10.1016/s0092-8674(02)01134-0. [DOI] [PubMed] [Google Scholar]
- 12.Lang K, Rieder R, Micura R. Ligand-induced folding of the thiM TPP riboswitch investigated by a structure-based fluorescence spectroscopic approach. Nucleic Acids Res. 2007;35:5370–5378. doi: 10.1093/nar/gkm580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miranda-Rios J, Navarro M, Soberon M. A conserved RNA structure (thi box) is involved in regulation of thiamin biosynthetic gene expression in bacteria. Proc Natl Acad Sci U S A. 2001;98:9736–9741. doi: 10.1073/pnas.161168098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Greenleaf WJ, Frieda KL, Foster DA, Woodside MT, Block SM. Direct observation of hierarchical folding in single riboswitch aptamers. Science. 2008;319:630–633. doi: 10.1126/science.1151298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thore S, Leibundgut M, Ban N. Structure of the eukaryotic thiamine pyrophosphate riboswitch with its regulatory ligand. Science. 2006;312:1208–1211. doi: 10.1126/science.1128451. [DOI] [PubMed] [Google Scholar]
- 16.Edwards TE, Ferre-D’Amare AR. Crystal structures of the thi-box riboswitch bound to thiamine pyrophosphate analogs reveal adaptive RNA-small molecule recognition. Structure. 2006;14:1459–1468. doi: 10.1016/j.str.2006.07.008. [DOI] [PubMed] [Google Scholar]
- 17.Serganov A, Polonskaia A, Phan AT, Breaker RR, Patel DJ. Structural basis for gene regulation by a thiamine pyrophosphate-sensing riboswitch. Nature. 2006;441:1167–1171. doi: 10.1038/nature04740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Winkler WC, Cohen-Chalamish S, Breaker RR. An mRNA structure that controls gene expression by binding FMN. Proc Natl Acad Sci U S A. 2002;99:15908–15913. doi: 10.1073/pnas.212628899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee JM, Zhang S, Saha S, Santa AS, Jiang C, Perkins J. RNA expression analysis using an antisense Bacillus subtilis genome array. J Bacteriol. 2001;183:7371–7380. doi: 10.1128/JB.183.24.7371-7380.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nahvi A, Sudarsan N, Ebert MS, Zou X, Brown KL, Breaker RR. Genetic control by a metabolite binding mRNA. Chem Biol. 2002;9:1043. doi: 10.1016/s1074-5521(02)00224-7. [DOI] [PubMed] [Google Scholar]
- 21.Lipfert J, Das R, Chu VB, Kudaravalli M, Boyd N, Herschlag D, Doniach S. Structural transitions and thermodynamics of a glycine-dependent riboswitch from Vibrio cholerae. J Mol Biol. 2007;365:1393–1406. doi: 10.1016/j.jmb.2006.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mandal M, Lee M, Barrick JE, Weinberg Z, Emilsson GM, Ruzzo WL, Breaker RR. A glycine-dependent riboswitch that uses cooperative binding to control gene expression. Science. 2004;306:275–279. doi: 10.1126/science.1100829. [DOI] [PubMed] [Google Scholar]
- 23.Welz R, Breaker RR. Ligand binding and gene control characteristics of tandem riboswitches in Bacillus anthracis. RNA. 2007;13:573–582. doi: 10.1261/rna.407707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen SJ. RNA folding: conformational statistics, folding kinetics, and ion electrostatics. Annu Rev Biophys. 2008;37:197–214. doi: 10.1146/annurev.biophys.37.032807.125957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dill KA, Chan HS. From Levinthal to pathways to funnels. Nat Struct Biol. 1997;4:10–19. doi: 10.1038/nsb0197-10. [DOI] [PubMed] [Google Scholar]
- 26.Chan HS, Dill KA. Protein folding in the landscape perspective: chevron plots and non-Arrhenius kinetics. Proteins. 1998;30:2–33. doi: 10.1002/(sici)1097-0134(19980101)30:1<2::aid-prot2>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 27.Dill KA, Ozkan SB, Shell MS, Weikl TR. The protein folding problem. Annu Rev Biophys. 2008;37:289–316. doi: 10.1146/annurev.biophys.37.092707.153558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ding Y. Statistical and Bayesian approaches to RNA secondary structure prediction. RNA. 2006;12:323–331. doi: 10.1261/rna.2274106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gultyaev AP, van Batenburg FH, Pleij CW. The computer simulation of RNA folding pathways using a genetic algorithm. J Mol Biol. 1995;250:37–51. doi: 10.1006/jmbi.1995.0356. [DOI] [PubMed] [Google Scholar]
- 30.Whitford PC, Schug A, Saunders J, Hennelly SP, Onuchic JN, Sanbonmatsu KY. Nonlocal helix formation is key to understanding S-adenosylmethionine-1 riboswitch function. Biophys J. 2009;96:L7–L9. doi: 10.1016/j.bpj.2008.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Komar AA. A pause for thought along the co-translational folding pathway. Trends Biochem Sci. 2009;34:16–24. doi: 10.1016/j.tibs.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 32.Hamlin J, Zabin I. -Galactosidase: immunological activity of ribosome-bound, growing polypeptide chains. Proc Natl Acad Sci U S A. 1972;69:412–416. doi: 10.1073/pnas.69.2.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kudlicki W, Kitaoka Y, Odom OW, Kramer G, Hardesty B. Elongation and folding of nascent ricin chains as peptidyl-tRNA on ribosomes: the effect of amino acid deletions on these processes. J Mol Biol. 1995;252:203–212. doi: 10.1006/jmbi.1995.0488. [DOI] [PubMed] [Google Scholar]
- 34.Guo Z, Thirumalai D. Kinetics and thermodynamics of folding of a de novo designed four-helix bundle protein. J Mol Biol. 1996;263:323–343. doi: 10.1006/jmbi.1996.0578. [DOI] [PubMed] [Google Scholar]
- 35.Wong TN, Sosnick TR, Pan T. Folding of noncoding RNAs during transcription facilitated by pausing-induced nonnative structures. Proc Natl Acad Sci U S A. 2007;104:17995–18000. doi: 10.1073/pnas.0705038104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pan T, Sosnick T. RNA folding during transcription. Annu Rev Biophys Biomol Struct. 2006;35:161–175. doi: 10.1146/annurev.biophys.35.040405.102053. [DOI] [PubMed] [Google Scholar]
- 37.Shapiro BA, Bengali D, Kasprzak W, Wu JC. RNA folding pathway functional intermediates: their prediction and analysis. J Mol Biol. 2001;312:27–44. doi: 10.1006/jmbi.2001.4931. [DOI] [PubMed] [Google Scholar]
- 38.Tang X, Thomas S, Tapia L, Giedroc DP, Amato NM. Simulating RNA folding kinetics on approximated energy landscapes. J Mol Biol. 2008;381:1055–1067. doi: 10.1016/j.jmb.2008.02.007. [DOI] [PubMed] [Google Scholar]
- 39.Liu F, Ou-Yang ZC. Monte Carlo simulation for single RNA unfolding by force. Biophys J. 2005;88:76–84. doi: 10.1529/biophysj.104.049239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schmitz M, Steger G. Description of RNA folding by “simulated annealing”. J Mol Biol. 1996;255:254–266. doi: 10.1006/jmbi.1996.0021. [DOI] [PubMed] [Google Scholar]
- 41.Xayaphoummine A, Bucher T, Thalmann F, Isambert H. Prediction and statistics of pseudoknots in RNA structures using exactly clustered stochastic simulations. Proc Natl Acad Sci U S A. 2003;100:15310–15315. doi: 10.1073/pnas.2536430100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wieland M, Benz A, Klauser B, Hartig JS. Artificial ribozyme switches containing natural riboswitch aptamer domains. Angew Chem Int Ed Engl. 2009;48:2715–2718. doi: 10.1002/anie.200805311. [DOI] [PubMed] [Google Scholar]
- 43.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hofacker IL, Fontana W, Stadler PF, Bonhoeffer LS, Tacker M, Schuster P. Fast folding and comparison of RNA secondary structures. Monatshefte fnr Chemie/Chemical Monthly. 1994;125:167–188. [Google Scholar]
- 45.Zuker M. Computer prediction of RNA structure. Methods Enzymol. 1989;180:262–288. doi: 10.1016/0076-6879(89)80106-5. [DOI] [PubMed] [Google Scholar]
- 46.Mathews DH, Sabina J, Zuker M, Turner DH. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J Mol Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- 47.Xia T, SantaLucia J, Jr, Burkard ME, Kierzek R, Schroeder SJ, Jiao X, Cox C, Turner DH. Thermodynamic parameters for an expanded nearest-neighbor model for formation of RNA duplexes with Watson-Crick base pairs. Biochemistry. 1998;37:14719–14735. doi: 10.1021/bi9809425. [DOI] [PubMed] [Google Scholar]
- 48.Brion P, Westhof E. Hierarchy and dynamics of RNA folding. Annu Rev Biophys Biomol Struct. 1997;26:113–137. doi: 10.1146/annurev.biophys.26.1.113. [DOI] [PubMed] [Google Scholar]
- 49.Tinoco I, Jr, Bustamante C. How RNA folds. J Mol Biol. 1999;293:271–281. doi: 10.1006/jmbi.1999.3001. [DOI] [PubMed] [Google Scholar]
- 50.Flamm C, Hofacker IL, Maurer-Stroh S, Stadler PF, Zehl M. Design of multistable RNA molecules. RNA. 2001;7:254–265. doi: 10.1017/s1355838201000863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shapiro BA, Yingling YG, Kasprzak W, Bindewald E. Bridging the gap in RNA structure prediction. Curr Opin Struct Biol. 2007;17:157–165. doi: 10.1016/j.sbi.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 52.Zuker M, Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981;9:133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Eddy SR. How do RNA folding algorithms work? Nat Biotechnol. 2004;22:1457–1458. doi: 10.1038/nbt1104-1457. [DOI] [PubMed] [Google Scholar]
- 54.Hofacker IL. Vienna RNA secondary structure server. Nucleic Acids Res. 2003;31:3429–3431. doi: 10.1093/nar/gkg599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bonhoeffer S, McCaskill JS, Stadler PF, Schuster P. RNA multi-structure landscapes. A study based on temperature dependent partition functions. Eur Biophys J. 1993;22:13–24. doi: 10.1007/BF00205808. [DOI] [PubMed] [Google Scholar]
- 56.Fontana W, Konings DA, Stadler PF, Schuster P. Statistics of RNA secondary structures. Biopolymers. 1993;33:1389–1404. doi: 10.1002/bip.360330909. [DOI] [PubMed] [Google Scholar]
- 57.R Development Core Team. R: A Language and Environment for Statistical Computing. Vienna: 2007. [Google Scholar]
- 58.Kaufman L, Rousseeuw PJ. Finding Groups in Data: An Introduction to Cluster Analysis. Wiley; New York: 1990. [Google Scholar]
- 59.Nahvi A, Barrick JE, Breaker RR. Coenzyme B12 riboswitches are widespread genetic control elements in prokaryotes. Nucleic Acids Res. 2004;32:143–150. doi: 10.1093/nar/gkh167. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.