Abstract
Turnip yellow mosaic virus (TYMV) contains a tRNA-like structure (TLS) in its 3′ untranslated region (3′ UTR). This highly structured element induces valylation of the viral RNA by host cell enzymes and is important for virus proliferation. Directly upstream of the TYMV TLS is an upstream pseudoknot domain (UPD) that has been considered to be structurally distinct from the TLS. However, using a combination of functional, biochemical, and biophysical assays, we show that the entire 3′ UTR of the viral genome is a single structured element in the absence of cellular protein. This packing architecture stabilizes RNA structure and creates a better substrate for aminoacylation, and thus the UPD and TLS are functionally and structurally coupled. It has been proposed that the TYMV TLS acts as a molecular switch between translation and replication. Our results suggest that this putative switch could be based on structural changes within the global architecture the UTR induced by interactions with the ribosome. The TYMV·TLS UPD may demonstrate how RNA structural plasticity can play a role in regulation of biological processes.
Introduction
In positive-strand RNA viruses that do not replicate via a DNA intermediate, the viral genomes perform many roles. In addition to serving as both the message for translation and the template for negative-strand RNA synthesis (replication), a universal feature of positive-stranded RNA viral (and many other viral) genomes is the presence of structured RNA elements that control diverse and critical events in the viral infection process. These elements can be found both in the 5′ and 3′ untranslated regions (UTRs) of the viral RNA as well as within the protein-encoding regions of the RNA, and they are involved in processes as diverse as translation initiation,1 RNA replication,2 catalysis,3 packaging,4 and suppression of the antiviral response.5 Steric incompatibility dictates that two of these processes, translation and replication, cannot occur concomitantly; thus they are both temporally and spatially regulated and this regulation could depend on RNA structures.2 Despite the presence of this regulation in many medically and economically important viruses, the molecular details of this mechanistic switch and the function of RNA structural elements remain poorly understood. It is likely that different viruses use different strategies to affect this regulation, but also possible that there are underlying RNA-dependent principles shared between different viruses.
An interesting example of RNA-structure based regulation is found in the Turnip Yellow Mosaic Virus (TYMV), a positive-strand ssRNA plant pathogen belonging to the Tymovirus family. While the TYMV genome resembles a classic cellular messenger RNA (mRNA) with a 5′ cap and coding region, it lacks a typical 3′ poly-(A) tail. Instead its 3′ UTR terminates with a structured RNA that drives aminoacylation of the 3′ end of the viral RNA (in the case of TYMV, the amino acid is valine).6–9 This RNA sequence was thus named a tRNA-like structure (TLS) (Figure 1A). The TYMV TLS enhances translation of its viral genome by an unknown mechanism, and includes cis-acting signals required for negative-strand RNA synthesis.10–13 The TYMV TLS has been reported to deliver the first amino acid in cis during translation of the viral polyprotein,14 but this remains controversial,15,16 and there are no reports of direct interaction between the TYMV TLS and the ribosome. The RNA sequence considered the minimal aminoacylateable TYMV TLS has been the subject of intense mutational analysis.10,11,13,16–27 These studies show that mutations that inhibit aminoacylation also inhibit virus replication and reduce translation enhancement. The central role of the TYMV TLS in these processes led to the proposal that the TLS acts as the physical switch between translation and replication of the viral genome.2
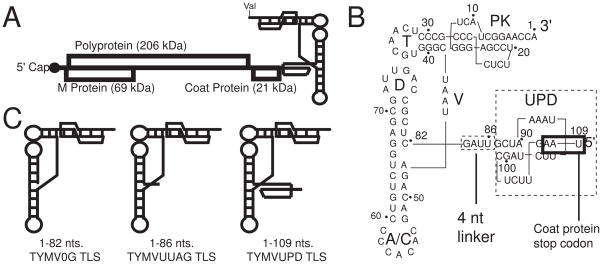
Figure 1. The TYMV TLS.
A. Schematic diagram of TYMV genome, with a 5′ cap, 3′ TLS, and overlapping reading frames labeled. B. Nucleotide sequence and secondary structure of the TYMV TLS and UPD. T, D and anticodon (A/C) loops, and acceptor stem pseudoknot (PK) are labeled. Stop codon for the CP is boxed in black. Note that numbering for the TLS is 3′ to 5′. C. Schematic of constructs used in this study. TYMV0G TLS includes nts 1–82, TYMVUUAG TLS includes nts 1–86, and TYMVUPD TLS includes nts 1–109.
The idea that the TYMV TLS is a physical switch that controls multiple viral processes presents a hypothesis that the three-dimensional folded structure, and alterations to the stability of this structure, are critical components of regulation.9,11,28–30 However, in most cases direct interrogation of changes to the folded TYMV TLS structure has not been conducted and hence key aspects of this hypothesis remain largely untested. The sequence generally considered to contain the TYMV TLS is found in the last 82 nucleotides (nts) of the viral RNA and is comprised of structural elements analogous to a tRNA’s anticodon (AC), D, T and acceptor stems (Figure 1B); we refer to them using this nomenclature.26 However, the TYMV TLS’s secondary structure differs from a classic tRNA in that the 5′ and 3 ends of the RNA do not pair in the acceptor stem; rather this stem contains a pseudoknot. Structural modeling based on chemical and enzymatic probing suggested a TYMV TLS architecture similar to a classical tRNA structure.9,24,31,32 Directly upstream of the 82 nt “core” structure is a 4 nt linker that is preceded by a 24 nt upstream pseudoknot domain (UPD). The full functional role of the UPD within the TYMV 3′UTR remains relatively unexplored but has generally been considered a discrete structural element folded independently of the TYMV TLS core (Figure 1B). In vitro valylation assays of RNAs containing the core TLS and upstream sequences show increased valylation activities compared to just the core TLS, but these upstream nucleotides have no effect on eukaryotic elongation factor 1A (eEF1A) binding or on the action of CCA-nucleotidyltransferase (CCA-NTase) on the TLS.33 In addition, the presence of the UPD in cis with the TLS in reporter constructs leads to an increase in translation; it was proposed that the UPD accomplishes this by allowing proper spacing of the TLS from the TYMV genome, which would allow the 82 nt TLS core to fold correctly and hence lead to an increase in aminoacylation efficiency.10
There has been relatively little analysis of the structural relationship between RNA upstream of the last 82 nt and the structure of the TLS core. Previous experiments using in vitro transcribed TYMV TLS species that contained 5′ nonviral nucleotides showed that this RNA was not an optimal substrate for aminoacylation, likely because these nonviral nucleotides promote an alternative secondary structure.30,34 For technical reasons that study did not test a construct containing exactly 82 nts. More recently, we used chemical probing and biophysical approaches to show that folding of the TYMV TLS into its native three-dimensional structure requires the presence of 3 nucleotides upstream of the 82 nt core, that this effect did not strictly require the viral sequence, and that this was likely due to tertiary interactions between these additional nucleotides and the elbow region of the TYMV TLS.28 This observation yielded a model for how the TYMV TLS could readily unfold and refold in some sort of structural switch.
The direct involvement of nucleotides immediately 5′ of the 82 nt TYMV TLS core (in the 4 nt linker) in folding of the TYMV TLS and to the operation of a putative structural switch led us to question whether additional RNA within the 3′ UTR would be part of a larger structural switch important for the overall success of the virus. That is, are the TYMV TLS core and UPD regions truly independent structural entities or are they part of a larger, conformationally dynamic global fold with functional importance? Using a combination of biophysical analyses, structural probing, and functional assays, we examined the direct link between the aminoacylability of TYMV TLS RNAs of various lengths and their tertiary structures. Consistent with previous studies,33 we have found the 82 nt core to be an inefficient substrate for aminoacylation but addition of upstream sequences dramatically increases aminoacylation efficiency. Furthermore, we find that this change in valylation efficiency correlates to a more stable RNA fold and a decrease in the conformational dynamics of the molecule overall. The most efficient aminoacylation substrate we tested contains 109 nucleotides, encompassing the 82 nts core, 4 nt linker, and 23 nt UPD packed together in a relatively compact two-domain higher-order structure comprising the entire 3′ UTR as well as some coding RNA. The global structure of this 3′ UTR likely changes during different viral events and this allows speculation on the role of this global structure in regulating multiple viral processes.
Results
5′ upstream nucleotides affect aminoacylation of the TYMV TLS
We used TYMV TLS constructs of varying lengths generated using 5′ and 3′ ribozymes (that self-cleaved during transcription and were removed during purification) to resolve ambiguities regarding the role of nucleotides located upstream of the 82 nt TYMV TLS core in aminoacylation efficiency (Figure 1C). These constructs have the advantage of precisely demarcated 5′ and 3′ ends but this strategy leaves a 2′-3′ cyclic phosphate. To ensure that these RNAs were suitable for aminoacylation, the 2′-3′ cyclic phosphate was enzymatically removed, which was a critical step as a cyclic phosphate substantially reduced the initial rates of aminoacylation (data not shown). These RNAs were used in aminoacylation assays using total wheat germ extract depleted of endogenous tRNAs as described.35 All of the TYMV TLS substrates we tested plateau close to 100% aminoacylation, indicating that most molecules are competent substrates (data not shown).
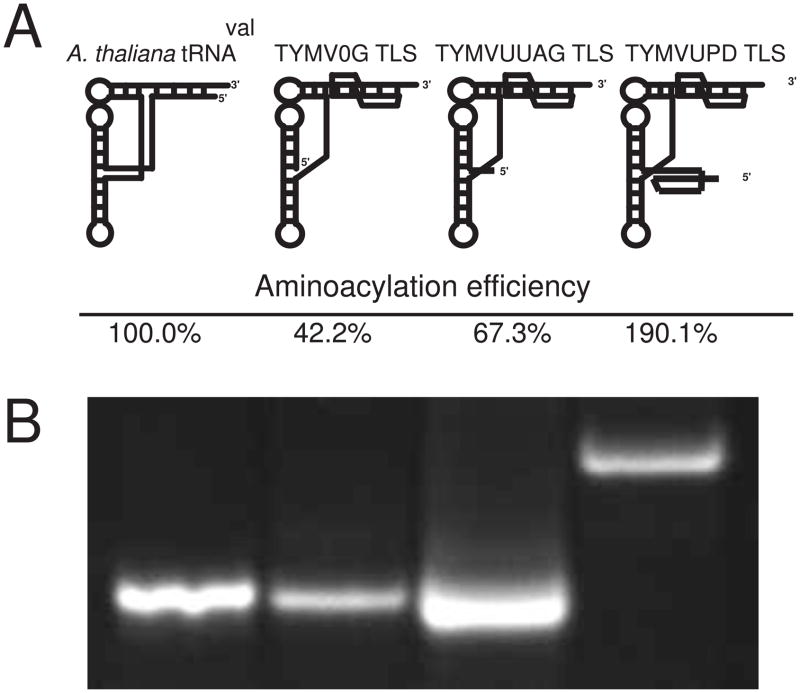
The tested RNAs exhibited marked differences in aminoacylation efficiencies in wheat germ extract, where we report aminoacylation efficiency as the ratio of the Vmax to Km, setting in vitro transcribed A. thaliana tRNAval to 100%. The TYMV TLS 82 nt core (TYMV0G TLS) exhibited an aminoacylation efficiency of 42.2% compared to the A. thaliana tRNAval (Figure 2A). Addition of the 4 nt upstream linker UUAG (TYMVUUAG TLS) increased efficiency by 25% (to 67.3%). These data indicate that 4 upstream nts, previously shown to effect forming the stable global fold of the TYMV TLS,28 also have a significant effect on its ability to be aminoacylated. We next assayed a 109 nt construct containing the TYMV TLS core as well as the linker and the UPD (TYMVUPD TLS), which in fact comprises the entire 3′ UTR of the viral RNA. This construct was nearly five times as efficient as TYMV0G TLS, and twice as efficient as the tRNAval at being aminoacylated (190.1%). Thus, in vitro assays indicated that the presence of the UPD significantly affects the aminoacylation of the TLS core; they are functionally coupled elements. Our results using these targeted and specifically designed RNAs are consistent with previous studies in which the presence of upstream RNA increased TLS aminoacylation.33 In those studies, the TLS was roughly identical in aminoacylation efficiency to a tRNA standard, a 264 nt RNA was a 2.45-fold more efficient substrate, and the entire virion RNA was 8.03-fold better. Direct quantitative comparison of Km and Vmax values between that study and ours is confounded by the use of different synthetases (purified vs. total extract), different tRNA standards (lupine vs. Arabidopsis), and different buffer/salt conditions, but the overall trends between the two studies agree.
Figure 2. Aminoacylation of tRNAval and TYMV TLS constructs.
A. Schematic representation of each construct used in the aminoacylation assay (above) and aminoacylation rate given by Vmax/Km relative to tRNAval (below). Experiments were performed in triplicate, and initial triplicate data numbers were used to graph rates in a Lineweaver-Burk plot, with all R values at 0.93 or higher. B. Native gel electrophoresis of each RNA used demonstrating single species’ presence in these assays.
Differences in aminoacylation efficiency are not due to misfolding
A previous study reported that nonviral 5′ upstream nucleotides can affect the secondary structure of the TYMV TLS, creating multiple conformations upon the addition of divalent magnesium ions.30 To determine if the reduction of aminoacylation efficiency of TYMV0G and TYMVUUAG TLSs is due to formation of a polydisperse sample, we used native gel electrophoresis, where the RNA’s migration rate is based on a combination of molecular weight and global conformation. RNAs that form a single folded species in solution (monodisperse) or that rapidly interconvert between multiple species will migrate as a single tight band in the gel. Both TYMV0G and TYMVUUAG TLSs migrated as a single band and at a similar rate as A. thaliana tRNAval, suggesting both adopt a single species with a fold similar to the tRNA (Figure 2B). It is worth noting that TYMVUUAG appeared to migrate slightly faster than TYMV0G despite being 4 nts larger, consistent with TYMVUUAG adopting a somewhat more compact structure. Likewise TYMVUPD ran as a single species but at a substantially slower rate. This is consistent with its larger molecular mass and perhaps an alternative tertiary architecture. Sedimentation velocity analytical ultracentrifugation (SV/AUC) data showed similar trends to the native gel electrophoresis and these data fit well with single-species models (data not shown). Taken together, these results suggest that the differential aminoacylation efficiencies of these RNAs is not due to global misfolding or formation of polydisperse RNA populations.
5′ upstream nucleotides do not affect the overall structure of the TLS core, but do play a role in the TLS structure
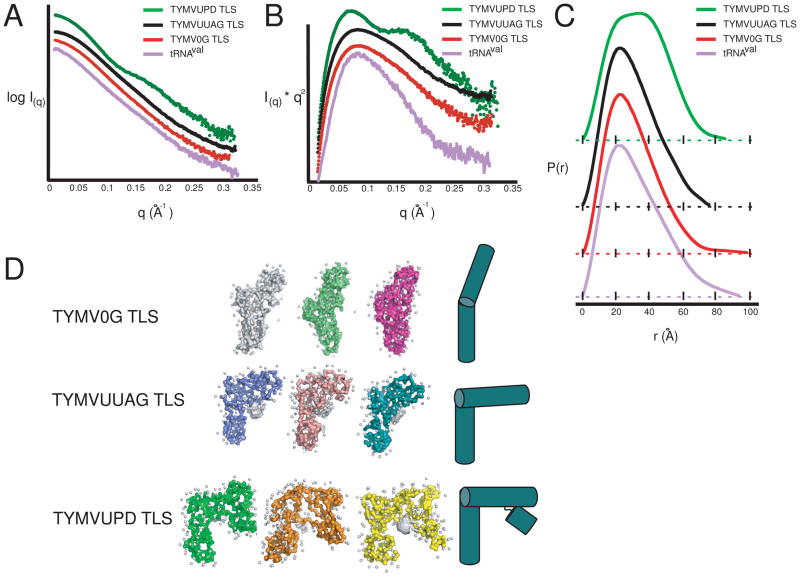
Native gel electrophoresis and SV/AUC experiments indicated that the TYMV TLS fold depends on 5′ upstream nucleotides. However, these methods do not give details about the RNA molecules global structure. Therefore we employed small angle x-ray scattering (SAXS), a powerful tool to describe the global architecture of monodisperse samples in solution to better understand the overall architecture of these individual constructs.28,36 In general, a macromolecule’s SAXS profile will be principally determined by the shape of the scattering particle, thus particles of differing masses but with similar shapes will demonstrate similar SAXS profiles. We previously showed that a TYMV TLS containing 3Gs at its 5′ end had a scattering pattern similar to a tRNAs, and ab inito reconstructions using these scattering data revealed a reproducible L-shaped architecture strongly reminiscent of in vivo produced S. cerevisiae tRNAphe.28
The TYMV0G and TYMVUUAG TLSs showed nearly identical scattering profiles suggesting these two TLSs share a common shape (Figure 3A). In addition, both had SAXS profiles that are very similar to that of A. thaliana tRNAval. Because similar SAXS profiles suggest similar folds, we can conclude that both TYMV0G and TYMVUUAG TLSs have a tRNA-like global shape, paralleling SV/AUC and native gel electrophoresis data. In contrast, the TYMVUPD TLS’ SAXS profile differed significantly from the previous two TLS constructs; additional features in the SAXS profile appeared between the low and mid range scattering angles (q = 0.075 and 0.12 A−1, and a shoulder at q = 0.12 to 0.17 A−1). These data also allowed us to determine and compare the radius of gyration (RG) values for these RNAs, which were 24.8 Å for A. thaliana tRNAval, 23.1 Å for TYMV0G, 23.3 Å for TYMVUUAG, and 26.07 Å for TYMVUPD. The similarities of the tRNA, TYMV0G and TYMVUUAG values may coincide with roughly similar overall architectures of these three RNAs. Interestingly, although the TYMVUPD RNA is 23 nts longer than the TYMVUUAG RNA and therefore contains ~27% more mass, its RG was less than 3 Å larger; this may indicate that the UPD element packs closely with the TLS core elements.
Figure 3. SAXS data and ab initio reconstructions.
Raw scattering data (A), Kratky plot (B), and Pairwise distribution function (C) for each RNA species. tRNAval is in purple throughout, with TYMVUPD TLS in green, TYMVUUAG TLS in black, and TYMV0G TLS in red. In each case, the x-axis is a solid black line, except in (C), where the traces have been offset for clarity, and the x-axis for each sample is displayed directly beneath the associated graph in the form of a dotted line. D. Three distinct reconstructions are shown for a representative sample of the over 25 reconstructions generated. White spheres represent modeled water molecules. Simple architectural models of each TLS are shown to the right.
The SAXS profile similarities and differences can be illustrated more clearly in a Kratky Plot analysis, a useful tool for determining if molecules used in SAXS are structured. Kratky plot of all curves display an inverted parabolic character, indicating structured RNA molecules (Figure 3B). While tRNAval, TYMVUUAG TLS, and TYMV0G TLS displayed a single maxima, the TYMVUPD TLS displayed a double-maxima, with the left maxima coinciding with the other three species. This indicates that TYMVUPD TLS is folded, but has additional structural features not present in the shorter RNAs.
A Fourier transform of the SAXS data yielded the pair-distance, P(r), distribution function of the scattering particle, and was used to reveal real-space architectural similarities and differences between macromolecules. For the TLS RNAs, the P(r)-distribution plots show that all TLSs share a maxima with tRNA at 20Å, consistent with the diameter of A-form helices found in this RNA structure species (Figure 3C). However, the TYMV0G TLS’s maximum dimension (Dmax, the largest possible and probable distance between scattering electron pairs) was nearly 20Å larger than that for TYMVUUAG TLS. This difference in the maximum dimension could be due to either conformational differences or to aggregation in the sample. The latter is highly unlikely, as neither sample showed characteristic aggregation patterns at lower scattering angles, there was no evidence of aggregation in our native gels, and each sample was purified and analyzed on a size exclusion column immediately before SAXS data collection. Therefore, this measured difference in Dmax was due to a conformational difference, which might involve differences in the degree to which the TLSs sample different conformational states. Interestingly, TYMV0G TLS Dmax was larger than tRNAval and tRNAphe, both of which display a larger solution Dmax than crystal structures would suggest.28 We also note that Dmax of our in vitro transcribed A. thaliana tRNAval was larger than that reported for tRNAval purified from E. coli,37 likely due to different sequences and chemical modifications present in the E. coli tRNA and also to how the RNA was purified immediately prior to SAXS data collection.36 For the purposes of this study, in vitro transcribed A. thaliana tRNAval was a more appropriate comparison molecule, because like the TLS RNAs it is of eukaryotic origin and is unmodified. This allows direct comparison of sequence effects unconfounded with differences due to post-transcriptional modifications.
In contrast to the other three species, the P(r)-distribution plot of the TYMVUPD TLS contained a broad peak with two maxima located at 20 and 40Å (Figure 3C). This type of distribution is indicative of a structure formed from A-form helices packed next to each other. Although the Dmax of TYMVUPD TLS is larger than TYMVUUAG TLS, it is still nearly 13 Å smaller than TYMV0G TLS, despite being 27 nts longer. Structurally, this can be interpreted as the UPD-containing TYMVUPD TLS (109 nt long) is more compact than the TYMV0G TLS (82 nt), and is consistent with the idea that the two domains directly interact within a higher order fold.
Further structural insights into the TLS architecture was obtained by modeling the experimental SAXS profiles for each TLS using an ab initio modeling algorithm. The reconstructed models are low-resolution space filling models of a molecule that define the shape of the scattering macromolecule in solution. For both TYMV0G TLS and TYMVUUAG TLS, multiple independently generated reconstructions showed a clear pattern for both TYMV0G TLS and TYMVUUAG TLS (Figure 3D). Perhaps unsurprising, TYMVUUAG TLS reconstructions were very similar to TYMV3G TLS reconstructions published previously, matching ·OH probing data indicating that 5′ upstream nucleotides are necessary to form the stable tight fold, and that the folds of these two RNAs are similar.28 TYMV0G TLS shows a similar architecture in that the reconstruction appears to have two helical regions meeting at an elbow, but in this 82 nt RNA the putative interhelical angle between the acceptor stem and anticodon stem is now nearly 180°. This suggests that in the absence of nts 5′ of the last 82, the TLS structure is conformationally dynamic and not locked into the L-shaped structure associated with TYMVUUAG. In contrast, TYMVUPD TLS showed an architecture similar to TYMVUUAG TLS, with a distinctive sharp bend in the elbow region. However, an additional region of density, presumably representing the UPD, was now packed against one of the ends of the TLS consistent with the helical packing suggested in the P(r)-distribution. With these data resolution, it cannot be established whether this additional density is packing against the AC or acceptor stem. However, this packing position is likely stable as the Kratky plot pattern showed a distinctive structured inverted parabola shape (Figure 3B), and the position of the putative UPD density was maintained in all the reconstructions. Therefore this UPD packing is likely specific and architecturally relevant to the entire structure of the TYMV 3′UTR.
The TYMV 3′ UTR UPD stabilizes the TYMV TLS architecture
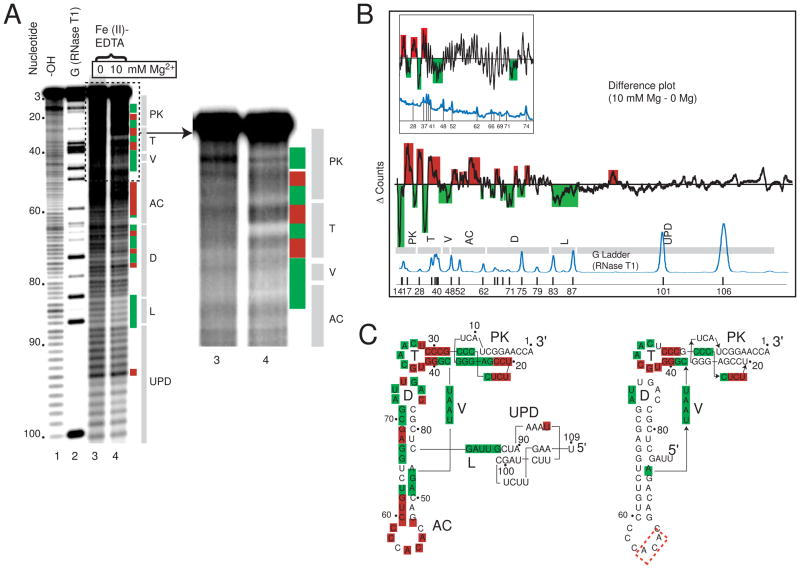
·OH probing was useful in understanding the tight backbone packing of the 82 nt TYMV TLS core and the effect of 5′ nucleotides.28 This method is not a probe of base-paring but is a direct probe of the solvent accessibility of the RNA backbone, and thus areas which pack closely together upon the addition of magnesium will be protected from cleavage, while solvent exposed structured regions will show increased cleavage. To interrogate the local molecular interactions involved in the global architecture of the entire TYMV 3′UTR we used ·OH probing on the TYMVUPD TLS. The probing was conducted as previously described,28 in the absence of Mg2+ and in the presence of 10 mM Mg2+. The probed RNA was resolved on denaturing gels (Figure 4A) and each lane was quantitated to yield a plot of position in the gel vs. intensity. These data were normalized to adjust for loading differences, aligned, and then a difference plot was generated by subtracting the 0 Mg2+ data from the 10 mM Mg2 data (Figure 4B). In this difference plot, areas above the zero line correspond to parts of the RNA with increased cleavage upon addition of magnesium, those below the line are decreased areas of cleavage. In this assay, effects can be subtle and therefore multiple experiments were used to determine which parts of the RNA showed reproducible changes in accessibility upon folding, and these were mapped onto the secondary structure (Figure 4C).
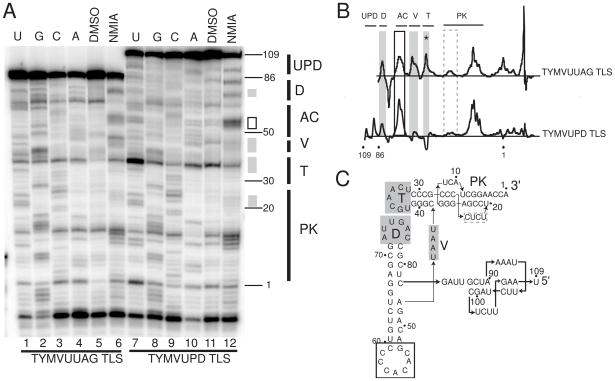
Figure 4. ·OH radical probing of TYMVUPD TLS.
A. Representative sequencing gel from an ·OH probing experiment. Nucleotide position is labeled on the left. Base hydrolysis ladder and RNase T1 ladder are lanes 1 and 2 respectively, while lanes 3 and 4 are the ·OH probing data without and with magnesium present. Colored boxes represent cleavage protection (green) and cleavage enhancement (red) consistently and significantly observed over multiple experiments. At right, the upper portion of the gel is shown with slightly adjusted contrast to show the changes in the 3′ part of the RNA. Labeled grey bars to the right indicate the locations of different TLS structural elements on the gel. UPD = upstream pseudoknot domain; L = linker; D = D stem and loop; AC = anticodon stem and loop, V = variable arm; T = T stem and loop; PK = pseudoknot. B. A difference plot of data from the gel shown in panel A resulting from the subtraction of sample with no MgCl2 from sample with 10mM MgCl2. Gray bars below indicate the locations of secondary structure elements, labeled as in panel A. Reproducible peaks above the baseline are cleavange enhancements, while those below are protections. The locations of G nucleotides are shown by a trace of an RNase T1 ladder below in blue. The inset difference plot is previously published data of the TYMV3G TLS, included for comparison.28 C. Secondary structures of the TYMVUPD TLS (left) and TYMVUUAG TLS (right), with cleavage protections (green boxes) and enhancements (red boxes) overlaid. The protections and enhancements are the result of analysis of several independent probing experiments. The red dashed box represents areas of weak cleavage enhancement.
Protected areas of the folded TYMVUPD TLS were located in the D, T, and V loops, as well as the acceptor stem pseudoknot, and many of these protected areas match those observed in the TYMVUUAG and TYMV3G TLS constructs (Figure 4C).28 However, comparison of a previously published difference plot of a TYMV TLS RNA lacking the UPD and the TYMVUPD TLS shows that parts of the RNA with no clear change upon addition of magnesium now exhibited changes (Figure 4B). In other words, the amount of the molecule that showed an increase or decrease in cleavage upon folding was more extensive for TYMVUPD TLS than for the TYMVUUAG TLS (Figure 4C). Specifically, nearly 30% more of the nucleotides found within the total structure now were either consistently protected from or enhanced for cleavages. Only C36 changed from an enhancement to a protection upon the addition of the UPD. Therefore, these probing data show that inclusion of the UPD results in a folded RNA in which the core elbow interactions are likely still intact, but the UPD also introduces additional stabilizing interactions that are indicated by the additional changes in the ·OH probing difference map. Addition of the UPD also caused the anticodon stem to change from containing light cleavage enhancements to stronger cleavage enhancement, suggesting its solvent-accessible fold is stabilized by the addition of this domain. In addition, protections were observed for the first time in the 5′ upstream UUAG sequence (linker), validating earlier suggestions that these nucleotides pack against the elbow and mimic tertiary interactions found in tRNA D/acceptor stems (Figure 4B&C). Determining the precise packing location of the UPD against the TLS is difficult as new protections are found both in the acceptor stem and AC stem. However, the extreme end of the AC stem exhibits mostly enhancements as opposed to protections, making it a less likely candidate. Overall, the ·OH probing data provide compelling evidence that the UPD and TLS core regions pack against one another and that this interaction stabilizes a specific conformation of the TLS that is an efficient substrate for aminoacylation.
Addition of the 5′ UPD decreases conformational dynamics
Hydroxyl radical probing demonstrates the structure-stabilizing role of nts upstream of the 82 nt core, but does not directly test the conformational dynamics of specific regions of these TLS constructs. Previous studies indicated tRNAs are conformationally dynamic in solution, sampling a variety of interhelical angles between the acceptor and anticodon stems.38–40 These conformational dynamics can be probed using Selective 2′-Hydroxyl Acylation Analyzed by Primer Extension (SHAPE) in which the 2′-OH moieties are selectively acylated based on the local pKa, which is in turn heavily affected by structural constraints.41 Sugars in conformationally dynamic regions will sample a reactive state and will be modified by the reagent (N-methylisotoic anhydride; NMIA, in this case), as will sugars constrained in a highly reactive state (e.g. C2′-endo).42 Thus, a change in NMIA reactivity indicates either a change in the local structure of that sugar/base, or a change in its dynamic character. Previously published SHAPE data demonstrated a striking resemblance between tRNA and TYMV3G TLS, both showing reactivity in the D, T, V, and AC stems.28
SHAPE was performed on the TYMVUUAG and TYMVUPD TLSs to detect changes in dynamic nature and local conformational architecture that could be attributed directly to the addition of the UPD (Figure 5A). The TYMVUPD TLS was modified significantly less in the V and T loops when compared to the TYMVUUAG TLS, although the signal in the T loop was confounded by the fact that in the TYMVUPD TLS the DMSO control lane showed a stop at this position that is as strong as in the NMIA lane. Nonetheless, the effect in the D loop was clear. Likewise, a slight but reproducible decrease in modification was seen in the UPD-containing RNA in the acceptor and pseudoknot regions compared to the TYMVUUAG RNA (Figure 5B&C). Interestingly, the AC stem showed increased modification, likely due to an increase in the overall conformational dynamics of the stem. Overall, regions of the 82 nt TLS core that are involved in tertiary structure in the elbow region (D,T, and V) appeared to become structurally constrained upon the addition of the UPD. When this result is combined with our other probing and biophysical analyses, this suggests that the UPD packs directly against the TLS core and is involved in stabilizing a specific conformation that includes the entire 3′UTR and that positively affects aminoacylation.
Figure 5. SHAPE analysis of TYMVUUAG TLS and TYMVUPD TLS.
A. Representative sequencing gel of a SHAPE experiment. Lanes 1–6 represent TYMVUUAG TLS, and lanes 7–12 represent TYMVUPD TLS. Lanes 1–4 and 7–10 are sequencing lanes U, G, C, and A sequentially. Lanes 5 and 11 are DMSO control lanes without NMIA reaction, and lanes 6 and 12 are NMIA-reacted RNA lanes. Grey boxes represent regions of decreased modification of the TYMVUPD TLS compared to TYMUUAG TLS, and black boxes represent increased modification. Nucleotide position is annotated on the right, as is the approximate location of structural elements, which are labeled as in figure 4. B. Difference maps of TYMVUUAG TLS (top) and TYMVUPD TLS (bottom), obtained by subtraction of the DMSO control lanes form the NMIA lane. Areas where the TYMV UPD TLS showed decreased reactivity with NMIA compared to the TYMVUUAG RNA are shaded grey, and areas of increased reactivity are boxed black. The grey dashed box indicates an area of slight change that was nonetheless reproducible across several experiments. The asterisk represents an area of possible protection that is complicated by a band in the DMSO only control lane (discussed in text). Secondary structure motifs are labeled on top. C. Secondary structure diagram of the TYMVUPD TLS with modification differences overlaid.
Discussion
The 3′ UTR of the Turnip Yellow Mosaic Virus, like many viral RNAs, contains structured sequences that are important for successful replication by the virus. In the case of the TYMV and some related viruses, this structure drives aminoacylation of the 3′ terminus of the viral RNA and must interact with several different proteins.6,12,43 These features suggest that the UTR is important for regulating processes critical to the virus. In fact, mutations to the 3′UTR that affect aminoacylation affect viral accumulation as well as translational enhancement of reporter constructs in vivo, but detailed molecular mechanisms for these important phenomena remain unclear.10,19 Working under the hypothesis that the three-dimensional structure of the TYMV UTR drives function, we employed biochemical, biophysical, and functional assays to link folding to the function of this RNA.
The TYMV 3′UTR has generally been considered to comprise several different structural entities. This includes the TLS, which contains the last 82 nts of the viral RNA, a -nt linker, and the UPD that includes the remainder of the UTR and a few nts of the coding region (Figure 1B). Previously, we showed that nucleotides in the linker region were important for stabilizing the tightly folded tRNA-like architecture of the TLS and this suggested the possibility that the UPD may also interact with the TLS.28 Here we report that the three structural entities of the TYMV 3′UTR pack together in a specific higher-order structure consisting of 109 nucleotides, three of which are actually part of the coat protein (CP) open reading frame. Although a previous study concluded that the TLS was more efficiently aminoacylated when more RNA was included upstream33, that study did not correlate aminoacylation with the thermodynamic stability or multi-domain packing of the TLS with the UPD. Functional and mechanistic consequences of this stable multi-domain higher-order packing include both measurable changes in the efficiency of aminoacylation of the RNA as well as implied changes in the architecture of the RNA that could be used in regulation.
The 3′UTR of the TYMV exhibits different folding characteristics depending on the amount of RNA included in the test construct, and these characteristics correlate with aminoacylation efficiency. Specifically, the more tightly packed and stably folded the RNA construct, the better a substrate for aminoacylation. The most efficient substrate we tested contained the entire 3′UTR with its multi-domain packing and the least efficient was the RNA containing only the last 82 nucleotides of the 3′UTR (Figure 2A). Because aminoacylation is important for virus replication, this suggests that the higher-order fold we have discovered has real implications for the viral life cycle and also raises the question of how the higher-order fold to promote aminoacylation.
One possible explanation for the measured increase in aminoacylation efficiency of the RNA that includes the entire 3′UTR is that the higher-order fold stabilizes a specific conformation that is recognized by the valine aminoacyl tRNA synthetase (ValRS). Evidence for this lies in the fact that combined SAXS, SV/AUC, and SHAPE data show an RNA containing only the TLS core (TYMV0G TLS) is flexible at its elbow and is likely sampling many interhelical angles between its acceptor and AC stems. As such, the TLS spends only a portion of its time in a conformation optimized for efficient interaction with the AARS and aminoacylation. In contrast, an RNA with the 4-nt linker (TYMVUUAG TLS) and one containing the entire 3′UTR (TYMVUPD TLS) exhibit stabilization of an interhelical angle matching the canonical L-shaped tRNA interhelical angles found in tRNA/AARS co-crystal structures (Figure 3D). Presumably this angle arranges portions of the TLS RNA responsible for making direct contact with ValRS in a favorable conformation (Figure 6A). In other words, the linker and UPD stabilize the conformation of the TLS core 82 nts that interacts with the enzyme. This interpretation is also consistent with our aminoacylation data in that all of these RNAs plateau at 95–100% aminoacylation (data not shown), as well as data from SV/AUC and native gel electrophoresis showing one RNA species, making alternate stable conformational dead ends an unlikely explanation for the observed differences.
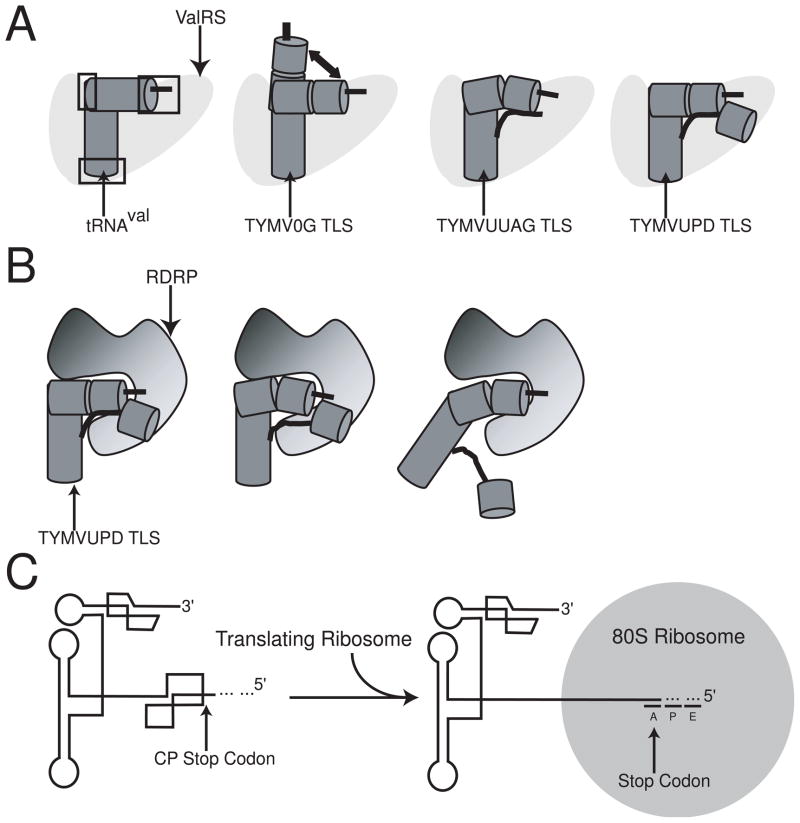
Figure 6. Aminoacylation, replication, and molecular switch models of the TYMV TLS.
A. A schematic model for aminoacylation of tRNA and TLS species. tRNAval (dark grey left figure) makes specific contacts with ValRS (light grey) in its acceptor stem, anticodon loop, and elbow region (boxed in black).54 TYMV0G TLS is less efficient at making these contacts as its architecture samples a variety of states due to flexibility in the elbow region (second image). TYMVUUAG TLS is more constrained than the TYMV0G TLS, making it a relatively better substrate, but still not optimal (third image). TYMVUPD TLS’ architecture is stabilized and hence is able to make each contact with ValRS efficiently, as well as possibly several new ones, increasing aminoacylation efficiency overall (right image). B. Model for how binding of the RDRP (shaded from black to grey) could recognize the CCA motif in the acceptor stem and disrupt interactions between the acceptor stem and the UPD/linker region. This might allow enough destabilization of the TLS overall to make replication more efficient through a previously highly structured region. C. The TYMV TLS as a molecular switch. The TYMV TLS folds into a stable global structure. Upon translation of the coat protein, the UPD is unfolded due to steric constraints of the ribosome placing the stop codon in the A site during translation.
If the UPD increases aminoacylation by stabilizing the 82-nt TLS core L-shaped structure, it could be by stabilizing the acceptor stem pseudoknot. Previous work by Matsuda et al. has shown that mutations which disrupt the UPD formation also decrease aminoacylation efficiency in a partially-purified synthetase system.10 Our hydroxyl radical probing data suggests that a likely location for UPD packing against the TLS is near the acceptor stem pseudoknot. If true, the UPD could stabilize the pseudoknot structure, which we and others previously showed to be relatively unstable and dynamic, and this leads to an increase in the ability of the RNA to serve as a good substrate for the ValRS. It is interesting to note that it has also been demonstrated that the UPD can be replaced by a poly-(A) sequence and valylation remains high.10 This might indicate that the stabilization effect of the UPD can be achieved by more than one structure or sequence; that is, the UPD structure is not a unique evolutionary solution. Indeed, other members of the Tymoviridae have other structures upstream of their TLS core elements.44
In addition to stabilization of a certain native conformation, an alternate though not mutually exclusive explanation for the increased rates of aminoacylation observed with the entire 3′UTR is that the UPD directly interacts with the enzyme. In fact, previous studies have shown a direct interaction between the UPD and the yeast ValRS in the presence of the TLS core.45 This interaction is likely to differ from the canonical interactions that occur between a tRNA and synthetase, since the UPD does not appear to mimic a portion of a classic L-shape tRNA and there are no obvious identity elements within the UPD sequence.17,20 Overall it appears that the UPD acts to increase the aminoacylation efficiency of the TLS through a combination of stabilizing an active conformation and also though direct interaction with the synthetase (Figure 6A).
Our studies help to define the extent of RNA in the 3′ UTR that affects aminoacylation. Previous studies aimed at defining the minimal TLS sequence capable of being aminoacylated tended to conclude that it consisted of the last 82 nt of the RNA, which we refer to here as the TLS core. Using purified E. coli ValRS, it was shown that the minimal TYMV TLS was between 82 and 86 nucleotides long, but the exact boundary was difficult to discern.46 Additional work demonstrated that nucleotides located 5′ upstream of the TLS core can affect aminoacylation rates, though further work suggested the addition of these non-viral nucleotides can lead to alternative secondary structure formation.30,34 The cumulative effect of these studies was that the TLS was generally defined as being 82 nt in length and the positive effects of upstream structural elements was not fully recognized. Our data suggests that the true aminoacylation substrate recognized by the ValRS likely comprises the entire 3′UTR.
The realization that the TLS aminoacylation depends on a higher-order fold involving not only the minimal animoacylateable RNA but also peripheral elements involved in stabilization is similar in some ways to what was recently observed with a catalytic RNA. While the hammerhead ribozyme (HH Rz) minimal element is able to perform catalysis and many studies were conducted using this as the standard construct, subsequent X-ray structures of that element failed to reconcile structural data with function.47,48 It was later found that addition of two additional domains to the HH Rz increased its catalytic efficiency by over 1000-fold, and a subsequent high-resolution structure of the full element reconciled previously contradictory functional data.49,50 So, while a minimal unit is a useful tool in the study of any RNA molecule, it is also critical to understand how additional elements can affect its structure and function. Our data suggest that future studies exploring the function and mechanism of TLS action should include the entire 3′ UTR folded element, and studies using mutations or deletions should consider the structural effect on the entire 3′UTR structure, not just on the specific element (TLS core, linker, UPD) that is altered.
In addition to its implications in terms of aminoacylation of the 3′ end of the viral RNA, the multidomain packing of the UTR also suggests how this element could be used to regulate or organize different viral processes. We previously proposed a “zipper model” for how the TLS can unfold by disrupting only a few tertiary interactions,28 and we can now expand on that model by combining our data with previous observations. First, the 3′UTR of the TYMV interacts not only with the ValRS, but also with eEF1A, the CCA-NTase and the viral RNA-dependent RNA polymerase (RDRP).8,23,43,51 In the case of RDRP binding, the TLS serves as the starting point for negative-strand RNA synthesis by the RDRP. Second, the presence of sequence upstream of the 82 nt core does not affect binding of eEF1A or the enzymatic activity of CCA-NTase.51 Put together, it seems plausible that the CCA-NTase operates on the 3′ end of the RNA independent of the UPD, then the stabilization effect of the UPD is felt during subsequent aminoacylation, followed by binding of eEF1A. eEF1A would then remain on the RNA until displaced by the RDRP. No data detailing the role of the UPD in RNA replication exists, but our structural model suggests the RDRP could bind in close proximity to the CCA promoter element start site of RNA replication, and this binding could disrupt the UPD-TLS interaction, leading to a change in the thermodynamic stability of the 3′UTR as a whole and allowing the RNA to sample conformations more amenable to reverse transcription (Figure 6B). Hence, by disrupting only a few RNA-RNA tertiary contacts the 3′UTR would be switched from a stable element that is an excellent substrate for aminoacylation and eEF1A binding to a less stable element that is readily replicated.
In addition to possible destabilization induced by RDRP binding, the overall architecture of the TYMV 3′UTR suggests that its structure must be altered when the CP sequence is translated. Specifically, the first nucleotides of the UPD are the stop codon for the CP open reading frame on the subgenomic RNA. Thus, an actively translating ribosome on this open reading frame would reach the stop codon and this would place ~12 downstream nucleotides within the ribosome, by necessity unfolding the UPD structure (Figure 6C). Although the nucleotides directly upstream of the TLS core might still be able to interact with the TLS and provide some structural stabilization, the larger multi-domain architecture of the 3′UTR would be disrupted, causing a change in the thermodynamic landscape of folding and allow the RNA to sample additional conformations, including one that is more akin to a classic tRNA. Note that these thermodynamic changes probably would not affect eEF1A binding, but might affect other processes such as replication of the genome, packaging, encapsidation, or even mRNA translation directly.
Valine-accepting TLS structures are found in a variety of positive-strand RNA viruses, often associated with members of the Tymoviridae family. While the presence of these structures seems to be a common theme, the sequences associated with them, and the 3′ UTRs in general, do not seem to be as highly conserved.44 As previously noted, the role performed by the UPD in the TYMV could be performed by other structures or sequences within other viruses. In fact, only some Tymovirus family members contain a UPD in their 3′ UTR at all, while others seem to possess a hairpin or other structured element. Hence, each virus likely evolved to use its TLS core element and any upstream domains in a manner specific for the need of that virus within its specific host.
Materials and Methods
DNA Cloning and RNA transcription
All constructs were prepared by overlapping primer polymerase chain reaction (PCR), creating (from the 5′ end) an EcoRI restriction site, T7 RNA polymerase promoter, Hammerhead Ribozyme (HH Rz), TLS/tRNA sequence, hepatitis delta virus ribozyme (HDV Rz), and a BamHI restriction site. DNA products were digested using EcoRI and BamHI restriction enzymes, phenol/chloroform/isoamyl alcohol (PCIAA) extracted, ethanol (EtOH) precipitated, then ligated into a pUC19 plasmid. All constructs were verified by DNA sequencing.
DNA for transcriptions was produced by PCR. A 1 mL PCR reaction contained 1.2 nmol of each 5′ and 3′ primer (M13F and M13R), Taq polymerase buffer (10 mM Tris-HCL pH 8.3 at 25°C, 50 mM KCl, 1.5 mM MgCl2, 5% glycerol, 0.5 mM DTT, and 0.08% NP-40, and 0.05% Tween-20), 25 units/mL Taq DNA polymerase, and 0.2 mM each dNTPs. The resulting product was then used in in vitro transcriptions, as described previously.52
RNA 3′ dephosphorylation
For those molecules that were generated using a HDV Rz on the 3′ end, it was necessary to remove the 2′–3′ cyclic phosphate before aminoacylation assays. 100 μg of RNA was incubated in 1X PNK/CIP buffer (40 mM Na-MES pH 6.0, 100 mM MgCl2, 50 mM dithiothreitol (DTT)), 100 units T4 polynucleotide kinase (PNK), and 50 units calf intestinal phosphatase (CIP) in 100 μL solution. The reaction was incubated at 37°C for 3 hours, followed by PCIAA extraction and EtOH precipitation.
DNA/RNA radiolabeling
RNA constructs and DNA oligonucleotides were radiolabeled in a similar manner. 100–150 μg RNA or 10–15 μg DNA were added to 1X PNK buffer (70 mM Tris-HCl pH 7.6, 10 mM MgCl2, 5 mM DTT), 20 units PNK enzyme, and 1 μL γ-32P-ATP in a 10 μL solution. The reaction was then incubated at 37°C for 1 hour, followed by desalting using a BioRad Micro Bio-Spin 30 Chromatography Column. Resulting solution was then added to equal volume RNA urea loading buffer and purified on a 10% denaturing polyacrylamide gel, then eluted in buffer (500 mM NaAcetate pH 5.2, 0.1% SDS) using the crush and soak method. Samples were then ethanol precipitated, and resuspended in RNase-free water.
SAXS data collection and analysis
All SAXS data were collected at beamline 12.3.1 of the Advanced Light Source (Berkeley, CA). RNA samples were heated at 85°C for 1 minute, and cooled to room temperature for 10 minutes, then diluted 50-fold into gel-filtration buffer (20 mM MOPS pH 6.5, 50 mM KCl, and 7.6 mM MgCl2). The samples were concentrated to 5 mg/mL using 3000 MWCO spin concentrators, and purified through a Superose 6 PC 3.2 column on an Ettan LC system (GE Healthcare) configured with a Rheodyne in-line vacuum degasser and a 0.01 μm Millipore solvent filter. Following the analytical chromatography, preparative-scale samples were prepared at a final injection concentration of ~10 mg/mL in 50 μL total volume. The peak corresponding to the monodisperse RNA sample (2 to 3 mg/mL) was collected for SAXS using 1 and 10 second exposures as described.53 Two sets of buffer measurements were collected for each RNA sample measured where each set consisted of matching 1 and 10 second exposures that were collected immediately before and after SAXS measurements of the RNA sample. Integration, scaling and buffer subtraction were performed with the program OgreNew (Greg Hura, Lawrence Berkeley National Laboratory). For each RNA sample, a set of 4 SAXS curves were collected as a 2/3rds dilution series and a direct overlay of all four curves was used to monitor for inter-particle interference within the Guinier region of the data. All collected data was judged free of inter-particle interference and processed further using GNOM for indirect transformation to real space and GASBOR for ab initio model reconstruction. GASBOR reconstructions were performed with adjusted residues with the equation: # Residues = # Nucleotides * 2.6.
Hydroxyl Radical Probing
For hydroxyl radical probing, RNA had a minimum radioactivity of 200,000 c.p.m./μL RNA, and was heat denatured at 85°C for 1 min, followed by slow cooling at room temperature. Reactions contained RNA, 30 mM HEPES-KOH pH 7.5, 0–10 mM MgCl2, 0.1μg/μL whole yeast tRNA, and DEPC H2O to a total volume of 7 μL. Solution was incubated at 30°C for 15 min to come to folding equilibrium. 1 μL Fe(II)-EDTA solution (10 mM (NH4)2Fe(SO4)2 and 18 mM EDTA pH 8.0), 1 μL 1% H2O2, and 1 μL 10 mM NaAscorbate were then added and incubated at 30°C for 2 min. Reactions were quenched with 1 μL 100 mM ThioUrea. Samples were ethanol precipitated by addition of 1 μL 3M NaAcetate and 40 μL 100% ethanol, then resuspended in 20 μL 7 M urea loading buffer. RNA samples were run on a 10% 7 M urea polyacrylamide sequencing gel, exposed to a phosphorscreen overnight, and imaged on a STORM phosphorimager. Data were analyzed as previously described.52
SHAPE analysis of RNA
SHAPE construct DNA templates were created using a primer internal to the delta hammerhead ribozyme (CCAGCGAGGAGGCTGGGACCATGC) and M13F, followed by in vitro transcription. Two samples, each containing 1–6 pmol of SHAPE RNA, were resuspended in 9 μL of RNase-free water, heat-cooled as above, and incubated at 35°C for 15 min. 6 μL folding mix (333 mM HEPES pH 8.0, 20 mM MgCl2, 333 mM NaCl) was then added and incubated at 37°C for 15 min. 1 μL arid 100% DMSO was added to one sample, and 1 μL freshly made 260 mM NMIA resuspended in arid 100% DMSO to another. The reaction was incubated at 37°C for 1 hr (approximately 5 half-lives of NMIA in H2O) and placed through a Biorad Micro Bio-Spin 30 Chromatography Column, eluting with a final volume of 9 μL. 1 μL of 5′-end labeled internal delta primer was added to each reaction and incubated at 65°C for 5 min followed by 35°C for 20 min. 6 μL of SHAPE enzyme mix (250 mM KCl, 167 mM Tris- HCl H 8.3, 1.67 mM each NTP, and 17 mM DTT) was then added to each tube and slightly agitated to gently mix solution. Reaction was incubated at 52°C for 1 min followed by the addition of 1 μL of RT Superscript III (Invitrogen) and 5 min incubation. 1 μL 4M NaOH was added to each tube and incubated at 95°C for 5 min. Finally, 29 μL acid stop mix was added to each sample and incubated for 5 min. The resultant cDNA were run on a 10% 7 M urea polyacrylamide sequencing gel, exposed to a phosphorscreen overnight, and imaged on a STORM phosphorimager, and analyzed as previous described.28
TLS and tRNA valylation assay
A tRNA-dependent wheat germ extract was prepared exactly as described.35 For valylation assay,1 μL aminoacylation buffer (500 mM Tris-HCl pH 7.5, 100 mM MgCl2, 100 mM DTT, and 15 mM ATP), 1 μL RNasin, 0.5 μL cell-free extract from wheat germ, 1 μL 3H-valine (40–50 Ci/mmol), 1 μL RNA (at 50, 10, 7.5, 5, 2.5, 1, and 0.5 μM), and 6 μL RNA were combined and incubated at 25°C. 10 μL sample was incubated for appropriate time (0.5, 1, 2, 3, 4, 5, 20, and 60 min) and added to 50 μL TCA carrier solution (10 mg/mL Baker’s yeast tRNA, 3 M Sodium Acetate), precipitated by 500 μL 50/50 10% TCA/ethanol (ice cold), and incubated on ice for 10 min. Solution was placed through vacuum manifold and RNA collected on Whatmann brand glass filter paper. Filters were washed with 1 mL 10% TCA and 2 mL ethanol. Filters were then added to Fisher Brand Scinti-Safe (5mL per sample), and the amount of radioactivity was measured by scintillation counting. Each experiment was done in triplicate for all RNA concentrations and time points. Data were combined for initial rate graphs and initial rates were used to determine Vmax and Km values according to Lineweaver-Burk plots. Data fits were R>0.9.
Acknowledgments
The authors would like to acknowledge members of the Kieft lab (especially David Costantino, Marisa Ruehle, Grant Ruehle, and Megan Filbin) for critical reading of this manuscript and useful discussions. We also thank David Barton, Linda VanDyk, and Rui Zhao for critical reading of this manuscript. This work was supported by NIH grants R01 GM081346 and R01 GM072560 to JSK. JSK is a Howard Hughes Medical Institute Early Career Scientist
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Doudna JA, Sarnow P. Translation initiation by viral internal ribosome entry sites. In: Mathews MB, Sonenberg N, Hershey JWB, editors. Translational Control in Biology and Medicine. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2007. pp. 129–153. [Google Scholar]
- 2.Dreher TW. Role of tRNA-like structures in controlling plant virus replication. Virus Res. 2009;139:217–229. doi: 10.1016/j.virusres.2008.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Talini G, Gallori E, Maurel MC. Natural and unnatural ribozymes: back to the primordial RNA world. Res Microbiol. 2009;160:457–465. doi: 10.1016/j.resmic.2009.05.005. [DOI] [PubMed] [Google Scholar]
- 4.Annamalai P, Rao AL. In vivo packaging of brome mosaic virus RNA3, but not RNAs 1 and 2, is dependent on a cis-acting 3′ tRNA-like structure. J Virol. 2007;81:173–181. doi: 10.1128/JVI.01500-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Han JQ, Townsend HL, Jha BK, Paranjape JM, Silverman RH, Barton DJ. A phylogenetically conserved RNA structure in the poliovirus open reading frame inhibits the antiviral endoribonuclease RNase L. J Virol. 2007;81:5561–5572. doi: 10.1128/JVI.01857-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yot P, Pinck M, Haenni AL, Duranton HM, Chapeville F. Valine-specific tRNA-like structure in turnip yellow mosaic virus RNA. Proc Natl Acad Sci U S A. 1970;67:1345–1352. doi: 10.1073/pnas.67.3.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Litvak S, Carr DS, Chapeville F. TYMV RNA As a substrate of the tRNA nucleotidyltransferase. FEBS Lett. 1970;11:316–319. doi: 10.1016/0014-5793(70)80557-9. [DOI] [PubMed] [Google Scholar]
- 8.Pinck M, Yot P, Chapeville F, Duranton HM. Enzymatic binding of valine to the 3′ end of TYMV-RNA. Nature. 1970;226:954–956. doi: 10.1038/226954a0. [DOI] [PubMed] [Google Scholar]
- 9.Rietveld K, Van Poelgeest R, Pleij CW, Van Boom JH, Bosch L. The tRNA-like structure at the 3′ terminus of turnip yellow mosaic virus RNA. Differences and similarities with canonical tRNA. Nucleic Acids Res. 1982;10:1929–1946. doi: 10.1093/nar/10.6.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matsuda D, Dreher TW. The tRNA-like structure of Turnip yellow mosaic virus RNA is a 3′-translational enhancer. Virology. 2004;321:36–46. doi: 10.1016/j.virol.2003.10.023. [DOI] [PubMed] [Google Scholar]
- 11.Weiland JJ, Dreher TW. Infectious TYMV RNA from cloned cDNA: effects in vitro and in vivo of point substitutions in the initiation codons of two extensively overlapping ORFs. Nucleic Acids Res. 1989;17:4675–4687. doi: 10.1093/nar/17.12.4675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gargouri-Bouzid R, David C, Haenni AL. The 3′ promoter region involved in RNA synthesis directed by the turnip yellow mosaic virus genome in vitro. FEBS Lett. 1991;294:56–58. doi: 10.1016/0014-5793(91)81342-6. [DOI] [PubMed] [Google Scholar]
- 13.Singh RN, Dreher TW. Turnip yellow mosaic virus RNA-dependent RNA polymerase: initiation of minus strand synthesis in vitro. Virology. 1997;233:430–439. doi: 10.1006/viro.1997.8621. [DOI] [PubMed] [Google Scholar]
- 14.Barends S, Bink HH, van den Worm SH, Pleij CW, Kraal B. Entrapping ribosomes for viral translation: tRNA mimicry as a molecular Trojan horse. Cell. 2003;112:123–129. doi: 10.1016/s0092-8674(02)01256-4. [DOI] [PubMed] [Google Scholar]
- 15.Matsuda D, Dreher TW. Cap- and initiator tRNA-dependent initiation of TYMV polyprotein synthesis by ribosomes: evaluation of the Trojan horse model for TYMV RNA translation. RNA. 2007;13:129–137. doi: 10.1261/rna.244407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cho TJ, Dreher TW. Encapsidation of genomic but not subgenomic Turnip yellow mosaic virus RNA by coat protein provided in trans. Virology. 2006;356:126–135. doi: 10.1016/j.virol.2006.06.038. [DOI] [PubMed] [Google Scholar]
- 17.Giege R, Rudinger J, Dreher T, Perret V, Westhof E, Florentz C, Ebel JP. Search of essential parameters for the aminoacylation of viral tRNA-like molecules. Comparison with canonical transfer RNAs. Biochim Biophys Acta. 1990;1050:179–185. doi: 10.1016/0167-4781(90)90163-v. [DOI] [PubMed] [Google Scholar]
- 18.Mans RM, Guerrier-Takada C, Altman S, Pleij CW. Interaction of RNase P from Escherichia coli with pseudoknotted structures in viral RNAs. Nucleic Acids Res. 1990;18:3479–3487. doi: 10.1093/nar/18.12.3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsai CH, Dreher TW. Turnip yellow mosaic virus RNAs with anticodon loop substitutions that result in decreased valylation fail to replicate efficiently. J Virol. 1991;65:3060–3067. doi: 10.1128/jvi.65.6.3060-3067.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dreher TW, Tsai CH, Florentz C, Giege R. Specific valylation of turnip yellow mosaic virus RNA by wheat germ valyl-tRNA synthetase determined by three anticodon loop nucleotides. Biochemistry. 1992;31:9183–9189. doi: 10.1021/bi00153a010. [DOI] [PubMed] [Google Scholar]
- 21.Rudinger J, Florentz C, Dreher T, Giege R. Efficient mischarging of a viral tRNA-like structure and aminoacylation of a minihelix containing a pseudoknot: histidinylation of turnip yellow mosaic virus RNA. Nucleic Acids Res. 1992;20:1865–1870. doi: 10.1093/nar/20.8.1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bransom KL, Weiland JJ, Tsai CH, Dreher TW. Coding density of the turnip yellow mosaic virus genome: roles of the overlapping coat protein and p206-readthrough coding regions. Virology. 1995;206:403–412. doi: 10.1016/s0042-6822(95)80056-5. [DOI] [PubMed] [Google Scholar]
- 23.Deiman BA, Kortlever RM, Pleij CW. The role of the pseudoknot at the 3′ end of turnip yellow mosaic virus RNA in minus-strand synthesis by the viral RNA-dependent RNA polymerase. J Virol. 1997;71:5990–5996. doi: 10.1128/jvi.71.8.5990-5996.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kolk MH, van der Graaf M, Wijmenga SS, Pleij CW, Heus HA, Hilbers CW. NMR structure of a classical pseudoknot: interplay of single- and double-stranded RNA. Science. 1998;280:434–438. doi: 10.1126/science.280.5362.434. [DOI] [PubMed] [Google Scholar]
- 25.Filichkin SA, Bransom KL, Goodwin JB, Dreher TW. The infectivities of turnip yellow mosaic virus genomes with altered tRNA mimicry are not dependent on compensating mutations in the viral replication protein. J Virol. 2000;74:8368–8375. doi: 10.1128/jvi.74.18.8368-8375.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Smit MH, Gultyaev AP, Hilge M, Bink HH, Barends S, Kraal B, Pleij CW. Structural variation and functional importance of a D-loop-T-loop interaction in valine-accepting tRNA-like structures of plant viral RNAs. Nucleic Acids Res. 2002;30:4232–4240. doi: 10.1093/nar/gkf539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matsuda D, Bauer L, Tinnesand K, Dreher TW. Expression of the two nested overlapping reading frames of turnip yellow mosaic virus RNA is enhanced by a 5′ cap and by 5′ and 3′ viral sequences. J Virol. 2004;78:9325–9335. doi: 10.1128/JVI.78.17.9325-9335.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hammond JA, Rambo RP, Filbin ME, Kieft JS. Comparison and functional implications of the 3D architectures of viral tRNA-like structures. RNA. 2009;15:294–307. doi: 10.1261/rna.1360709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rietveld K, Pleij CW, Bosch L. Three-dimensional models of the tRNA-like 3′ termini of some plant viral RNAs. Embo J. 1983;2:1079–1085. doi: 10.1002/j.1460-2075.1983.tb01549.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mans RM, Verlaan PW, Pleij CW, Bosch L. Aminoacylation of 3′ terminal tRNA-like fragments of turnip yellow mosaic virus RNA: the influence of 5′ nonviral sequences. Biochim Biophys Acta. 1990;1050:186–192. doi: 10.1016/0167-4781(90)90164-w. [DOI] [PubMed] [Google Scholar]
- 31.Kolk MH, van der Graaf M, Fransen CT, Wijmenga SS, Pleij CW, Heus HA, Hilbers CW. Structure of the 3′-hairpin of the TYMV pseudoknot: preformation in RNA folding. Embo J. 1998;17:7498–7504. doi: 10.1093/emboj/17.24.7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Giege R, Florentz C, Dreher TW. The TYMV tRNA-like structure. Biochimie. 1993;75:569–582. doi: 10.1016/0300-9084(93)90063-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dreher TW, Goodwin JB. Transfer RNA mimicry among tymoviral genomic RNAs ranges from highly efficient to vestigial. Nucleic Acids Res. 1998;26:4356–4364. doi: 10.1093/nar/26.19.4356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dreher TW, Florentz C, Giege R. Valylation of tRNA-like transcripts from cloned cDNA of turnip yellow mosaic virus RNA demonstrate that the L-shaped region at the 3′ end of the viral RNA is not sufficient for optimal aminoacylation. Biochimie. 1988;70:1719–1727. doi: 10.1016/0300-9084(88)90030-2. [DOI] [PubMed] [Google Scholar]
- 35.Pfitzinger H, Weil JH, Pillay DTN, Guillemaut P. Preparation of a tRNA-dependent wheat germ protein-synthesizing system. Plant Mol Biol. 1989;12:301–301. doi: 10.1007/BF00043207. [DOI] [PubMed] [Google Scholar]
- 36.Rambo RP, Tainer JA. Improving small-angle X-ray scattering data for structural analyses of the RNA world. RNA. 2010;16:638–646. doi: 10.1261/rna.1946310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grishaev A, Ying J, Canny MD, Pardi A, Bax A. Solution structure of tRNAVal from refinement of homology model against residual dipolar coupling and SAXS data. J Biomol NMR. 2008;42:99–109. doi: 10.1007/s10858-008-9267-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Friederich MW, Gast FU, Vacano E, Hagerman PJ. Determination of the angle between the anticodon and aminoacyl acceptor stems of yeast phenylalanyl tRNA in solution. Proc Natl Acad Sci U S A. 1995;92:4803–4807. doi: 10.1073/pnas.92.11.4803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Friederich MW, Vacano E, Hagerman PJ. Global flexibility of tertiary structure in RNA: yeast tRNAPhe as a model system. Proc Natl Acad Sci U S A. 1998;95:3572–3577. doi: 10.1073/pnas.95.7.3572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ishitani R, Nureki O, Nameki N, Okada N, Nishimura S, Yokoyama S. Alternative tertiary structure of tRNA for recognition by a posttranscriptional modification enzyme. Cell. 2003;113:383–394. doi: 10.1016/s0092-8674(03)00280-0. [DOI] [PubMed] [Google Scholar]
- 41.Wilkinson KA, Merino EJ, Weeks KM. RNA SHAPE chemistry reveals nonhierarchical interactions dominate equilibrium structural transitions in tRNA(Asp) transcripts. J Am Chem Soc. 2005;127:4659–4667. doi: 10.1021/ja0436749. [DOI] [PubMed] [Google Scholar]
- 42.Gherghe CM, Mortimer SA, Krahn JM, Thompson NL, Weeks KM. Slow conformational dynamics at C2′-endo nucleotides in RNA. J Am Chem Soc. 2008;130:8884–8885. doi: 10.1021/ja802691e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Matsuda D, Yoshinari S, Dreher TW. eEF1A binding to aminoacylated viral RNA represses minus strand synthesis by TYMV RNA-dependent RNA polymerase. Virology. 2004;321:47–56. doi: 10.1016/j.virol.2003.10.028. [DOI] [PubMed] [Google Scholar]
- 44.Hellendoorn K, Mat AW, Gultyaev AP, Pleij CW. Secondary structure model of the coat protein gene of turnip yellow mosaic virus RNA: long, C-rich, single-stranded regions. Virology. 1996;224:43–54. doi: 10.1006/viro.1996.0505. [DOI] [PubMed] [Google Scholar]
- 45.Florentz C, Giege R. Contact areas of the turnip yellow mosaic virus tRNA-like structure interacting with yeast valyl-tRNA synthetase. J Mol Biol. 1986;191:117–130. doi: 10.1016/0022-2836(86)90427-4. [DOI] [PubMed] [Google Scholar]
- 46.Joshi S, Chapeville F, Haenni AL. Length requirements for tRNA-specific enzymes and cleavage specificity at the 3′ end of turnip yellow mosaic virus RNA. Nucleic Acids Res. 1982;10:1947–1962. doi: 10.1093/nar/10.6.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Prody GA, Bakos JT, Buzayan JM, Schneider IR, Bruening G. Autolytic Processing of Dimeric Plant Virus Satellite RNA. Science. 1986;231:1577–1580. doi: 10.1126/science.231.4745.1577. [DOI] [PubMed] [Google Scholar]
- 48.Pley HW, Flaherty KM, McKay DB. Three-dimensional structure of a hammerhead ribozyme. Nature. 1994;372:68–74. doi: 10.1038/372068a0. [DOI] [PubMed] [Google Scholar]
- 49.Khvorova A, Lescoute A, Westhof E, Jayasena SD. Sequence elements outside the hammerhead ribozyme catalytic core enable intracellular activity. Nat Struct Biol. 2003;10:708–712. doi: 10.1038/nsb959. [DOI] [PubMed] [Google Scholar]
- 50.Martick M, Scott WG. Tertiary contacts distant from the active site prime a ribozyme for catalysis. Cell. 2006;126:309–320. doi: 10.1016/j.cell.2006.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Goodwin JB, Dreher TW. Transfer RNA mimicry in a new group of positive-strand RNA plant viruses, the furoviruses: differential aminoacylation between the RNA components of one genome. Virology. 1998;246:170–178. doi: 10.1006/viro.1998.9193. [DOI] [PubMed] [Google Scholar]
- 52.Kieft JS, Costantino DA, Filbin ME, Hammond J, Pfingsten JS. Structural methods for studying IRES function. Methods Enzymol. 2007;430:333–371. doi: 10.1016/S0076-6879(07)30013-X. [DOI] [PubMed] [Google Scholar]
- 53.Hura GL, Menon AL, Hammel M, Rambo RP, Poole FL, 2nd, Tsutakawa SE, Jenney FE, Jr, Classen S, Frankel KA, Hopkins RC, Yang SJ, Scott JW, Dillard BD, Adams MW, Tainer JA. Robust, high-throughput solution structural analyses by small angle X-ray scattering (SAXS) Nat Methods. 2009;6:606–612. doi: 10.1038/nmeth.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fukai S, Nureki O, Sekine S, Shimada A, Tao J, Vassylyev DG, Yokoyama S. Structural basis for double-sieve discrimination of L-valine from L-isoleucine and L-threonine by the complex of tRNA(Val) and valyl-tRNA synthetase. Cell. 2000;103:793–803. doi: 10.1016/s0092-8674(00)00182-3. [DOI] [PubMed] [Google Scholar]