Abstract
Odoribacter splanchnicus (Werner et al. 1975) Hardham et al. 2008 is the type species of the genus Odoribacter, which belongs to the family Porphyromonadaceae in the order ‘Bacteroidales’. The species is of interest because members of the Odoribacter form an isolated cluster within the Porphyromonadaceae. This is the first completed genome sequence of a member of the genus Odoribacter and the fourth sequence from the family Porphyromonadaceae. The 4,392,288 bp long genome with its 3,672 protein-coding and 74 RNA genes and is a part of the Genomic Encyclopedia of Bacteria and Archaea project.
Keywords: strictly anaerobic, non-motile, Gram-negative, opportunistic pathogen, mesophilic, chemoorganotrophic, Porphyromonadaceae, GEBA
Introduction
Strain 1651/6T (= DSM 20712 = ATCC 29572 = JCM 15291) is the type strain of Odoribacter splanchnicus [1,2]. Currently, there are three species placed in the genus Odoribacter [1]. The generic name derives from the Latin noun odor meaning smell and the Neo-Latin word bacter meaning a rod, referring to a rod of (bad) smell [2]. The species epithet is derived from the Greek plural noun splanchna meaning innards, referring to the internal organs as the site of isolation [2]. O. splanchnicus strain 1651/6T was isolated as Bacteroides splanchnicus from a human, abdominal abscess by Werner and Reichertz in 1971 [3] and described in 1975 [4]. The species was first validly published as B. splanchnicus due to a number of shared characteristics with the members of the genus Bacteroides. However, the organism differs from other Bacteroides species in a number of important biochemical characteristics [5] and shows less than 20% relatedness in the homology of 16S rRNA genes compared to the B. fragilis group [6]. In 1994, through further studies of the phylogenetic structure of the bacteroides subgroup it became clear that B. splanchnicus did not belong to the genera Bacteroides, Prevotella or Porphyromonas, but fell just outside these three major clusters [7]. Finally, in 2008, the new genus Odoribacter was described and B. splanchnicus was reclassified as its new type species [2]. Additional isolates of O. splanchnicus have been obtained from stool specimens and surgically removed appendices [2]; in one case of pelviperitonitis the organism was isolated from a blood sample and peritoneal pus [8]. In general, O. splanchnicus can be described as an inhabitant of the human intestine that has the potential to become an opportunistic pathogen. Here we present a summary classification and a set of features for O. splanchnicus 1651/6T, together with the description of the complete genomic sequencing and annotation.
Classification and features
A representative genomic 16S rRNA sequence of strain 1651/6T was compared using NCBI BLAST under default settings (e.g., considering only the high-scoring segment pairs (HSPs) from the best 250 hits) with the most recent release of the Greengenes database [9] and the relative frequencies of taxa and keywords (reduced to their stem [10]) were determined, weighted by BLAST scores. The most frequently occurring genera were Bacteroides (43.5%), Odoribacter (37.9%), Alistipes (15.2%) and Brumimicrobium (3.4%) (21 hits in total). Regarding the two hits to sequences from members of the species, the average identity within HSPs was 99.7%, whereas the average coverage by HSPs was 97.9%. Regarding the two hits to sequences from other members of the genus, the average identity within HSPs was 93.4%, whereas the average coverage by HSPs was 42.5%. The highest-scoring environmental sequence was EF401000 ('human fecal clone SJTU D 04 48'), which showed an identity of 99.8% and an HSP coverage of 98.2%. The most frequently occurring keywords within the labels of environmental samples which yielded hits were 'human' (13.4%), 'biopsi' (7.4%), 'mucos' (7.1%), 'fecal' (6.1%) and 'colon' (5.3%) (229 hits in total). The most frequently occurring keyword within the labels of environmental samples which yielded hits of a higher score than the highest scoring species was 'fecal/human' (50.0%) (27 hits in total).
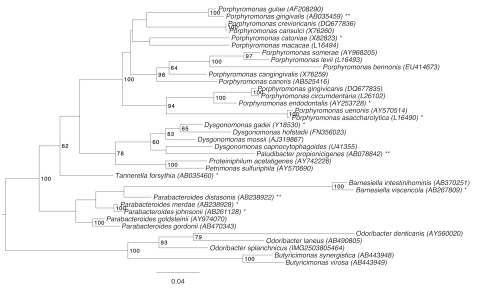
Figure 1 shows the phylogenetic neighborhood of O. splanchnicus in a 16S rRNA based tree. The sequences of the four 16S rRNA gene copies in the genome differ from each other by up to eight nucleotides, and differ by up to nine nucleotides from the previously published 16S rRNA sequence (L16496), which contains nine ambiguous base calls
Figure 1.
Phylogenetic tree highlighting the position of O. splanchnicus relative to the other type strains within the family Porphyromonadaceae. The tree was inferred from 1,401 aligned characters [11,12] of the 16S rRNA gene sequence under the maximum likelihood criterion [13]. Rooting was done initially using the midpoint method [14] and then checked for its agreement with the current classification (Table 1). The branches are scaled in terms of the expected number of substitutions per site. Numbers to the right of bifurcations are support values from 250 bootstrap replicates [15] if larger than 60%. Lineages with type strain genome sequencing projects registered in GOLD [16] are labeled by one asterisk, published genomes by two asterisks [17-19].
The cells of O. splanchnicus generally have the shape of short rods (0.7 × 1.0-5.0 µm) which occur singly or in lightly associated groups (Figure 2). They can also be pleomorphic. O. splanchnicus is a Gram-negative, non-pigmented and non spore-forming bacterium (Table 1). The organism is described as non-motile and only ten genes associated with motility have been found in the genome (see below). O. splanchnicus grows well at 37°C, is strictly anaerobic, chemoorganotrophic and is able to ferment glucose, fructose, galactose, arabinose, lactose and mannose but does not utilize sucrose, rhamnose, trehalose or salicin [4,5]. The organism does not reduce nitrate but it produces indole from tryptophan and hydrolyzes esculin [28]. O. splanchnicus does not require hemin for growth but is highly stimulated by its presence and does not show hemolysis on blood agar. Growth is enhanced by the addition of 20% bile. Major fermentation products are acetic acid, propionic acid and succinic acid; butyric acid, isovaleric acid and isobutyric acid are produced in small amounts [4,29]. When amino acids are used as carbon sources, only lysine enables butyrate production [4]. It is known that O. splanchnicus possesses highly active pentose phosphate pathway enzymes such as glucose-6-phosphate dehydrogenase and 6-phosphogluconate dehydrogenase as well as active malate dehydrogenase and glutamate dehydrogenase [30]. The organism produces large amounts of hydrogen and H2S. Strain 1651/6T is phosphatase, α- and β-galactosidase, α-fucosidase, N-acetylglucosaminidase and glutamic acid decarboxylase active and urease and catalase inactive [2]. The organism produces arginine arylamidase, leucyl glycine arylamidase, leucine arylamidase, alanine arylamidase (own, unpublished data) and glycylprolyl arylamidase [31]. O. splanchnicus is reported to grow in the presence of aminoglycosides and polymyxins (minimum inhibitory concentration (MIC) value greater than 60 µg/ml); chloramphenicol, penicillins and cephalosporins show bacteriostatic activity (5-40 µg/ml). The organism is susceptible to tetracyclines, lincomycin, clindamycin, rifampicin and erythromycin (MIC values less than 0.5 µg/ml) [4,28].
Figure 2.
Scanning electron micrograph of O. splanchnicus 1651/6T
Table 1. Classification and general features of O. splanchnicus 1651/6T according to the MIGS recommendations [20].
| MIGS ID | Property | Term | Evidence code |
|---|---|---|---|
| Current classification | Domain Bacteria | TAS [21] | |
| Phylum 'Bacteroidetes' | TAS [22] | ||
| Class 'Bacteroidia' | TAS [23,24] | ||
| Order 'Bacteroidales' | TAS [25] | ||
| Family 'Porphyromonadaceae' | TAS [25] | ||
| Genus Odoribacter | TAS [2] | ||
| Species Odoribacter splanchnicus | TAS [2] | ||
| Type strain 1651/6 | TAS [4] | ||
| Gram stain | negative | TAS [4] | |
| Cell shape | rod-shaped | TAS [4] | |
| Motility | non-motile | TAS [4] | |
| Sporulation | none | TAS [4] | |
| Temperature range | mesophile | TAS [4] | |
| Optimum temperature | 37°C | TAS [4] | |
| Salinity | normal | TAS [4] | |
| MIGS-22 | Oxygen requirement | strictly anaerobic | TAS [4] |
| Carbon source | carbohydrates; nitrogenous substrates | TAS [4] | |
| Energy metabolism | chemoorganotroph | TAS [4] | |
| MIGS-6 | Habitat | Homo sapiens, gastrointestinal tract | TAS [4] |
| MIGS-15 | Biotic relationship | free-living | NAS |
| MIGS-14 | Pathogenicity | opportunistic pathogen | TAS [4] |
| Biosafety level | 2 | TAS [26] | |
| Isolation | abdominal abscess | TAS [3] | |
| MIGS-4 | Geographic location | Germany | TAS [3] |
| MIGS-5 | Sample collection time | 1971 or before | TAS [3] |
| MIGS-4.1 | Latitude | not reported | NAS |
| MIGS-4.2 | Longitude | not reported | NAS |
| MIGS-4.3 | Depth | not reported | NAS |
| MIGS-4.4 | Altitude | not reported | NAS |
Evidence codes - IDA: Inferred from Direct Assay (first time in publication); TAS: Traceable Author Statement (i.e., a direct report exists in the literature); NAS: Non-traceable Author Statement (i.e., not directly observed for the living, isolated sample, but based on a generally accepted property for the species, or anecdotal evidence). These evidence codes are from of the Gene Ontology project [27]. If the evidence code is IDA, then the property was directly observed by one of the authors or an expert mentioned in the acknowledgements.
Chemotaxonomy
Little chemotaxonomic information is available for strain 1651/6T. It possesses meso-diaminopimelic acid in its peptidoglycan [30], sphingophospholipids as polar lipids [32] and the sole menaquinone present is MK-9 [30]. The major fatty acids found are iso-C15:0, C14:0, anteiso-C15:0 and C16:03-OH [30].
Genome sequencing and annotation
Genome project history
This organism was selected for sequencing on the basis of its phylogenetic position [33], and is part of the Genomic Encyclopedia of Bacteria and Archaea project [34]. The genome project is deposited in the Genomes On Line Database [16] and the complete genome sequence is deposited in GenBank. Sequencing, finishing and annotation were performed by the DOE Joint Genome Institute (JGI). A summary of the project information is shown in Table 2.
Table 2. Genome sequencing project information.
| MIGS ID | Property | Term |
|---|---|---|
| MIGS-31 | Finishing quality | Finished |
| MIGS-28 | Libraries used | Three genomic libraries: one 454 pyrosequence standard library, one 454 PE library (8 kb insert size), one Illumina library |
| MIGS-29 | Sequencing platforms | Illumina GAii, 454 GS FLX Titanium |
| MIGS-31.2 | Sequencing coverage | 521.0 × Illumina; 31.5 × pyrosequence |
| MIGS-30 | Assemblers | Newbler version 2.3-PreRelease-10-21-2009, Velvet version 0.7.63, phrap version 4.24 |
| MIGS-32 | Gene calling method | Prodigal 1.4, GenePRIMP |
| INSDC ID | CP002544 | |
| Genbank Date of Release | February 28, 2011 | |
| GOLD ID | Gc01667 | |
| NCBI project ID | 43469 | |
| Database: IMG-GEBA | 2503754021 | |
| MIGS-13 | Source material identifier | DSM 20712 |
| Project relevance | Tree of Life, GEBA |
Growth conditions and DNA isolation
O. splanchnicus 1651/6T, DSM 20712, was grown anaerobically in DSMZ medium 110 (Chopped meat medium with carbohydrates) [35] at 37°C. DNA was isolated from 0.5-1 g of cell paste using Jetflex Genomic DNA Purification kit (GENOMED 600100) following the standard protocol as recommended by the manufacturer, but adding 20 µL proteinase K for 45 min lysis at 58ºC. DNA is available through the DNA Bank Network [36].
Genome sequencing and assembly
The genome was sequenced using a combination of Illumina and 454 sequencing platforms. All general aspects of library construction and sequencing can be found at the JGI website [37]. Pyrosequencing reads were assembled using the Newbler assembler version 2.3-PreRelease-10-21-2009 (Roche). The initial Newbler assembly consisting of 57 contigs in eight scaffolds was converted into a phrap [38] assembly by making fake reads from the consensus, to collect the read pairs in the 454 paired end library. Illumina GAii sequencing data (2,241.8 Mb) was assembled with Velvet, version 0.7.63 [39] and the consensus sequences were shredded into 1.5 kb overlapped fake reads and assembled together with the 454 data. The 454 draft assembly was based on 138 Mb 454 draft data and all of the 454 paired end data. Newbler parameters are -consed -a 50 -l 350 -g -m -ml 20. The Phred/Phrap/Consed software package [38] was used for sequence assembly and quality assessment in the subsequent finishing process. After the shotgun stage, reads were assembled with parallel phrap (High Performance Software, LLC). Possible mis-assemblies were corrected with gapResolution [37], Dupfinisher, or sequencing cloned bridging PCR fragments with subcloning or transposon bombing (Epicentre Biotechnologies, Madison, WI) [40]. Gaps between contigs were closed by editing in Consed, by PCR and by Bubble PCR primer walks (J.-F.Chang, unpublished). A total of 65 additional reactions were necessary to close gaps and to raise the quality of the finished sequence. Illumina reads were also used to correct potential base errors and increase consensus quality using a software Polisher developed at JGI [41]. The error rate of the completed genome sequence is less than 1 in 100,000. Together, the combination of the Illumina and 454 sequencing platforms provided 552.5 × coverage of the genome. The final assembly contained 389,415 pyrosequence and 33,128,505 Illumina reads.
Genome annotation
Genes were identified using Prodigal [42] as part of the Oak Ridge National Laboratory genome annotation pipeline, followed by a round of manual curation using the JGI GenePRIMP pipeline [43]. The predicted CDSs were translated and used to search the National Center for Biotechnology Information (NCBI) nonredundant database, UniProt, TIGR-Fam, Pfam, PRIAM, KEGG, COG, and InterPro databases. Additional gene prediction analysis and functional annotation was performed within the Integrated Microbial Genomes - Expert Review (IMG-ER) platform [44].
Genome properties
The genome consists of a 4,392,288 bp long chromosome with a G+C content of 43.4% (Table 3 and Figure 3). Of the 3,746 genes predicted, 3,672 were protein-coding genes, and 74 RNAs; 175 pseudogenes were also identified. The majority of the protein-coding genes (61.2%) were assigned with a putative function while the remaining ones were annotated as hypothetical proteins. The distribution of genes into COGs functional categories is presented in Table 4.
Table 3. Genome Statistics.
| Attribute | Value | % of Total |
|---|---|---|
| Genome size (bp) | 4,392,288 | 100.00% |
| DNA coding region (bp) | 3,824,553 | 87.07% |
| DNA G+C content (bp) | 1,904,432 | 43.36% |
| Number of replicons | 1 | |
| Extrachromosomal elements | 0 | |
| Total genes | 3,746 | 100.00% |
| RNA genes | 74 | 1.98% |
| rRNA operons | 4 | |
| Protein-coding genes | 3,672 | 98.02% |
| Pseudo genes | 175 | 4.67% |
| Genes with function prediction | 2,291 | 61.16% |
| Genes in paralog clusters | 734 | 19.59% |
| Genes assigned to COGs | 2,252 | 60.12% |
| Genes assigned Pfam domains | 2,523 | 67.35% |
| Genes with signal peptides | 909 | 24.27% |
| Genes with transmembrane helices | 823 | 21.97% |
| CRISPR repeats | 1 |
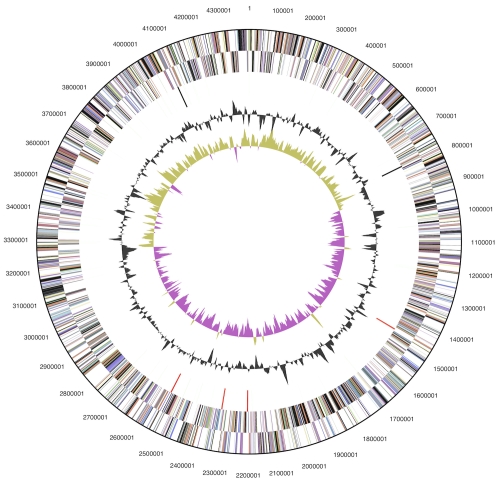
Figure 3.
Graphical circular map of the chromosome. From outside to the center: Genes on forward strand (color by COG categories), Genes on reverse strand (color by COG categories), RNA genes (tRNAs green, rRNAs red, other RNAs black), GC content, GC skew.
Table 4. Number of genes associated with the general COG functional categories.
| Code | value | %age | Description |
|---|---|---|---|
| J | 149 | 5.9 | Translation, ribosomal structure and biogenesis |
| A | 0 | 0.0 | RNA processing and modification |
| K | 188 | 7.5 | Transcription |
| L | 161 | 6.4 | Replication, recombination and repair |
| B | 0 | 0.0 | Chromatin structure and dynamics |
| D | 23 | 0.9 | Cell cycle control, cell division, chromosome partitioning |
| Y | 0 | 0.0 | Nuclear structure |
| V | 67 | 2.7 | Defense mechanisms |
| T | 144 | 5.7 | Signal transduction mechanisms |
| M | 215 | 8.6 | Cell wall/membrane/envelope biogenesis |
| N | 10 | 0.4 | Cell motility |
| Z | 0 | 0.0 | Cytoskeleton |
| W | 0 | 0.0 | Extracellular structures |
| U | 48 | 2.1 | Intracellular trafficking, secretion, and vesicular transport |
| O | 134 | 5.3 | Posttranslational modification, protein turnover, chaperones |
| C | 164 | 6.5 | Energy production and conversion |
| G | 111 | 4.4 | Carbohydrate transport and metabolism |
| E | 175 | 7.0 | Amino acid transport and metabolism |
| F | 62 | 2.5 | Nucleotide transport and metabolism |
| H | 126 | 5.2 | Coenzyme transport and metabolism |
| I | 62 | 2.5 | Lipid transport and metabolism |
| P | 216 | 8.6 | Inorganic ion transport and metabolism |
| Q | 24 | 1.0 | Secondary metabolites biosynthesis, transport and catabolism |
| R | 280 | 11.2 | General function prediction only |
| S | 149 | 5.9 | Function unknown |
| - | 1,494 | 39.9 | Not in COGs |
Acknowledgements
We would like to gratefully acknowledge the help of Sabine Welnitz (DSMZ) for growing O. splanchnicus cultures. This work was performed under the auspices of the US Department of Energy Office of Science, Biological and Environmental Research Program, and by the University of California, Lawrence Berkeley National Laboratory under contract No. DE-AC02-05CH11231, Lawrence Livermore National Laboratory under Contract No. DE-AC52-07NA27344, and Los Alamos National Laboratory under contract No. DE-AC02-06NA25396, UT-Battelle and Oak Ridge National Laboratory under contract DE-AC05-00OR22725, as well as German Research Foundation (DFG) INST 599/1-2.
References
- 1.Garrity G. NamesforLife. BrowserTool takes expertise out of the database and puts it right in the browser. Microbiol Today 2010; 7:1 [Google Scholar]
- 2.Hardham JM, King KW, Dreier K, Wong J, Strietzel C, Eversole RR, Sfintescu C, Evans RT. Transfer of Bacteroides splanchnicus to Odoribacter gen. nov. as Odoribacter splanchnicus comb. nov., and description of Odoribacter denticanis sp. nov., isolated from the crevicular spaces of canine periodontitis patients. Int J Syst Evol Microbiol 2008; 58:103-109 10.1099/ijs.0.63458-0 [DOI] [PubMed] [Google Scholar]
- 3.Werner H, Reichertz C. Buttersäurebildende Bacteroides-Kulturen. Zentralbl Bakteriol Parasitenkd Infektionskr Hyg Abt 1 Orig Reihe A 1971; 217:206-216. [PubMed]
- 4.Werner H., Rintelen G, Kunstek-Santos H. A new butyric acid-producing Bacteroides species: B. splanchnicus n. sp. Zentralbl Bakteriol Parasitenkd Infektionskr Hyg Abt 1 Orig Reihe A 1975; 231:133-144. [PubMed]
- 5.Shah HN. 1992. The genus Bacteroides and related species, p.3593-3607. In Balows A, Trüper HG, Dworkin M, Harder W, Schleifer K.-H (ed.), The prokaryotes, 2nd ed. Springer, New York. [Google Scholar]
- 6.Johnson JL, Harich B. Ribosomal ribonucleic acid homology among species of the genus Bacteroides. Int J Syst Bacteriol 1986; 36:71-79 10.1099/00207713-36-1-71 [DOI] [Google Scholar]
- 7.Paster BJ, Dewhirst FE, Olsen I, Fraser GJ. Phylogeny of Bacteroides, Prevotella, and Porphyromonas spp. and related bacteria. J Bacteriol 1994; 176:725-732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Labbe M, Mertens A, Schoutens E. Pelviperitonitis and bacteremia due to Bacteroides splanchnicus. Report of a case. Zentralbl Bakteriol 1977; 238:251-254 [PubMed] [Google Scholar]
- 9.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 2006; 72:5069-5072 10.1128/AEM.03006-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Porter MF. An algorithm for suffix stripping. Program: electronic library and information systems 1980; 14:130-137.
- 11.Lee C, Grasso C, Sharlow MF. Multiple sequence alignment using partial order graphs. Bioinformatics 2002; 18:452-464 10.1093/bioinformatics/18.3.452 [DOI] [PubMed] [Google Scholar]
- 12.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 2000; 17:540-552 [DOI] [PubMed] [Google Scholar]
- 13.Stamatakis A, Hoover P, Rougemont J. A rapid bootstrap algorithm for the RAxML Web servers. Syst Biol 2008; 57:758-771 10.1080/10635150802429642 [DOI] [PubMed] [Google Scholar]
- 14.Hess PN, De Moraes Russo CA. An empirical test of the midpoint rooting method. Biol J Linn Soc Lond 2007; 92:669-674 10.1111/j.1095-8312.2007.00864.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pattengale ND, Alipour M, Bininda-Emonds ORP, Moret BME, Stamatakis A. How many bootstrap replicates are necessary? Lect Notes Comput Sci 2009; 5541:184-200 10.1007/978-3-642-02008-7_13 [DOI] [PubMed] [Google Scholar]
- 16.Liolios K, Chen IM, Mavromatis K, Tavernarakis N, Hugenholtz P, Markowitz VM, Kyrpides NC. The Genomes On Line Database (GOLD) in 2009: status of genomic and metagenomic projects and their associated metadata. Nucleic Acids Res 2010; 38:D346-D354 10.1093/nar/gkp848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu J, Mahowald MA, Ley RE, Lozupone CA, Hamady M, Martens EC, Henrissat B, Coutinho PM, Minx P, Latreille P, et al. Evolution of symbiotic bacteria in the distal human intestine. PLoS Biol 2007; 5:e156 10.1371/journal.pbio.0050156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Naito M, Hirakawa H, Yamashita A, Ohara N, Shoji M, Yukitake H, Nakayama K, Toh H, Yoshimura F, Kuhara S, et al. Determination of the genome sequence of Porphyromonas gingivalis strain ATCC 33277 and genomic comparison with strain W83 revealed extensive genome rearrangements in P. gingivalis. DNA Res 2008; 15:215-225 10.1093/dnares/dsn013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gronow S, Munk C, Lapidus A, Nolan M, Lucas S, Hammon N, Deshpande S, Cheng JF, Tapia R, Han C, et al. Complete genome sequence of Paludibacter propionicigenes type strain (WB4T). Stand Genomic Sci 2011; 4:36-44 10.4056/sigs.1503846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Field D, Garrity G, Gray T, Morrison N, Selengut J, Sterk P, Tatusova T, Thomson N, Allen MJ, Angiuoli SV, et al. The minimum information about a genome sequence (MIGS) specification. Nat Biotechnol 2008; 26:541-547 10.1038/nbt1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci USA 1990; 87:4576-4579 10.1073/pnas.87.12.4576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garrity GM, Holt JG. The Road Map to the Manual. In: Garrity GM, Boone DR, Castenholz RW (eds), Bergey's Manual of Systematic Bacteriology, Second Edition, Volume 1, Springer, New York, 2001, p. 119-169. [Google Scholar]
- 23.Ludwig W, Euzeby J, Whitman WG. Draft taxonomic outline of the Bacteroidetes, Planctomycetes, Chlamydiae, Spirochaetes, Fibrobacteres, Fusobacteria, Acidobacteria, Verrucomicrobia, Dictyoglomi, and Gemmatimonadetes http://www.bergeys.org/outlines/Bergeys_Vol_4_Outline.pdf. Taxonomic Outline 2008
- 24.Abt B, Tashima H, Lucas S, Lapidus A, Glavina Del Rio T, Nolan M, Tice H, Cheng JF, Pitluck S, Liolios K, et al. Complete genome sequence of Leadbetterella byssophila type strain (4M15T). Stand Genomic Sci 2011; 4:2-12 10.4056/sigs.1413518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garrity GM, Holt JG. Taxonomic outline of the Archaea and Bacteria, In Garrity GM, Boone DR, Castenholz RW (eds), Bergey's Manual of Systematic Bacteriology, 2nd ed, vol. 1. Springer, New York, 2001, p. 155-166. [Google Scholar]
- 26.Classification of bacteria and archaea in risk groups. http://www.baua.de TRBA 466.
- 27.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene Ontology: tool for the unification of biology. Nat Genet 2000; 25:25-29 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Holdeman LV, Kelley RW, Moore WEC. Anaerobic Gram-negative straight, curved and helical rods. In: Krieg NR, Holt JG (eds), Bergey's Manual of Systematic Bacteriology, 1st ed, vol. 1 Williams & Wilkins, Baltimore, USA,1984, p. 602-662. [Google Scholar]
- 29.Holdeman LV, Cato EP, Moore WEC. 1974. Anaerobe laboratory manual, 4th ed, Southern Printing Co., Blacksburg, Virginia, USA. [Google Scholar]
- 30.Shah HN, Collins MD. Genus Bacteroides. A chemotaxonomical perspective. J Appl Bacteriol 1983; 55:403-416 [DOI] [PubMed] [Google Scholar]
- 31.MacFarlane S, Macfarlane GT. Formation of a dipeptidyl arylamidase by Bacteroides splanchnicus NCTC 10825 with specificities towards glycylprolyl-x and valylalanine-x substrates. J Med Microbiol 1997; 46:547-555 10.1099/00222615-46-7-547 [DOI] [PubMed] [Google Scholar]
- 32.Miyagawa T, Azuma R, Suto T. Distribution of sphingolipids in Bacteroides species. J Gen Appl Microbiol 1978; 24:341-348 10.2323/jgam.24.341 [DOI] [Google Scholar]
- 33.Klenk HP, Goeker M. En route to a genome-based classification of Archaea and Bacteria? Syst Appl Microbiol 2010; 33:175-182 10.1016/j.syapm.2010.03.003 [DOI] [PubMed] [Google Scholar]
- 34.Wu D, Hugenholtz P, Mavromatis K, Pukall R, Dalin E, Ivanova NN, Kunin V, Goodwin L, Wu M, Tindall BJ, et al. A phylogeny-driven genomic encyclopaedia of Bacteria and Archaea. Nature 2009; 462:1056-1060 10.1038/nature08656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.List of growth media used at DSMZ: http//www.dsmz.de/microorganisms/media_list.php
- 36.Gemeinholzer B, Dröge G, Zetzsche H, Haszprunar G, Klenk HP, Güntsch A, Berendsohn WG, Wägele JW. The DNA Bank Network: the start from a German initiative. Biopreservation and Biobanking 2011; 9:51-55 10.1089/bio.2010.0029 [DOI] [PubMed] [Google Scholar]
- 37.DOE Joint Genome Institute http://www.jgi.doe.gov
- 38.Phrap and Phred for Windows. MacOS, Linux, and Unix. http://www.phrap.com
- 39.Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 2008; 18:821-829 10.1101/gr.074492.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sims D, Brettin T, Detter JC, Han C, Lapidus A, Copeland A, Glavina Del Rio T, Nolan M, Chen F, Lucas S, et al. Complete genome sequence of Kytococcus sedentarius type strain (541T). Stand Genomic Sci 2009; 1:12-20 10.4056/sigs.761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lapidus A, LaButti K, Foster B, Lowry S, Trong S, Goltsman E. POLISHER: An effective tool for using ultra short reads in microbial genome assembly and finishing. AGBT, Marco Island, FL, 2008. [Google Scholar]
- 42.Hyatt D, Chen GL, LoCascio PF, Land ML, Larimer FW, Hauser LJ. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 2010; 11:119 10.1186/1471-2105-11-119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pati A, Ivanova NN, Mikhailova N, Ovchinnikova G, Hooper SD, Lykidis A, Kyrpides NC. GenePRIMP: a gene prediction improvement pipeline for prokaryotic genomes. Nat Methods 2010; 7:455-457 10.1038/nmeth.1457 [DOI] [PubMed] [Google Scholar]
- 44.Markowitz VM, Ivanova NN, Chen IMA, Chu K, Kyrpides NC. IMG ER: a system for microbial genome annotation expert review and curation. Bioinformatics 2009; 25:2271-2278 10.1093/bioinformatics/btp393 [DOI] [PubMed] [Google Scholar]