Abstract
Vibrio tubiashii NCIMB 1337 is a major and increasingly prevalent pathogen of bivalve mollusks, and shares a close phylogenetic relationship with both V. orientalis and V. coralliilyticus. It is a Gram-negative, curved rod-shaped bacterium, originally isolated from a moribund juvenile oyster, and is both oxidase and catalase positive. It is capable of growth under both aerobic and anaerobic conditions. Here we describe the features of this organism, together with the draft genome and annotation. The genome is 5,353,266 bp long, consisting of two chromosomes, and contains 4,864 protein-coding and 86 RNA genes.
Introduction
The genus Vibrio is both numerous and ubiquitous within marine environments, with Vibrio species harbored within many diverse marine organisms, such as mollusks, shrimps, fishes, cephalopods and corals [1]. Comparative genome analysis has revealed a huge genetic diversity within this genus, which is driven by mutations, chromosomal rearrangements, loss of genes by decay or deletion, and gene acquisitions through duplication or horizontal transfer (e.g. the acquisition of bacteriophages, pathogenicity islands, and super-integrons), the combination of which presumably stimulates genetic and functional diversity and allows this group to colonize a wide variety of ecological niches and hosts [1,2].
Vibrio tubiashii was first described as three strains of Vibrio anguillarum by Tubiash et al [3] in 1965. The organisms were isolated from bivalve mollusks during an outbreak of bacillary necrosis in Milford, Connecticut, and deposited in the American Type Culture Collection as ATCC 19105, 19106 and 19109. These three strains were further elucidated and formally named as V. tubiashii by Hada et al [4] in 1984. Subsequently, several virulence factors have been identified [5,6] and the organism is increasingly implicated in major disease outbreaks in bivalve mollusks [1].
V. tubiashii is closely related to the proposed coral pathogen V. coralliilyticus, as well as V. orientalis, a bacterium associated with penaeid shrimps [7]. Indeed, V. coralliilyticus was initially designated as a V. tubiashii strain [8,9] due to their close similarity.
Classification and features
Vibrio tubiashii 1337 belongs to the Gammaproteobacteria and are contained within the family, Vibrionaceae [Table 1]. Cells of Vibrio tubiashii are Gram-negative curved-rods of approximately 0.5 by 1.5 µm, which are motile in liquid media by means of a single sheathed, polar flagellum [3,4] These cells are facultative anaerobes, [3,4,22]. It is catalase and oxidase positive, capable of splitting indole from tryptophan, and can use glucose, xylose, mannitol, rhamnose, sucrose, arabinose and acetate as sole carbon sources, and has β-galactosidase activity, despite an apparent inability to ferment lactose. V. tubiashii is capable of dissimilatory nitrate and nitrite reduction under anaerobic conditions, can use organic phosphorus during phosphate limitation, and can utilize 2-aminoethylphosphonate as a sole phosphorus source.
Table 1. Classification and general features of V. tubiashii according to the MIGS recommendations.
| MIGS ID | Property | Term | Evidence code |
|---|---|---|---|
| Domain Bacteria | TAS [10] | ||
| Phylum Proteobacteria | TAS [11] | ||
| Class Gammaproteobacteria | TAS [12,13] | ||
| Current classification | Order Vibrionales | TAS [14] | |
| Family Vibrionaceae | TAS [15,16] | ||
| Genus Vibrio | TAS [15,17-19] | ||
| Species Vibrio tubiashii NCIMB 1337 | TAS [4] | ||
| Gram stain | negative | IDA | |
| Cell shape | Curved rods (vibroid) | IDA | |
| Motility | motile via single polar flagellum | IDA | |
| Sporulation | Non-sporulating | IDA | |
| Temperature range | Mesophile 12-30oC | IDA | |
| Optimum temperature | 25oC | IDA | |
| MIGS 6.3 | Salinity | Slightly halophylic, optimum 1-3% NaCl | IDA |
| MIGS-22 | Oxygen requirement | Aerobic/ facultative anaerobic | IDA |
| Carbon source | Highly diverse | IDA | |
| Energy source | Highly diverse | IDA | |
| MIGS-6 | Habitat | Marine invertebrates | TAS [20] |
| MIGS-16 | Biotic relationship | Parasitic | TAS [3] |
| MIGS-14 | Biosafety level | 2 | TAS [4] |
| Isolation | Moribund juvenile oyster (Crassostrea virginica) | TAS [3,4] | |
| MIGS-4 | Geographical location | Milford, Connecticut, USA | TAS [3] |
| MIGS-5 | Sample collection time | 01/02/1965 | TAS [3] |
| MIGS 4.1 | latitude | 41.22 N | TAS [3] |
| MIGS 4.2 | longitude | -73.06 W | TAS [3] |
| MIGS 4.3 | Depth | Not reported | |
| MIGS 4.4 | Altitude | Marine | TAS [3] |
Evidence codes - IDA: Inferred from Direct Assay (first time in publication); TAS: Traceable Author Statement (i.e., a direct report exists in the literature); NAS: Non-traceable Author Statement (i.e., not directly observed for the living, isolated sample, but based on a generally accepted property for the species, or anecdotal evidence). These evidence codes are from the Gene Ontology project [21]. If the evidence code is IDA, then the property was directly observed, for a live isolate by one of the authors, or an expert or reputable institution mentioned in the acknowledgements.
V. tubiashii has an absolute requirement for sodium and chloride ions, and is incapable of growth on media containing less than 0.5% W/V NaCl. The temperature optimum for growth is 25oC, but growth does occur in the range of 12-30oC. The organism is killed at 37oC. V. tubiashii has a biphasic pH response and grows optimally at both pH 8.0 and 6.5, but displays weakened growth at pH 7.0 and 7.5. The bacterium shows rapid growth on marine broth and produces buff colored, opaque, irregular, slightly convex colonies on marine agar, and yellow colonies, characteristic of the Vibrionaceae, on Thiosulfate-Citrate-Bile-Sucrose Agar (TCBS).
Growth conditions and DNA isolation
Vibrio tubiashii NCIMB 1337 (ATCC19106) was grown in marine broth (seawater + 1 gl-1 yeast extract and 0.5 gl-1 tryptone) at 25oC for 24 hours. DNA was extracted using the Qiagen DNAeasy blood and tissue kit, without modification of the manufacturer’s protocol.
Genome sequencing and annotation
Genome sequencing
The genome was sequenced using the Illumina sequencing platform. All general aspects of library construction and sequencing performed at the NERC Biomolecular analysis facility can be found on the NBAF website [23]. SOLEXA Illumina reads were assembled using VELVET Large Newbler contigs that were broken into 4,074 overlapping fragments of 1,000 bp and entered into the assembly as pseudo-reads. The sequences were assigned quality scores based on consensus q-scores with modifications to account for overlap redundancy and to adjust inflated q-scores. The error rate of the completed genome sequence is less than 1 in 100,000. Overall sequencing provided 131 × coverage of the genome.
Genome annotation
Genes were identified using the RAST server The predicted CDSs were translated and used to search the National Center for Biotechnology Information (NCBI) nonredundant database, UniProt, TIGRFam, Pfam, PRIAM, KEGG, COG, and InterPro databases. The tRNAScanSE tool [24] was used to find tRNA genes, whereas ribosomal RNAs were found by using BLASTn against the ribosomal RNA databases. The RNA components of the protein secretion complex and the RNaseP were identified by searching the genome for the corresponding Rfam profiles using INFERNAL [25]. Additional gene prediction analysis and manual functional annotation was performed within the Integrated Microbial Genomes (IMG) platform developed by the Joint Genome Institute, Walnut Creek, CA, USA [26,27].
Genome project information
This organism was selected for sequencing on the basis of its increasing impact as a bivalve pathogen, and was funded by i-G Peninsula. The genome project is deposited in the IMG database and the complete genome sequence in GenBank (CP001643). Sequencing, finishing and annotation were performed by the GenePool Team at NERC Biomolecular Analysis Facility (NBAF) Edinburgh. A summary of the project information is shown in Table 2.
Table 2. Project information.
| MIGS ID | Property | Term |
|---|---|---|
| MIGS-31 | Finishing quality | Draft |
| MIGS-28 | Libraries used | Illumina |
| MIGS-29 | Sequencing platforms | Illumina SOLEXA GAIIx |
| MIGS-31.2 | Fold coverage | 131× |
| MIGS-30 | Assemblers | Velvet |
| MIGS-32 | Gene calling method | RAST |
| Genome Database release | 181 | |
| Genbank ID | 866909 | |
| Genbank Date of Release | December 12, 2010 | |
| GOLD ID | Gi07317 |
Genomic properties
The genome was assembled into 335 contigs and includes two circular chromosomes combining to give a total size of 5,353,266 bp (44.84% GC content). A total of 4,950 genes were predicted, 4,864 of which are protein-coding genes. 74.22% of protein coding genes were assigned to a putative function with the remaining annotated as hypothetical proteins. 658 protein coding genes belong to paralogous families in this genome corresponding to a gene content redundancy of 13.29%. The properties and the statistics of the genome are summarized in Tables 3-5.
Table 3. Summary of genome*.
| Label | Size (Mb) |
|---|---|
| Chromosome 1 | 3.4 |
| Chromosome 2 | 1.9 |
* Two chromosomes with no plasmids. Approximate chromosome size estimated by Pulse field gel electrophoresis
Table 5. Number of genes associated with the 25 general COG functional categories.
| Code | Value | %age | Description |
|---|---|---|---|
| J | 200 | 4.86 | Translation |
| A | 1 | 0.02 | RNA processing and modification |
| K | 369 | 8.96 | Transcription |
| L | 154 | 3.74 | Replication, recombination and repair |
| B | 1 | 0.02 | Chromatin structure and dynamics |
| D | 37 | 0.9 | Cell cycle control, mitosis and chromosome partitioning |
| Y | Nuclear structure | ||
| V | 75 | 1.82 | Defense mechanisms |
| T | 432 | 8.31 | Signal transduction mechanisms |
| M | 227 | 5.51 | Cell wall/membrane biogenesis |
| N | 148 | 3.59 | Cell motility |
| U | 146 | 3.55 | Intracellular trafficking and secretion |
| O | 173 | 4.2 | Posttranslational modification, protein turnover, chaperones |
| C | 203 | 4.93 | Energy production and conversion |
| G | 248 | 6.02 | Carbohydrate transport and metabolism |
| E | 348 | 8.45 | Amino acid transport and metabolism |
| F | 105 | 2.55 | Nucleotide transport and metabolism |
| H | 159 | 3.86 | Coenzyme transport and metabolism |
| I | 119 | 2.89 | Lipid transport and metabolism |
| P | 188 | 4.57 | Inorganic ion transport and metabolism |
| Q | 77 | 1.77 | Secondary metabolites biosynthesis, transport and catabolism |
| R | 445 | 10.81 | General function prediction only |
| S | 356 | 8.65 | Function unknown |
| - | 1276 | 25.78 | Not in COGs |
a) The total is based on the total number of protein coding genes in the annotated genome.
Table 4. Nucleotide content and gene count levels of the genome.
| Attribute | Value | % of totala |
|---|---|---|
| Size (bp) | 5,353,266 | 100% |
| G+C content (bp) | 2,400,750 | 44.87% |
| Coding region (bp) | 4,627,782 | 86.45% |
| Total genesb | 4950 | 100% |
| RNA genes | 86 | 1.74% |
| Protein-coding genes | 4864 | 98.26% |
| Genes in paralog clusters | 658 | 13.29% |
| Genes assigned to COGs | 3674 | 74.22% |
| Genes with signal peptides | 1655 | 33.43% |
| Genes with transmembrane helices | 1167 | 23.58% |
| Paralogous groups | 658 | 13.29% |
a)The total is based on either the size of the genome in base pairs or the total number of protein coding genes in the annotated genome.
b)Also includes 54 pseudogenes and 5 other genes.
Genomic comparison
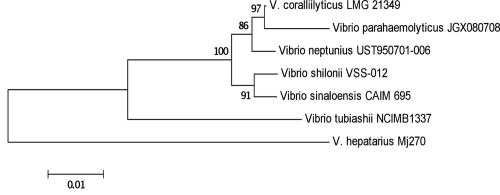
Based on COG I.D the Vibrio tubiashii genome shows most similarity to the genome of V coralliilyticus (R2 = 0.96) and to V. orientalis (R2 = 0.94), while showing less similarity to V. shilonii (R2= 0.86) [Table 6]. This is in contrast to the 16S-based analysis shown in Figure 1. However, it should be noted that 16S rRNA analysis often poorly discriminates vibrios due to low sequence heterogeneity in the 16S gene [28].
Table 6. Comparison of the genome of Vibrio tubiashii NCIMB 1337 with other sequenced Vibrios.
| Genome Name | Vibrio coralliilyticus ATCC BAA-450 | Vibrio orientalis CIP 102891 | Vibrio shilonii AK1 | Vibrio tubiashii NCIMB 1337 |
|---|---|---|---|---|
| Genes | 5,144 | 4,297 | 5,438 | 4,950 |
| RNA | 122 | 128 | 78 | 86 |
| w/ Func Pred | 3,687 | 3185 | 3,517 | 4,062 |
| w/ Func Pred % | 71.68% | 74.12% | 64.67% | 82.06% |
| Enzymes | 1,143 | 1,058 | 1,258 | 1,116 |
| Enzymes % | 22.22% | 24.62% | 23.13% | 22.55% |
| KEGG | 1397 | 1,257 | 1,511 | 1,354 |
| KEGG % | 27.16% | 29.25% | 27.79% | 27.35% |
| COG | 3815 | 3,302 | 4,093 | 3,674 |
| COG % | 74.16% | 76.84% | 75.27% | 74.22% |
| Pfam | 4127 | 3,520 | 4,379 | 3,976 |
| Pfam % | 80.23% | 81.92% | 80.53% | 80.32% |
| TIGRfam | 1,643 | 1,515 | 1,708 | 1,651 |
| TIGRfam % | 31.94% | 35.26% | 31.41% | 33.35% |
| Signal peptide | 1,733 | 1,408 | 1,214 | 1,655 |
| Signal peptide % | 33.69% | 32.77% | 22.32% | 33.43% |
| TransMb | 1,227 | 1,018 | 1,326 | 1,167 |
| TransMb Perc | 23.85% | 23.69% | 24.38% | 23.58% |
| Pfam Clusters | 2,183 | 2,091 | 2,163 | 2,186 |
| COG Clusters | 2,030 | 1,943 | 2,087 | 2,041 |
| TIGRfam Clusters | 1,310 | 1,246 | 1,300 | 1,323 |
| GC Perc | 0.46 | 0.45 | 0.44 | 0.45 |
| Bases | 5,680,628 | 4698244 | 5,701,826 | 5,353,266 |
Figure 1.
Phylogenetic tree highlighting the position of V. tubiashii NCIMB 1337 relative to other Vibrio strains. The tree was inferred from 1,159 aligned characters of the 16S rRNA gene sequence under the neighborhood joining criterion. Numbers above the branches are support values from 1,000 bootstrap replicates if greater than 60%.
Regulatory systems
The Vibrio tubiashii NCIMB 1337 genome contains multiple quorum sensing systems, most notably a luxM/N system which has two adjacent copies of the luxN gene. In addition, there is a luxS/PQ system, with the lux P and Q gene appearing consecutively. There is also a cqsA/S system. It is probable that these three systems converge on the phospho-relay transfer system encoded by the luxO/luxU/hapR genes. There are two additional lux genes (LuxT and LuxZ). The genome also contains the rpoN gene encoding for the sigma-54 factor, which may indicate the presence of the two-component phosphorylation-dephosphorylation cascade described in V. harveyi [29] (note: Vibrio harveyi is also known as Lucibacterium harveyi and Beneckea harveyi.)
Antibiotic resistance
There are six separate genes encoding for putative β-lactamases within the genome, but only two have homology at the protein levels with any know Vibrio β-lactamases. There is also a multi-antibiotic resistance protein MarC, associated with an operon containing a variety of multidrug resistance proteins. This operon is controlled by a MerR type transcriptional regulator, which is often associated with antibiotic resistance [30], and may account for the kanamycin resistance observed in this strain by the authors.
Acknowledgements
We wish to thank i-G Peninsula (Prospect Place, the Hoe, Plymouth, Devon, UK) for providing funding for this project, and NBAF Edinburgh for performing the sequencing.
References
- 1.Thompson FL, Iida T, Swings J. “Biodiversity of vibrios.,” Microbiology and molecular biology reviews. [Table of contents]. Microbiol Mol Biol Rev 2004; 68:403-431 10.1128/MMBR.68.3.403-431.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Colwell RR, Huq A. Environmental Reservoir of Vibrio cholerae The Causative Agent of Cholera. Ann N Y Acad Sci 1994; 740:44-54 10.1111/j.1749-6632.1994.tb19852.x [DOI] [PubMed] [Google Scholar]
- 3.Tubiash HS, Chanley PE, Leifson E. Bacillary necrosis, a disease of larval and juvenile bivalve mollusks. I. Etiology and epizootiology. J Bacteriol 1965; 90:1036-1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hada HS, West PA, Lee JV, Stemmler J, Colwell RR. Vibrio tubiashii sp. nov., a Pathogen of Bivalve Mollusks. Int J Syst Bacteriol 1984; 34:1-4 10.1099/00207713-34-1-1 [DOI] [Google Scholar]
- 5.Beaubrun JJG, Kothary MH, Curtis SK, Flores NC, Eribo BE, Tall BD. Isolation and characterization of Vibrio tubiashii outer membrane proteins and determination of a toxR homolog. Appl Environ Microbiol 2008; 74:907-911 10.1128/AEM.02052-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kothary MH, Delston RB, Curtis SK, McCardell BA, Tall BD. Purification and characterization of a vulnificolysin-like cytolysin produced by Vibrio tubiashii. Appl Environ Microbiol 2001; 67:3707-3711 10.1128/AEM.67.8.3707-3711.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abraham T. Distribution of luminous bacteria in semi-intensive penaeid shrimp hatcheries of Tamil Nadu, India. Aquaculture 2004; 232:81-90 10.1016/S0044-8486(03)00485-X [DOI] [Google Scholar]
- 8.Beaubrun JJG, Kothary MH, Curtis SK, Flores NC, Eribo BE, Tall BD. Isolation and characterization of Vibrio tubiashii outer membrane proteins and determination of a toxR homolog. Appl Environ Microbiol 2008; 74:907-911 10.1128/AEM.02052-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ben-Haim Y, Zicherman-Keren M, Rosenberg E. Temperature-Regulated Bleaching and Lysis of the Coral Pocillopora damicornis by the Novel Pathogen Vibrio coralliilyticus. Appl Environ Microbiol 2003; 69:4236-4242 10.1128/AEM.69.7.4236-4242.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci USA 1990; 87:4576-4579 10.1073/pnas.87.12.4576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garrity GM, Holt JG. The Road Map to the Manual. In: Garrity GM, Boone DR, Castenholz RW (eds), Bergey's Manual of Systematic Bacteriology, Second Edition, Volume 1, Springer, New York, 2001, p. 119-169. [Google Scholar]
- 12.List Editor Validation of publication of new names and new combinations previously effectively published outside the IJSEM. List no. 106. Int J Syst Evol Microbiol 2005; 55:2235-2238 10.1099/ijs.0.64108-0 [DOI] [PubMed] [Google Scholar]
- 13.Garrity GM, Bell JA, Lilburn T. Class III. Gammaproteobacteria class. nov. In: Garrity GM, Brenner DJ, Krieg NR, Staley JT (eds), Bergey's Manual of Systematic Bacteriology, Second Edition, Volume 2, Part B, Springer, New York, 2005, p. 1. [Google Scholar]
- 14.Garrity GM, Holt JG. Taxonomic Outline of the Archaea and Bacteria In: Garrity GM, Boone DR, Castenholz RW (eds), Bergey's Manual of Systematic Bacteriology, Second Edition, Volume 1, Springer, New York, 2001, p. 155-166. [Google Scholar]
- 15.Skerman VBD, McGowan V, Sneath PHA. Approved Lists of Bacterial Names. Int J Syst Bacteriol 1980; 30:225-420 10.1099/00207713-30-1-225 [DOI] [PubMed] [Google Scholar]
- 16.Véron M. La position taxonomique des Vibrio et de certaines bactéries comparables. C R Acad Sci Hebd Seances Acad Sci 1965; 261:5243-5246 [PubMed] [Google Scholar]
- 17.Pacini F. Osservazione microscopiche e deduzioni patologiche sul cholera asiatico. Gazette Medicale de Italiana Toscano Firenze 1854; 6:405-412 [Google Scholar]
- 18.Shewan J, Veron M. Genus I. Vibrio Pacini 1854, 411. In: Buchanan RE, Gibbons NE (eds), Bergey's Manual of Determinative Bacteriology, Eighth Edition, The Williams and Wilkins Co., Baltimore, 1974, p. 340-345. [Google Scholar]
- 19.Judicial Commission Opinion 31. Conservation of Vibrio Pacini 1854 as a Bacterial Generic Name, Conservation of Vibrio cholerae Pacini 1854 as the Nomenclatural Type Species of the Bacterial Genus Vibrio, and Designation of Neotype Strain of Vibrio cholerae Pacini. Int Bull Bacteriol Nomencl Taxon 1965; 15:185-186 10.1099/00207713-15-3-185 [DOI] [Google Scholar]
- 20.Hada HS, West PA, Lee JV, Stemmler J, Colwell RR. Vibrio tubiashii sp. nov., a Pathogen of Bivalve Mollusks. Int J Syst Bacteriol 1984; 34:1-4 10.1099/00207713-34-1-1 [DOI] [Google Scholar]
- 21.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene Ontology: tool for the unification of biology. Nat Genet 2000; 25:25-29 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pillidge CJ, Colwell RR. Nucleotide sequence of the 5S rRNA from Listonella (Vibrio) ordalii ATCC 33509 and Listonella (Vibrio) tubiashii ATCC 19105. Nucleic Acids Res 1988; 16:3111 10.1093/nar/16.7.3111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.NBAF website. http://nbaf.nerc.ac.uk
- 24.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 1997; 25:955-964 10.1093/nar/25.5.955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.INFERNAL http://infernal.janelia.org
- 26.Markowitz VM, Korzeniewski F, Palaniappan K, Szeto E, Werner G, Padki A, Zhao X, Dubchak I, Hugenholtz P, Anderson I, et al. The Integrated Microbial Genomes (IMG) system. Nucleic Acids Res 2006; 34:D344-D348 10.1093/nar/gkj024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.The DOE Joint Genome Institute. http://img.jgi.doe.gov
- 28.Thompson FL, Gevers D, Thompson CC, Dawyndt P, Naser S, Hoste B, Munn CB, Swings J. Phylogeny and molecular identification of vibrios on the basis of multilocus sequence analysis. Appl Environ Microbiol 2005; 71:5107-5115 10.1128/AEM.71.9.5107-5115.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lilley BN, Bassler BL. Regulation of quorum sensing in Vibrio harveyi by LuxO and sigma-54. Mol Microbiol 2000; 36:940-954 10.1046/j.1365-2958.2000.01913.x [DOI] [PubMed] [Google Scholar]
- 30.Brown NL, Stoyanov JV, Kidd SP, Hobman JL. The MerR family of transcriptional regulators. FEMS Microbiol Rev 2003; 27:145-163 10.1016/S0168-6445(03)00051-2 [DOI] [PubMed] [Google Scholar]