Abstract
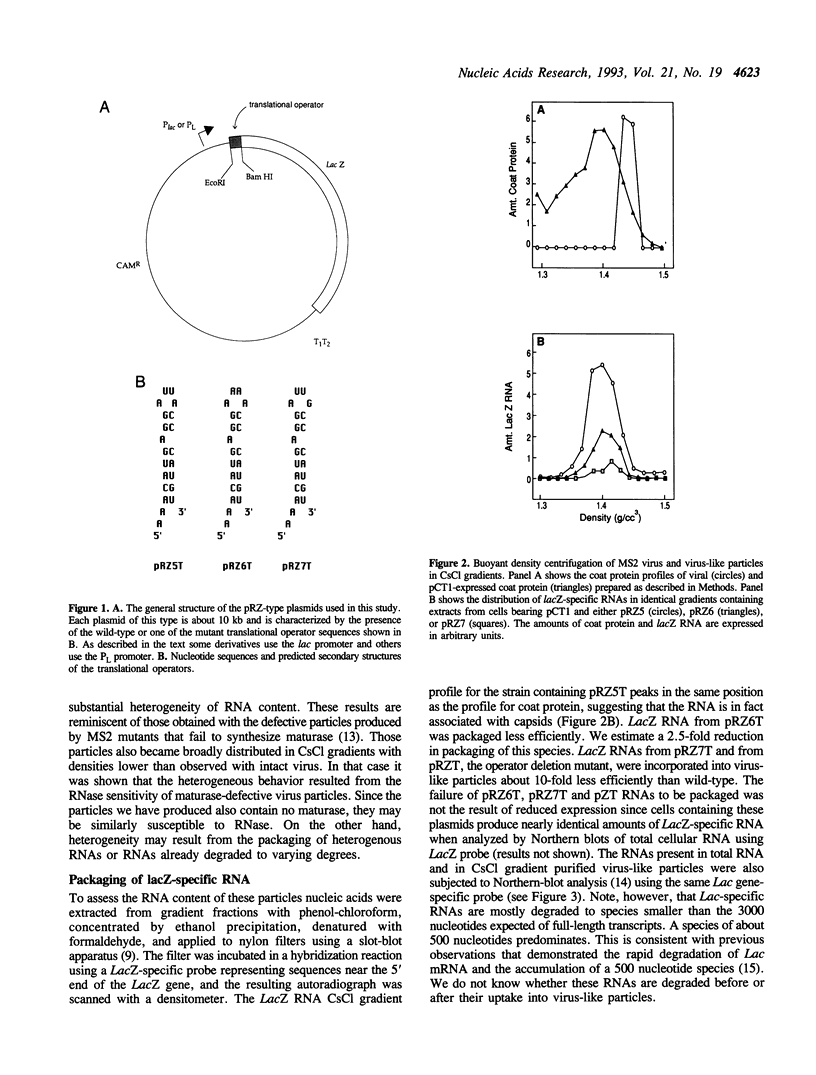
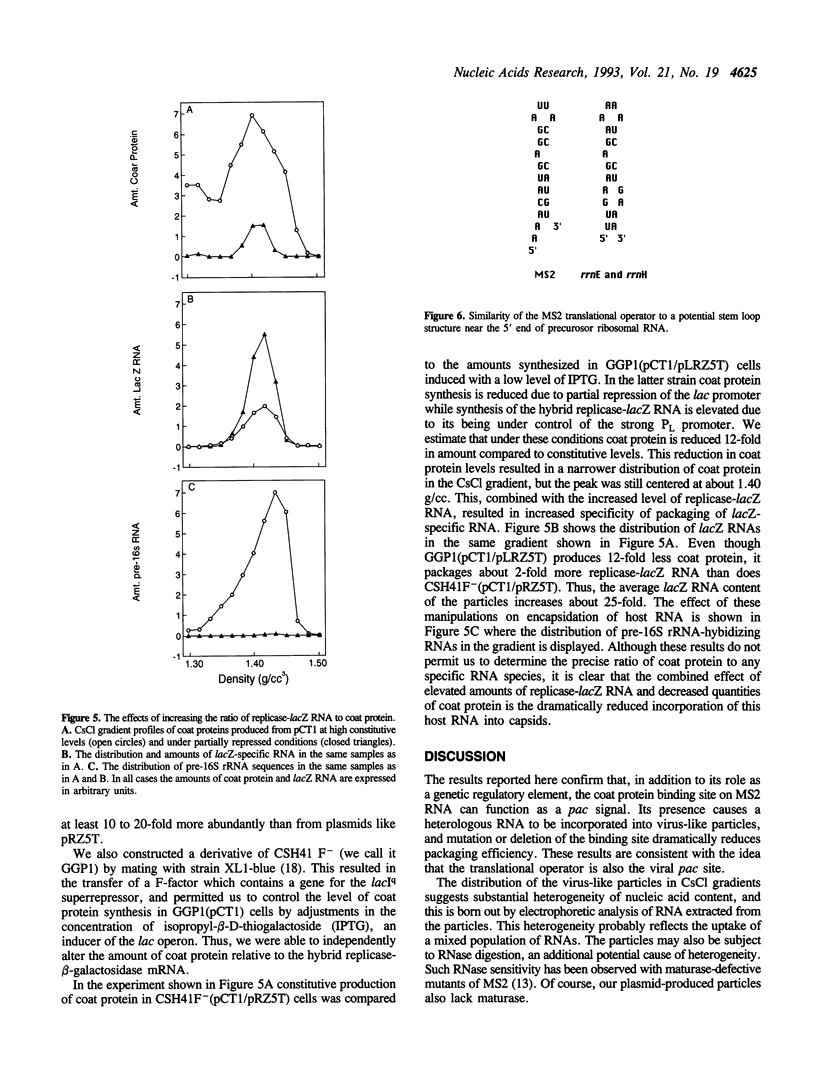
The RNA bacteriophages of E. coli specifically encapsidate a single copy of the viral genome in a protein shell composed mainly of 180 molecules of coat protein. Coat protein is also a translational repressor and shuts off viral replicase synthesis by interaction with a RNA stem-loop containing the replicase initiation codon. We wondered whether the translational operator also serves as the viral pac site, the signal which mediates the exclusive encapsidation of viral RNA by its interaction with coat protein. To test this idea we measured the ability of lacZ RNA fused to the translational operator to be incorporated into virus-like particles formed from coat protein expressed from a plasmid. The results indicate that the operator-lacZ RNA is indeed encapsidated and that nucleotide substitutions in the translational operator which reduce the tightness of the coat protein-operator interaction also reduce or abolish encapsidation of the hybrid RNA. When coat protein is expressed in excess compared to the operator-lacZ RNA, host RNAs are packaged as well. However, elevation of the level of operator-lacZ RNA relative to coat protein results in its selective encapsidation at the expense of cellular RNAs. Our results are consistent with the proposition that this single protein-RNA interaction accounts both for translational repression and viral genome encapsidation.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alwine J. C., Kemp D. J., Stark G. R. Method for detection of specific RNAs in agarose gels by transfer to diazobenzyloxymethyl-paper and hybridization with DNA probes. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5350–5354. doi: 10.1073/pnas.74.12.5350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckett D., Wu H. N., Uhlenbeck O. C. Roles of operator and non-operator RNA sequences in bacteriophage R17 capsid assembly. J Mol Biol. 1988 Dec 20;204(4):939–947. doi: 10.1016/0022-2836(88)90053-8. [DOI] [PubMed] [Google Scholar]
- Bernardi A., Spahr P. F. Nucleotide sequence at the binding site for coat protein on RNA of bacteriophage R17. Proc Natl Acad Sci U S A. 1972 Oct;69(10):3033–3037. doi: 10.1073/pnas.69.10.3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnette W. N. "Western blotting": electrophoretic transfer of proteins from sodium dodecyl sulfate--polyacrylamide gels to unmodified nitrocellulose and radiographic detection with antibody and radioiodinated protein A. Anal Biochem. 1981 Apr;112(2):195–203. doi: 10.1016/0003-2697(81)90281-5. [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Hohn T. Role of RNA in the assembly process of bacteriophage fr. J Mol Biol. 1969 Jul 14;43(1):191–200. doi: 10.1016/0022-2836(69)90088-6. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Meinkoth J., Wahl G. Hybridization of nucleic acids immobilized on solid supports. Anal Biochem. 1984 May 1;138(2):267–284. doi: 10.1016/0003-2697(84)90808-x. [DOI] [PubMed] [Google Scholar]
- Peabody D. S. Translational repression by bacteriophage MS2 coat protein expressed from a plasmid. A system for genetic analysis of a protein-RNA interaction. J Biol Chem. 1990 Apr 5;265(10):5684–5689. [PubMed] [Google Scholar]
- Romaniuk P. J., Lowary P., Wu H. N., Stormo G., Uhlenbeck O. C. RNA binding site of R17 coat protein. Biochemistry. 1987 Mar 24;26(6):1563–1568. doi: 10.1021/bi00380a011. [DOI] [PubMed] [Google Scholar]
- Scheuermann R. H., Echols H. A separate editing exonuclease for DNA replication: the epsilon subunit of Escherichia coli DNA polymerase III holoenzyme. Proc Natl Acad Sci U S A. 1984 Dec;81(24):7747–7751. doi: 10.1073/pnas.81.24.7747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiba T., Suzuki Y. Localization of A protein in the RNA-A protein complex of RNA phage MS2. Biochim Biophys Acta. 1981 Jul 27;654(2):249–255. doi: 10.1016/0005-2787(81)90179-9. [DOI] [PubMed] [Google Scholar]
- Srivastava A. K., Schlessinger D. Mechanism and regulation of bacterial ribosomal RNA processing. Annu Rev Microbiol. 1990;44:105–129. doi: 10.1146/annurev.mi.44.100190.000541. [DOI] [PubMed] [Google Scholar]
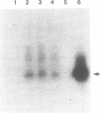
- Subbarao M. N., Kennell D. Evidence for endonucleolytic cleavages in decay of lacZ and lacI mRNAs. J Bacteriol. 1988 Jun;170(6):2860–2865. doi: 10.1128/jb.170.6.2860-2865.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valegård K., Liljas L., Fridborg K., Unge T. The three-dimensional structure of the bacterial virus MS2. Nature. 1990 May 3;345(6270):36–41. doi: 10.1038/345036a0. [DOI] [PubMed] [Google Scholar]