Abstract
Campylobacter is one of the leading causes of food-borne gastroenteritis and has a high prevalence in poultry. Campylobacter jejuni subsp. jejuni 327 is a subspecies of the genus Campylobacter of the family Campylobacteraceae in the phylum Proteobacteria. The microaerophilic, spiral shaped, catalase positive bacterium obtains energy from the metabolism of amino acids and Krebs cycle intermediates. Strain 327 was isolated from a turkey slaughter production line and is considered environmentally sensitive to food processing (cold, heat, drying) and storage conditions. The 327 whole genome shotgun sequence of 1,618,613 bp long consists of 1,740 protein-coding genes, 46 tRNA genes and 3 rRNA operons. A protein based BLAST analysis places the turkey isolate 327 close to the human clinical strain 81116 (NCTC 11828).
Introduction
Campylobacter is known worldwide as a common cause of human bacterial diarrhea; however, it is commensal in the gastrointestinal tract of many domestic and wild animals, especially birds. In 2008, campylobacteriosis remained the most frequently reported zoonotic disease in humans in the European Union with 190,566 confirmed cases [1]. Broiler meat and broiler flocks throughout the production chain in many EU-Member States, along with raw milk were reported as the most important food vehicles in food-borne Campylobacter outbreaks in 2008.
Classification and features
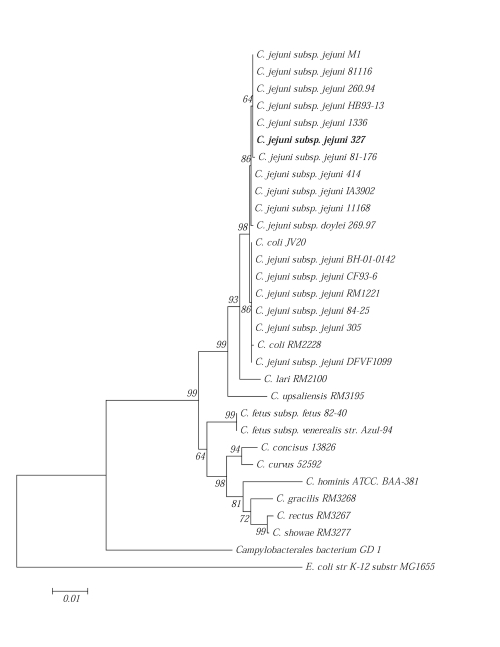
The genus Campylobacter belongs to the Epsilonproteobacteria [2] and is classified in the family Campylobacteraceae [3, Table 1], which includes the genera Campylobacter, Arcobacter, Dehalospirillum and Sulfurospirillum. The closest genetically related genera are Helicobacter and Wolinella, which together belong to the family Helicobacteraceae [7,22]. Currently, available genomes of the genus Campylobacter comprises 29 species and 4 subspecies (see phylogenetic tree, Figure 1). The most commonly isolated pathogenic species are C. jejuni, C. coli and C. fetus. All these species have small genomes (1.6–2.0 megabases) and can establish long-term associations with their hosts, sometimes with pathogenic consequences. Figure 1 shows the phylogenetic neighborhood of C. jejuni 327 in a 16S rRNA based tree.
Table 1. Classification and general features of C. jejuni 327 according to the MIGS recommendations [4].
| MIGS ID | Property | Term | Evidence code |
|---|---|---|---|
| Domain Bacteria | TAS [5] | ||
| Phylum Proteobacteria | TAS [6] | ||
| Class Epsilonproteobacteria | TAS [2,7,8] | ||
| Current classification | Order Campylobacterales | TAS [2,7] | |
| Family Campylobacteraceae | TAS [3] | ||
| Genus Campylobacter | TAS [3,9-15] | ||
| Species Campylobacter jejuni | TAS [9,10] | ||
| Strain 327 | TAS [16] | ||
| Gram stain | negative | TAS [9] | |
| Cell shape | Helical or curved rods can be observed in short or longer chains that can be V-, S-, or comma-shaped |
TAS [9] | |
| Motility | motile via lateral flagella | TAS [9] | |
| Sporulation | Non-sporulating | TAS [9] | |
| Temperature range | thermophilic, 37-42°C | TAS [17] | |
| Optimum temperature | 42°C | TAS [17] | |
| Salinity | sensitive to 1% NaCl | TAS [18] | |
| MIGS-22 | Oxygen requirement | microaerophilic (optimal concentrations of O2 [5–10%] and CO2 [3–5%]) |
TAS [9] |
| Carbon source | peptides and amino acids | TAS [3] | |
| Energy source | amino acids and Krebs cycle intermediates for energy production |
TAS [3] | |
| MIGS-6 | Habitat | Commensal organism in birds, primary enteric pathogen in humans |
TAS [17] |
| MIGS-15 | Biotic relationship | Free living | TAS [17] |
| MIGS-14 | Pathogenicity | pathogenic | TAS [17] |
| Biosafety level | 2 | TAS [4,19] | |
| Isolation | Poultry processing plant during slaughter of turkey batches Source of sampling: after killing |
TAS [20] | |
| MIGS-4 | Geographic location | Different poultry processing plants located in Germany | TAS [20] |
| MIGS-5 | Sample collection time | Autumn 2002 | TAS [20] |
Evidence codes - IDA: Inferred from Direct Assay (first time in publication); TAS: Traceable Author Statement (i.e., a direct report exists in the literature); NAS: Non-traceable Author Statement (i.e., not directly observed for the living, isolated sample, but based on a generally accepted property for the species, or anecdotal evidence). These evidence codes are from of the Gene Ontology project [21]. If the evidence code is IDA, then the property was directly observed for a live isolate by one of the authors, or an expert mentioned in the acknowledgements.
Figure 1.
Phylogenetic tree based on 16S rRNA highlighting the position of C. jejuni 327 relative to the other type and non-type strains within the species Campylobacter jejuni. Strains shown are those within Campylobacter jejuni having corresponding NCBI genome project IDs listed below. The strains and their corresponding GenBank accession numbers (and, when applicable, draft sequence coordinates) for 16S rRNA genes are (type=T): C. jejuni subsp. jejuni NCTC 11168T, AL11168; C. jejuni subsp. jejuni M1, CP001900; C. jejuni subsp. jejuni 81116, CP000814; C. jejuni subsp. jejuni 260.94, AANK01000000; C. jejuni subsp. jejuni HB93-13, AANQ01000000; C. jejuni subsp. jejuni 1336, ADGL01000000; C. jejuni subsp. jejuni 327, ADHM00000000; C. jejuni subsp. jejuni 81-176, CP000538; C. jejuni subsp. jejuni 414, ADGM01000000; C. jejuni subsp. jejuni IA3902, CP001876; C. jejuni subsp. doylei 269.97, CP000768; C. coli JV20, AEER01000000; C. jejuni subsp. jejuni BH-01-0142, ABKD01000000; C. jejuni subsp. jejuni CF93-6, AANJ01000000; C. jejuni subsp. jejuni RM1221, CP000025; C. jejuni subsp. jejuni 84-25, AANT02000000; C. jejuni subsp. jejuni 305, ADHL00000000; C. coli RM2228, AAFL01000000; C. jejuni subsp. jejuni DFVF1099, ADHK00000000; C. lari RM2100, CP000932; C. upsaliensis RM3195, AAFJ01000000; C. fetus subsp. fetus 82-40, CP000487; C. fetus subsp. venerealis str. Azul-94, ACLG01000000; C. concisus 13826, CP000792; C. curvus 52592, CP000767; C. hominis ATCC. BAA-381, CP000776; C. gracilis RM3268, ACYG01000000; C. rectus RM3267, ACFU01000000; C. showae RM3277, ACVQ01000000; Campylobacterales bacterium GD 1, ABXD01000000; E. coli str K-12 substr MG1655, U00096. The tree uses sequences aligned by PRANK [23-25], which by default uses the Hasegawa, Kishino and Yano (HKY) model with empirical base frequencies and kappa=2. The tree is built with MEGA4 [26] using Neighbor-Joining method and 1000 re-samplings to calculate bootstrap values. C. jejuni 327 is found in the C jejuni subsp. jejuni cluster that has a 100% bootstrap value along with the C. coli species. E. coli K-12 was used as an outgroup.
Chemotaxonomy
All Campylobacter species contained menaquinone-6 (2-methyl-3-farnesyl-farnesyl-1,4-naphthoquinone) and methyl-substituted menaquinone-6 (2,[5 or 8]-dimethyl-3-farnesyl-farnesyl-1,4-napthoquinone) as the major isoprenoid quinones. The latter menaquinone has not been reported in other bacteria and may prove to be a useful chemical marker of Campylobacter species. Campylobacter jejuni and most strains of Campylobacter coli were distinguished from other Campylobacter species by the presence of a Cl9 cyclopropane fatty acid acid in whole cell hydrolysates [21,27]
Genome sequencing and annotation
Genome project history
Campylobacter jejuni strain 327, one of the strains present in a turkey production line, was isolated from turkey skin surface swabs [20], and was selected for sequencing based on the sensitivity to environmental conditions in food-related environments [28]. Sequencing and finishing were performed by the Department of Biology (KU-NAT) and the Institute of Food Science (IFV) at the University of Copenhagen. The annotation was performed by the Institute for Genome Science (IGS, University of Maryland). The manual curation was completed by IFV and will be presented for public access with the publication of the Genome Announcement article. Genome analysis was performed by the Center for Biological Sequence Analysis (CBS) at the Technical University of Denmark (DTU). The Whole Genome Shotgun (WGS) project has been deposited at DDBJ/EMBL/GenBank under the project ID 41643 and accession number ADHM01000000. A summary of the project information is shown in Table 1 and Table 2 according to the Minimum Information about a Genomic Sequence (MIGS) recommendations [29].
Table 2. Genome sequencing project information.
| MIGS ID | Property | Term |
|---|---|---|
| MIGS-29 | Sequencing platform | 454 Life Sciences |
| MIGS-31 | Finishing quality | Improved high-quality draft |
| MIGS-31.2 | Fold coverage | 20 × |
| MIGS-32 | Genome database release | with SIGS publication |
| GenBank ID | 41643 | |
| Genbank Date of Release | with SIGS publication | |
| Project relevance | Food safety |
Growth conditions and DNA isolation
The turkey strain 327 was provided by Thomas Alter [20], and showed a sensitive phenotype to gentle food processing stresses [28]. C. jejuni cells were grown at 42 °C under microaerobic conditions (5% O2, 10% CO2, 85% N2). Stocks were stored at -80°C in Brain Heart Infusion broth (BHI) (Oxoid CM225, England) supplemented with 15% glycerol. The frozen stocks were transferred to Blood Agar Base No.2 (Oxoid CM271, England) amended with 5% horse blood and incubated in a microaerobic atmosphere (5% O2, 10% CO2, 85% N2) at 42 °C for 24–72 h. The respective cultures were subsequently re-streaked on Blood Agar Base No.2 plates. After 24 hours of growth, a 3/4 loop-full of bacteria was resuspended in 1 ml phosphate buffered saline (PBS, Oxoid BR0014, England) and vortexed to ensure no bacterial clumps. Cells were centrifuged at 14,000 × g using a benchtop Sartorius centrifuge (model Sigma 1-14) and the medium was decanted. The cells were resuspended in 200 µl PBS for genomic DNA isolation using the Easy-DNATM Kit (Invitrogen, K1800-01). The protocol was followed as described by the manufacturer. A yield of approximately 10 mg of total genomic DNA was obtained for each C. jejuni strain.
Genome sequencing and assembly
Pyrosequencing of C. jejuni strain 327 was performed on a Genome Sequencer GS FLX System (454 Life Sciences, Branford, CT, USA) at the Faculty of Biology, University of Copenhagen (KU-NAT). GS FLX sequencing was performed following the manufacturer’s protocol with minor modifications. Briefly, library preparations were done from 3μg of DNA using the shotgun library protocol with Multiplex Identifiers (MID) tags for each bacteria/sample, and DNA was released using heat instead of NaOH [30,31]. Libraries were quantified by qPCR as described in [32], and sequenced on a full GS FLX-LR70 plate. Genome sequences resulted in sequence reads which passed the length and quality criteria of the machine software. Draft assemblies were based on 134,679 total reads with 20-fold coverage of the genome. The 454 data files were loaded into the CLC Genomics Workbench version 3.7.1 (CLC Bio, Aarhus, Denmark). The initial reference was created using the human clinical strain 81116 [33] (NCTC 11828) as scaffold, yielding 133,175 matched reads (99% of match). For de novo assembly the 134,679 sequence reads were condensed to 48 contigs.
Genome annotation
The C. jejuni genome sequences were automatically annotated using the Annotation Engine and the initial GenBank files were generated at the Institute for Genome Sciences (IGS, University of Maryland). These annotations and the GenBank files were further refined and corrected at the Center for Biological Sequence Analysis (CBS) at the Technical University of Denmark (DTU) by reference to codon usage, positional base preference methods and comparisons to the non-redundant protein databases using BLAST [34]. In-house Perl scripts from CBS and the Sequin program provided by NCBI [35] were used in this refinement process. The entire DNA sequence was also compared in all six potential reading frames against UniProt. Furthermore, the RNAmmer 1.2 server was used for ribosomal RNA predictions of 5S, 16S, and 23S [36]. The outcome of all these predictions was corrected on September 14th 2010.
Genome properties
The C. jejuni 327 genome was found to be 1,618,613 bp long, and contains 1,740 protein coding genes as identified with the gene prediction program Prodigal version 1.20 [37], Table 3). The average G+C content is 30.4%, and there are 43 tRNAs and 5 rRNA genes found using the respective prediction server [36,40]. C. jejuni strain 327 does not contain any plasmids. Strain 327 contains 10 homopolymeric G tracts (HGTs, defined as tracts of >7 consecutive G-residues), fewer than the other complete genome sequences described to date (29 in NCTC 11168, 25 in RM1221 and 19 in 81-176 [41-43]). Variation in the length of homopolymeric G tracts may be produced by slipped-strand mispairing during replication [44], and can evolutionarily affect changes on the genome sequence. Thus, the number of hypervariable G tracts can give important hints on the genetic stability of the strain of C. jejuni studied. Of the 1,786 genes predicted, 1,740 were protein-coding genes, and 5 rRNA genes; 7 pseudogenes were identified. The majority of the protein-coding genes (97%) were assigned with a putative function while the remaining ones were annotated as hypothetical proteins. The distribution of genes into COGs functional categories is presented in Table 4.
Table 3. Genome Statistics.
| Attribute | Value | % of Total |
|---|---|---|
| Genome size (bp) | 1,618,613 | 100.00% |
| DNA coding region (bp) | 1,495,833 | 92.4% |
| DNA G+C content (bp) | 492,058 | 30.4% |
| Number of replicons | 1 | |
| Extrachromosomal elements | 0 | |
| Total genes | 1,786 | 100.00% |
| tRNA genes | 46 | 2.58% |
| rRNA genes | 5 | 0.28% |
| Protein-coding genes | 1,740 | 97.42% |
| Pseudo genes | 9 | 0.5% |
| Genes with function prediction | 1,383 | 77.43% |
| Genes in paralog clusters | 21 | 1.18% |
| Genes assigned to COGs | 1,280 | 71.67% |
| Genes assigned Pfam domains * | 1,350 | 75.59% |
| Genes with signal peptides [38] | 263 | 14.7% |
| Genes with transmembrane helices | 338 | 18.92% |
| CRISPR repeats [39] | 1 |
* E-value cutoff is 0.05.
Table 4. Number of genes associated with the general COG functional categories.
| Code | value | %age | Description |
|---|---|---|---|
| J | 259 | 7.4 | Translation, ribosomal structure and biogenesis |
| A | 0 | 0 | RNA processing and modification |
| K | 89 | 2.6 | Transcription |
| L | 134 | 3.8 | Replication, recombination and repair |
| B | 0 | 0 | Chromatin structure and dynamics |
| D | 33 | 0.9 | Cell cycle control, mitosis and meiosis |
| Y | 0 | 0 | Nuclear structure |
| V | 42 | 1.2 | Defense mechanisms |
| T | 101 | 2.9 | Signal transduction mechanisms |
| M | 237 | 6.8 | Cell wall/membrane biogenesis |
| N | 114 | 3.3 | Cell motility |
| Z | 2 | 0.1 | Cytoskeleton |
| W | 0 | 0 | Extracellular structures |
| U | 102 | 2.9 | Intracellular trafficking and secretion. and vesicular transport |
| O | 153 | 4.4 | Posttranslational modification. protein turnover. chaperones |
| C | 206 | 5.9 | Energy production and conversion |
| G | 110 | 3.2 | Carbohydrate transport and metabolism |
| E | 303 | 8.7 | Amino acid transport and metabolism |
| F | 95 | 2.7 | Nucleotide transport and metabolism |
| H | 168 | 4.8 | Coenzyme transport and metabolism |
| I | 67 | 1.9 | Lipid transport and metabolism |
| P | 210 | 6 | Inorganic ion transport and metabolism |
| Q | 38 | 1.1 | Secondary metabolites biosynthesis, transport and catabolism |
| R | 336 | 9.7 | General function prediction only |
| S | 176 | 5.1 | Function unknown |
| - | 506 | 14.5 | Not in COGs |
Genome Atlas construction
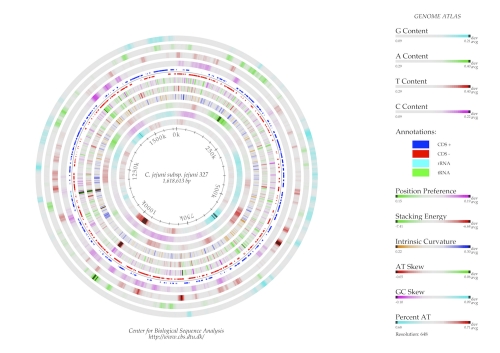
The genome atlas of C. jejuni subsp. jejuni 327 was generated using the Genewiz program (Figure 2). In order to create the atlas, a FASTA file containing the nucleotide sequence in one piece and an annotation file showing the position of the genes were used. The FASTA file was created by concatenating the nucleotide sequences of the contigs. In the atlas, gene annotation, base content, AT and GC skew, percent AT and some structural properties of the DNA were shown. The structural properties are Position Preference, Stacking Energy and Intrinsic curvature which are all related to the flexibility and strength of the DNA molecule [45].
Figure 2.
Genome Atlas of C. jejuni strain 327. The legend to the right explains what is represented from the outer to the inner circle. Shown are the fraction of each nucleotide along the genome (first four circles counting inwards), the coding sequences on the positive (clockwise strand), the AT and GC skew, and the percent AT.
Available sequence data from completed strains (NCTC 11168, 81116 (NCTC 11828), RM1221, 81–176, 269.97, M1) and ongoing C. jejuni sequencing projects (strains 84–25, 260-94, HB93-13, CF93-6, CG8421, CG8486) obtained from the NCBI database at the time of writing, were used for homology searching of genes in selected loci using the program BLASTP [34]. It revealed a high protein sequence homology with strain 81116 (NCTC 11828), first isolated from a case of campylobacteriosis associated with a human waterborne outbreak [46]. The initial reference assembly using strain 81116 [33] (NCTC 11828) as scaffold created 133,175 matched reads (99% of match). In addition, C. jejuni 327 genome contains only a single tonB gene as compared to 2 or 3 genes in other C. jejuni strains, and lacks the ferric enterobactin uptake receptor CfrA and TonB-dependent outer membrane receptor for iron uptake [47]. Strain 327 also lacks the transcriptional regulator mar A (multiple antibiotic resistance) locus, first described for E. coli [48]. The marA locus mediates global stress response and affects the expression of iron-sulfur cluster proteins involved in sensing O2 and iron. The lack of this gene could explain the phenotype of strain 327 observed under some environmental stresses [28].
Acknowledgements
We acknowledge the Danish Directorate for Food, Fisheries and Agriculture, grant number 93S-953-00090, for funding the whole genome shotgun sequence project of strain 327, and the Institute for Genome Sciences (IGS) Annotation Engine at the University of Maryland School of Medicine.
References
- 1.EFSA CSR Trends and sources of zoonoses and zoonotic agents and food-borne outbreaks in the European Union in 2008. EFSA 2010; 8:1-370 [Google Scholar]
- 2.Garrity G, Bell J, Lilburn T. Epsilonproteobacteria class. nov. In: Garrity G, Brenner D, Krieg N, Staley J, editors. Bergey's Manual of Systematic Bacteriology. Second Edition ed. Volume 2, Part C. New York: Springer; 2005. p 1145. [Google Scholar]
- 3.Vandamme P, Ley JD. Proposal for a New Family, Campylobacteraceae. Int J Syst Evol Microbiol 1991; 41:451-455 [Google Scholar]
- 4.Anonymous. Technical Rules for Biological Agents - Guideline for Risk Assessment and for the Instruction of Employees in relation to Activities with Biological Agents. Dortmund, Germany: Committee for Biological Agents (ABAS), Federal Ministry for Work and Social Order in the "Bundesarbeitsblatt"; 2006. 1-20 p. [Google Scholar]
- 5.Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci USA 1990; 87:4576-4579 10.1073/pnas.87.12.4576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garrity G, Holt J. The Road Map to the Manual. In: Garrity G, Boone D, Castenholz R, editors. Bergey's Manual of Systematic Bacteriology. Second ed. Volume 1. New York: Springer; 2001. p 119-169. [Google Scholar]
- 7.Validation list no. 107. List of new names and new combinations previously effectively, but not validly, published. Int J Syst Evol Microbiol 2006; 56:1-6 10.1099/ijs.0.64188-0 [DOI] [PubMed] [Google Scholar]
- 8.Garrity G, Brenner DJ, Krieg NR, Staley JT. The Proteobacteria, Part C: The Alpha-, Beta-, Delta-, and Epsilonproteobacteria In: Garrity G, editor. Bergey's Manual of Systematic Bacteriology. 2nd. ed. New York: Springer; 2005. p 2791. [Google Scholar]
- 9.Véron M, Chatelain R. Taxonomic Study of the genus Campylobacter Sebald and Veron and designation of the neotype strain for the type species, Campylobacter fetus (Smith and Taylor) Sebald and Veron. Int J Syst Bacteriol 1973; 23:122-134 10.1099/00207713-23-2-122 [DOI] [Google Scholar]
- 10.Skerman V, McGowan V, Sneath P. Approved Lists of Bacterial Names. Int J Syst Bacteriol 1980; 30:225-420 10.1099/00207713-30-1-225 [DOI] [PubMed] [Google Scholar]
- 11.Sebald M, Verón M. Teneur en bases de l'ADN et classificatiion des vibrions. Ann Inst Pasteur (Paris) 1963; 105:897-910 [PubMed] [Google Scholar]
- 12.Smibert R. Genus II. Campylobacter Sebald and Veron 1963, 907. In: Buchanan R, Gibbons N, editors. Bergey's Manual of Determinative Bacteriology. Eighth ed. Baltimore: The Williams and Wilkins Co.; 1974. p 207-212. [Google Scholar]
- 13.Tanner A, Badger S, Lai C, Listgarten M, Visconti R, Socransky S. Wolinella gen. nov., Wolinella succinogenes (Vibrio succinogenes Wolin et al.) comb. nov., and Description of Bacteroides gracilis sp. nov., Wolinella recta sp. nov., Campylobacter concisus sp. nov., and Eikenella corrodens from Humans with Periodontal Disease. Int J Syst Bacteriol 1981; 31:432-445 10.1099/00207713-31-4-432 [DOI] [Google Scholar]
- 14.Vandamme P, Debruyne L, De Brandt E, Falsen E. Reclassification of Bacteroides ureolyticus as Campylobacter ureolyticus comb. nov., and emended description of the genus Campylobacter. Int J Syst Evol Microbiol 2010; 60:2016-2022 10.1099/ijs.0.017152-0 [DOI] [PubMed] [Google Scholar]
- 15.Vandamme P, Falsen E, Rossau R, Hoste B, Segers P, Tytgat R, De Ley J. Revision of Campylobacter, Helicobacter, and Wolinella taxonomy: emendation of generic descriptions and proposal of Arcobacter gen. nov. Int J Syst Bacteriol 1991; 41:88-103 10.1099/00207713-41-1-88 [DOI] [PubMed] [Google Scholar]
- 16.Birk T, Ingmer H, Andersen MT, Jørgensen K, Brøndsted L. Chicken juice, a food-based model system suitable to study survival of Campylobacter jejuni. Lett Appl Microbiol 2004; 38:66-71 10.1046/j.1472-765X.2003.01446.x [DOI] [PubMed] [Google Scholar]
- 17.Wassenaar T, Newell D. The Genus Campylobacter In: Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E, editors. The Prokaryotes. New York: Springer; 2006. p 119-138. [Google Scholar]
- 18.Kelly AF, Park SF, Bovill R, Mackey BM. Survival of Campylobacter jejuni during stationary phase: Evidence for the absence of a phenotypic stationary-phase response. Appl Environ Microbiol 2001; 67:2248-2254 10.1128/AEM.67.5.2248-2254.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anonymous. Classification of biological agents in riskgroups 2, 3 and 4 - Appendix 8. The Danish Working Environment Authority; 1993. [Google Scholar]
- 20.Alter T, Gaull F, Froeb A, Fehlhaber K. Distribution of Campylobacter jejuni strains at different stages of a turkey slaughter line. Food Microbiol 2005; 22:345-351 10.1016/j.fm.2004.08.008 [DOI] [Google Scholar]
- 21.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene Ontology: Tool for the unification of biology. Nat Genet 2000; 25:25-29 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garrity G, Bell J, Lilburn T. Family II. Helicobacteraceae fam. nov. In: Garrity G, Brenner D, Krieg N, Staley J, editors. Bergey's Manual of Systematic Bacteriology. Second ed. Volume 2, Part C. New York: Springer; 2005. p 1168. [Google Scholar]
- 23.Löytynoja A, Goldman N. An algorithm for progressive multiple alignment of sequences with insertions. Proc Natl Acad Sci USA 2005; 102:10557-10562 10.1073/pnas.0409137102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Löytynoja A, Goldman N. Phylogeny-Aware Gap Placement Prevents Errors in Sequence Alignment and Evolutionary Analysis. Science 2008; 320:1632-1635 10.1126/science.1158395 [DOI] [PubMed] [Google Scholar]
- 25.Löytynoja A, Goldman N. webPRANK: a phylogeny-aware multiple sequence aligner with interactive alignment browser. BMC Bioinformatics 2010; 11:579 10.1186/1471-2105-11-579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 2007; 24:1596-1599 10.1093/molbev/msm092 [DOI] [PubMed] [Google Scholar]
- 27.Moss CW, Kai A, Lambert MA, Patton C. Isoprenoid quinone content and cellular fatty acid composition of Campylobacter species. J Clin Microbiol 1984; 19:772-776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Birk T, Grønlund AC, Christensen BB, Knøchel S, Lohse K, Rosenquist H. Effect of organic acids and marination ingredients on the survival of Campylobacter jejuni on meat. J Food Prot 2010; 73:258-265 [DOI] [PubMed] [Google Scholar]
- 29.Field D, Garrity G, Gray T, Morrison N, Selengut J, Sterk P, Tatusova T, Thomson N, Allen MJ, Angiuoli SV, et al. The minimum information about a genome sequence (MIGS) specification. Nat Biotechnol 2008; 26:541-547 10.1038/nbt1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maricic T, Paabo S. Optimization of 454 sequencing library preparation from small amounts of DNA permits sequence determination of both DNA strands. Biotechniques 2009; 46:51-52 10.2144/000113042 [DOI] [PubMed] [Google Scholar]
- 31.Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature 2005; 437:376-380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meyer M, Briggs AW, Maricic T, Hober B, Hoffner B, Krause J, Weihmann A, Paabo S, Hofreiter M. From micrograms to picograms: quantitative PCR reduces the material demands of high-throughput sequencing. Nucleic Acids Res 2007; 36:e5 10.1093/nar/gkm1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pearson BM, Gaskin DJH, Segers RPAM, Wells JM, Nuijten PJM, van Vliet AHM. The Complete Genome Sequence of Campylobacter jejuni Strain 81116 (NCTC11828). J Bacteriol 2007; 189:8402-8403 10.1128/JB.01404-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 1997; 25:3389-3402 10.1093/nar/25.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Benson DA, Boguski MS, Lipman DJ, Ostell J, Ouellette BF. GenBank. Nucleic Acids Res 1998; 26:1-7 10.1093/nar/26.1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lagesen K, Hallin P, Rødland EA, Stærfeldt HH, Rognes T, Ussery DW. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res 2007; 35:3100-3108 10.1093/nar/gkm160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hyatt D, Chen GL, LoCascio P, Land M, Larimer F, Hauser L. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 2010; 11:119 10.1186/1471-2105-11-119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved Prediction of Signal Peptides: SignalP 3.0. J Mol Biol 2004; 340:783-795 10.1016/j.jmb.2004.05.028 [DOI] [PubMed] [Google Scholar]
- 39.Grissa I, Vergnaud G, Pourcel C. CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res 2007; 35(suppl 2):W52-W57 10.1093/nar/gkm360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lowe TM, Eddy SR. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 1997; 25:955-964 10.1093/nar/25.5.955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hofreuter D, Tsai J, Watson RO, Novik V, Altman B, Benitez M, Clark C, Perbost C, Jarvie T, Du L, et al. Unique features of a highly pathogenic Campylobacter jejuni strain. Infect Immun 2006; 74:4694-4707 10.1128/IAI.00210-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Parker CT, Quiñones B, Miller WG, Horn ST, Mandrell RE. Comparative genomic analysis of Campylobacter jejuni strains reveals diversity due to genomic elements similar to those present in C. jejuni strain RM1221. J Clin Microbiol 2006; 44:4125-4135 10.1128/JCM.01231-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Parkhill J, Wren BW, Mungall K, Ketley JM, Churcher C, Basham D, Chillingworth T, Davies RM, Feltwell T, Holroyd S, et al. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 2000; 403:665-668 10.1038/35001088 [DOI] [PubMed] [Google Scholar]
- 44.Moxon ER, Rainey PB, Nowak MA, Lenski RE. Adaptive evolution of highly mutable loci in pathogenic bacteria. Curr Biol 1994; 4:24-33 10.1016/S0960-9822(00)00005-1 [DOI] [PubMed] [Google Scholar]
- 45.Hallin PF, Binnewies TT, Ussery DW. The genome BLASTatlas-a GeneWiz extension for visualization of whole-genome homology. Mol Biosyst 2008; 4:363-371 10.1039/b717118h [DOI] [PubMed] [Google Scholar]
- 46.Manning G, Duim B, Wassenaar T, Wagenaar JA, Ridley A, Newell DG. Evidence for a Genetically Stable Strain of Campylobacter jejuni. Appl Environ Microbiol 2001; 67:1185-1189 10.1128/AEM.67.3.1185-1189.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guerry P, Perez-Casal J, Yao R, McVeigh A, Trust TJ. A genetic locus involved in iron utilization unique to some Campylobacter strains. J Bacteriol 1997; 179:3997-4002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Barbosa TM, Levy SB. Differential Expression of over 60 Chromosomal Genes in Escherichia coli by Constitutive Expression of MarA. J Bacteriol 2000; 182:3467-3474 10.1128/JB.182.12.3467-3474.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]