Abstract
Objectives
To explore the role of topoisomerase I in gene activation and increased RecA levels during the bacterial SOS response, as well as the effect of antibiotic treatment and stress challenge on cell killing initiated by trapped topoisomerase I cleavage complex.
Methods
A mutant Escherichia coli strain with a ΔtopA mutation was used to investigate the role of topoisomerase I function in the SOS response to trimethoprim and mitomycin C. Induction of the recA and dinD1 promoters was measured using luciferase reporters of these promoters fused to luxCDABE. An increase in the RecA level following trimethoprim treatment was quantified directly by western blotting. The effect of stress challenge from trimethoprim and acidified nitrite treatments on cell killing by topoisomerase I cleavage complex accumulation was measured by the decrease in viability following induction of recombinant mutant topoisomerase I that forms a stabilized cleavage complex.
Results
Topoisomerase I function was found to be required for efficient transcriptional activation of the recA and dinD1 promoters during the E. coli SOS response to trimethoprim and mitomycin C. The role of topoisomerase I in the SOS response was confirmed with quantitative western blot analysis of RecA following trimethoprim treatment. The bactericidal effect from topoisomerase I cleavage complex accumulation was shown to be enhanced by stress challenge from trimethoprim and acidified nitrite.
Conclusions
Bacterial topoisomerase I function is actively involved in the SOS response to antibiotics and stress challenge. Cell killing initiated by the topoisomerase I cleavage complex would be enhanced by antibiotics and the host response. These findings provide further support for bacterial topoisomerase I as a therapeutic target.
Keywords: anti-infective development, drug targets, gene expression, killing, stress response
Introduction
DNA topoisomerases are ubiquitous enzymes that control the topology of DNA by catalysing the breaking and rejoining of DNA phosphodiester bonds coupled to DNA strand passage.1 In addition to their involvements in vital cellular processes,2 topoisomerases are also important clinical targets for anticancer and antibacterial drugs.3–6 Quinolones are broad-spectrum antibiotics that target bacterial topoisomerase II (gyrase) and IV by trapping the covalent complex formed between the enzyme and cleaved DNA.7,8 Bactericidal antibiotics, including quinolones, have been shown to increase the formation of reactive oxygen species (ROS) to initiate an oxidative cell death pathway in Escherichia coli.9,10 The widespread prevalence of multidrug-resistant bacterial pathogens presents an urgent need for the development of novel antibacterial therapy against new bacterial targets. Bacterial topoisomerase I is a promising target for novel antibacterial therapy, because type IA topoisomerase is present in all bacteria11 and would act as a potential target for cell killing via stabilization of its covalent cleavage complex.12 Our previous studies showed that accumulation of the bacterial topoisomerase I cleavage complex in E. coli leads to chromosomal fragmentation and an increase in hydroxyl radicals, resulting in bacterial cell death.13–15 New alkaloid antibiotics that target Streptococcus pneumoniae by inhibiting topoisomerase I were described recently.16 The structure of these compounds is similar to that of stephenanthrine, which was found previously to inhibit the activity of E. coli and Yersinia pestis topoisomerase I.17
E. coli topoisomerase I is encoded by the topA gene and plays a major role in preventing excessive negative supercoiling of DNA.18 In addition to being required for optimal bacterial functions under normal growth conditions,2,19 topoisomerases are also important for the adaptation to environmental challenges.20,21 The topA gene is controlled by four promoters22,23 recognized by σ32 and σs in addition to σ70. The presence of multiple promoters is probably due to the requirement of topA function for bacterial adaptation to different conditions. During rapid transcription at gene loci induced by the stress response, movement of the RNA polymerase complex leads to increased negative supercoiling behind the complex.24 Topoisomerase I is needed to prevent hypernegative supercoiling and R-loop formation, which would otherwise inhibit the expression of the stress response genes.18,25,26 The removal of transcription-driven supercoiling by topoisomerase I is facilitated by direct association with RNA polymerase via protein–protein interactions.27 Our previous studies showed that topA mutations increased the sensitivity to high temperatures,28,29 hydrogen peroxide and N-ethylmaleimide.30 Loss of topA function also affected acid resistance.31
The RecA-dependent SOS response has been implicated in the development of antibiotic resistance.32–35 The role of topoisomerase I function in gene transcription during the SOS response has not been evaluated previously. Antibiotic treatment during infection would activate the bacterial stress response, including the SOS response, and increase the interaction between topoisomerase I and DNA due to rapid transcription at the activated gene loci. Therefore, combination therapy that couples topoisomerase I poisons that trap the covalent complex with existing antibiotics could be a promising strategy for enhancing the bactericidal effect of treatment. In this report, we established the involvement of topA function in the E. coli SOS response and increase in the RecA protein level following treatment with the bacteriostatic antibiotic trimethoprim. A mutant topoisomerase I with defective DNA rejoining activity36 was utilized to investigate the bactericidal effect from the stabilized topoisomerase cleavage complex under stress conditions.
Materials and methods
Strains and plasmids
Luria–Bertani (LB) medium was used for culture. E. coli strain RFM475, with the genotype gyrB221(cour), gyrB203(Ts), Δ(topA cysB)204, was used along with its isogenic topA+ strain RFM44526 to study the involvement of topoisomerase I function in the SOS response. These two strains were transformed with plasmids pDEW237 and pDinlux containing the recA::luxCDABE37 or dinD1::luxCDABE17 fusions, respectively, as reporters for SOS induction of gene expression.
To monitor the extent of cell killing from accumulation of the topoisomerase I cleavage complex, E. coli strain BW117N with a mutant Y. pestis topoisomerase I (YpTOP1-D117N) gene under the control of the BAD promoter inserted into the chromosome36 was used. The recombinant topoisomerase gene was linked to the chloramphenicol resistance marker for selection. This recombinant mutant topoisomerase can cleave DNA to form a covalent complex, but is defective in DNA religation. Dominant lethal mutations in the recombinant Y. pestis topoisomerase I gene can be more stably maintained in E. coli than a comparable mutant E. coli topoisomerase I gene.
Luciferase assay
E. coli RFM445 and RFM475 transformed with luciferase reporter plasmids were grown in LB medium containing 100 mg/L ampicillin until OD600 = 0.4. Cells (100 μL aliquots) were dispensed into flat-bottomed 96-well white Microlite 1 plates. One microlitre of mitomycin C or trimethoprim at various concentrations was added to each well. Luminescence was measured using the Perkin Elmer HTS7000 Plus plate reader at 37°C and 10 min per cycle, with 30 s of shaking before every measurement. The luciferase response ratio was obtained by dividing the luciferase signal from the treated cultures by the luciferase signal from the culture with no treatment.
RecA quantification by western blotting
The procedures were similar to those described previously for the characterization of SOS constitutive mutants of E. coli.38 Strains RFM445 (topA+) and RFM475 (ΔtopA) were grown to exponential phase and treated with or without 0.5 mg/L trimethoprim for 3 h. The OD600 was measured before 200 μL aliquots of each culture were collected for western blot analysis. The cell pellets were resuspended in volumes of water adjusted according to the OD600 values obtained. They were further diluted to be in the linear range of detection by western blotting before mixing with an equal volume of 2× SDS sample buffer. The cells were then lysed by boiling for 5 min. The total protein in the lysates was separated by electrophoresis in a 10% SDS/polyacrylamide gel and transferred onto nitrocellulose membranes. Serial dilutions of pure RecA protein (New England Biolabs) were included on each gel to obtain a standard curve for quantification of the RecA protein. The RecA protein was detected using a 1:5000 dilution of primary monoclonal RecA antibody (ARM193; Enzo Life Sciences). The ECL Plus reagent (Amersham) was used for protein detection and the signal was visualized with a STORM phosphorimager. The bands were quantified using AlphaEaseFC (ChemiImager 5500). The RecA standard curve was used to determine the RecA protein level in each sample, which was further adjusted according to the dilution used.
Cell-killing assay
The E. coli strain BW117N was grown overnight in LB medium with 2% glucose and chloramphenicol (30 mg/L) for inoculation of fresh LB medium with chloramphenicol at 1:100 dilution. Experiments with acidified nitrite were conducted with 100 mM 4-morpholine-ethanesulphonate-buffered LB medium (pH 5). Exponential-phase cultures were treated with the indicated concentration of arabinose and/or trimethoprim or acidified sodium nitrite for the stated time at 37°C. Serial dilutions of the culture were then plated on LB plates containing 2% glucose and chloramphenicol. The viable counts were recorded after overnight incubation at 37°C. The number of viable colonies from the treated cultures was divided by the number of viable colonies from the control untreated cultures to obtain the percentage of viable colonies.
Western blot analysis of expression of mutant YpTOP1-D117N protein
Cells were collected by centrifugation, and resuspended in SDS gel sample buffer before boiling and electrophoresis. The same number of cells was loaded in each lane, and equal loading of cellular proteins was confirmed by Coomassie blue staining after SDS–PAGE. The expression of mutant YpTOP1-D117N was monitored using mouse monoclonal antibodies against E. coli topoisomerase I (EcTOP), which also recognize the highly homologous YpTOP expressed as a fusion protein with a thioredoxin tag.
Results
Topoisomerase I function is required for SOS induction by mitomycin C and trimethoprim
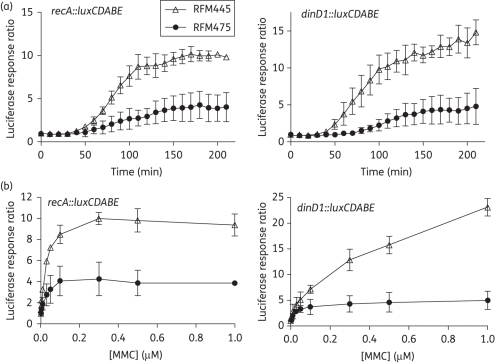
To investigate the role of topoisomerase I in the SOS response, mitomycin C, which has DNA cross-linking activity,39 was used to trigger the SOS response. The luciferase signal from the recA::luxCDABE and dinD1::luxCDABE reporters allowed the SOS induction to be monitored continuously over a period of time after the addition of different concentrations of inducers. As shown in Figure 1, the SOS response from the recA promoter resulted in up to 10-fold induction in luciferase activity from the recA::luxCDABE fusion in RFM445 treated with mitomycin C when compared with the non-treated culture. Induction of dinD1::luxCDABE could reach a higher ratio of >20-fold, but required a higher concentration of mitomycin C (Figure 1b). The induction of both recA::luxCDABE and dinD1::luxCDABE by this DNA damage agent was consistently lower in the isogenic ΔtopA strain RFM475.
Figure 1.
Effect of topA deletion on induction of recA::luxCDABE and dinD1::luxCDABE after mitomycin C treatment. E. coli RFM445 (topA+) and RFM475 (ΔtopA) transformed with either pDEW237 with the recA::luxCDABE fusion or pDinlux with the dinD1::luxCDABE fusion were grown to exponential phase (OD600 = 0.4) before the addition of mitomycin C. (a) Comparison of time course of luciferase response to 0.3 μM mitomycin C in RFM445 and RFM475. (b) Luciferase response ratio measured at 180 min after treatment with different concentrations of mitomycin C. The averages and standard deviations from three experiments are shown. MMC, mitomycin C.
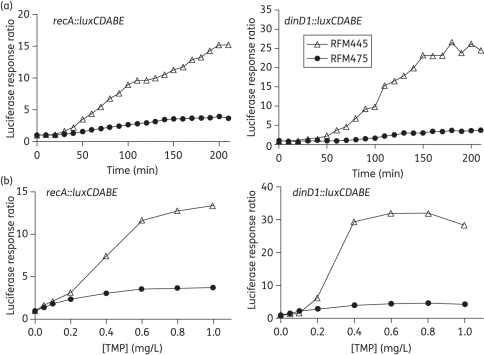
Trimethoprim is a bacteriostatic antibiotic that prevents DNA synthesis by inhibiting dihydrofolate reductase.40 In E. coli, trimethoprim can induce the bacterial SOS response and cause DNA filamentation by thymine starvation and replication fork blockage.40 As shown in Figure 2, induction of luciferase activity from recA::luxCDABE was as high as 13-fold in RFM445 after treatment with trimethoprim. The induction of dinD1::luxCDABE reached a greater ratio of up to 30-fold (Figure 2b). The induction of recA::luxCDABE and dinD1::luxCDABE after trimethoprim treatment was again much lower in the topA deletion strain RFM475. These results indicate that topoisomerase I function is required for efficient induction of the SOS response elicited by different mechanisms.
Figure 2.
Effect of topA mutation on SOS induction of recA::luxCDABE and dinD1::luxCDABE following trimethoprim treatment. E. coli strains RFM445 (topA+) and RFM475 (ΔtopA) transformed with pDEW237 or pDinlux plasmid were treated with different concentrations of trimethoprim. (a) Comparison of time course of response to 1 mg/L trimethoprim in RFM445 and RFM475. (b) Luciferase response ratio measured at 180 min after treatment with different concentrations of trimethoprim. TMP, trimethoprim.
Effect of topA mutation on RecA protein level following SOS induction by trimethoprim
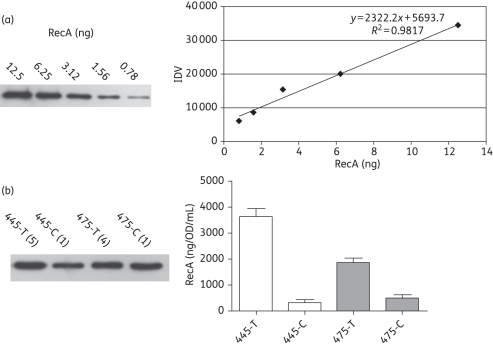
In order to eliminate the possibility that the effect observed with the topA mutation was due to the presence of the recA and dinD1 promoters on a plasmid, the effect of the topA mutation on the SOS response following trimethoprim treatment was also studied directly by quantitative western blotting of the RecA protein. A standard curve for the RecA protein was established, with good linearity within a 16-fold range of protein level (Figure 3a). The lysates from untreated cells (445-C and 475-C) and trimethoprim-treated cells (445-T and 475-T) were diluted to different extents, so that the RecA protein level fell within this linear range for quantification. The background levels of the RecA protein found in RFM445 (341 ng/OD/mL) and RFM475 (507 ng/OD/mL), where OD is the optical density of the culture at 600 nm, are within the range previously observed for other E. coli strains.38 The lesser extent of induction observed for RFM475 (Figure 3b) is in agreement with the results from the luciferase measurements. The RecA protein increased by 10.7-fold in RFM445 (topA+) following trimethoprim treatment, but only by 3.7-fold in RFM475 (ΔtopA).
Figure 3.
Quantitative western blot analysis of the RecA protein level. (a) Purified RecA protein visualized by phosphorimager after SDS–PAGE and western blotting. The resulting standard curve is shown. IDV, integrated density value. (b) Lysates of trimethoprim-treated RFM445 and RFM475 cells (445-T and 475-T) or control untreated RFM445 and RFM475 cells (445-C and 475-C) were diluted by the ratios shown in the parentheses so that they were in the linear range of the RecA signal detection, and were then analysed by SDS–PAGE and western blotting. The averages and standard deviations from three experiments are shown.
Bactericidal effect of stabilized topoisomerase I cleavage complex is enhanced by trimethoprim treatment
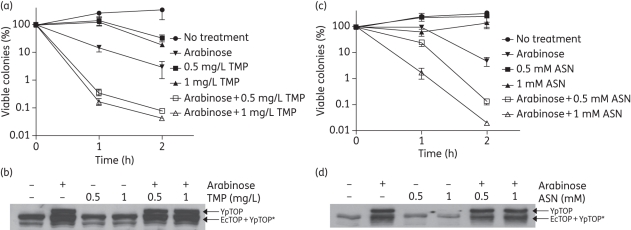
The induction of mutant YpTOP1-D117N topoisomerase I in E. coli strain BW117N with 0.002%–0.02% arabinose resulted in a 104–105-fold decrease in viability due to the lethal stabilized covalent complex.36 In order to observe enhancement of the bactericidal effect under stress conditions, a lower concentration of arabinose (0.00005%) was used to induce mutant YpTOP1-D117N. The results (Figure 4a) showed that the combination treatment of trimethoprim and arabinose significantly increased bacterial cell killing by up to 100-fold compared with treatment with arabinose alone, suggesting that cell killing mediated by the topoisomerase I cleavage complex can be enhanced during the SOS response. Western blot analysis with antibodies against topoisomerase I (Figure 4b) was performed to confirm that the induction of mutant YpTOP1-D117N topoisomerase by arabinose was similar in the absence or presence of trimethoprim.
Figure 4.
Trimethoprim (TMP) or acidified nitrite (ASN) treatment enhanced cell killing by the stabilized topoisomerase I cleavage complex. (a) BW117N was grown to exponential phase before the indicated treatment. Viable cell counts of BW117N at 1 and 2 h of incubation at 37°C with shaking after the addition of trimethoprim (0.5 or 1 mg/L) and/or induction of the topoisomerase I cleavage complex with 0.00005% arabinose were determined by serial dilutions and plating. The viable counts from each sample were then divided by the viable cell counts from the untreated culture. The averages and standard errors from four experiments are shown. (b) Western blot analysis of the expression of mutant YpTOP1-D117N protein in the presence of trimethoprim. Exponential-phase BW117N were treated with 0.00005% arabinose and/or trimethoprim at 37°C for 2 h. Some of the recombinant YpTOP proteins were degraded into the lower molecular weight form YpTOP*. (c) Arabinose (0.00005%) and/or ASN (0.5 or 1 mM) were added to the exponential-phase BW117N. Viable cell counts at 1 or 2 h after treatment and incubation at 37°C with shaking were determined. (d) Western blot analysis of YpTOP1-D117N induction with arabinose in the presence of ASN. Exponentially growing cells were treated with ASN and/or arabinose (0.00005%) at 37°C for 2 h.
Acidic nitrite enhanced the bactericidal effect of stabilized topoisomerase I cleavage complex
We have previously shown that topA deletion in E. coli is associated with increased sensitivity to oxidative challenges.30 It has been suggested that topA P1 promoter activation by Fis is an important component of the E. coli response to oxidative stress.41 The oxidative stress response is important for microbial survival during infection. We attempted to determine whether the lethal effect from mutant topoisomerase I cleavage complex stabilization is increased by hydrogen peroxide treatment. However, the addition of hydrogen peroxide decreased the level of arabinose-induced mutant topoisomerase I expression (data not shown), making it difficult to evaluate the effect of oxidative stress from hydrogen peroxide on cell killing by the stabilized topoisomerase I cleavage complex. The host defence system against bacterial pathogens includes the production of nitric oxide (NO) by activated macrophages.42 NO can be further converted into other reactive nitrogen species that are toxic to bacteria. To determine the effect of the stabilized topoisomerase cleavage complex during nitrosative stress response, an NO generator, acidified sodium nitrite (ASN), was used.43 No significant difference in viability was observed between E. coli BW117N cells treated only with ASN and the control cells, which did not receive any treatment. However, the viability of cells treated with a combination of arabinose and ASN was greatly decreased when compared with arabinose alone (Figure 4c). Western blot analysis with antibodies against topoisomerase I showed similar expression levels of mutant YpTOP1-D117N in the arabinose-treated cells, regardless of the ASN addition, confirming that ASN did not affect YpTOP1-D117N expression (Figure 4d). Therefore, it can be concluded that the bactericidal effect of the stabilized topoisomerase I cleavage complex could be enhanced under NO-induced nitrosative stress.
Discussion
Our previous study14 showed that cell killing by the stabilized topoisomerase I cleavage complex is synergistic with the bactericidal effect of quinolone treatment, which also increases the level of ROS and induces the SOS response. In this study, the involvement of topoisomerase I function in the SOS response was demonstrated using mutant strain RFM475 with a ΔtopA mutation. In this strain, mutations in the gyrase gene are present in the genetic background to compensate for the loss of topA function.44,45 Trimethoprim was therefore chosen as the antibiotic for inducing the SOS response along with the DNA cross-linking agent mitomycin C. Results from the luciferase reporter fused to the recA or dinD1 promoter, as well as direct measurement of the RecA protein level by quantitative western blot analysis, provided evidence that topoisomerase I function is important for gene transcription in the E. coli SOS response. It has been suggested that the RecA-induced SOS response is significant for the development of antibiotic resistance and persistence.32–35,46 The inhibition of topoisomerase I activity by a small molecule inhibitor similar to the alkaloids identified for S. pneumoniae topoisomerase I16 may limit the transcription of the SOS-inducible genes after antibiotic treatment, and have the added benefit of decreasing the frequencies of resistance to antibacterial drugs and persistence. The topoisomerase I inhibitors would also affect the bacterial response to high temperature and oxidative stress,20 resulting in greater vulnerability of the bacterial pathogen to the host defence. Topoisomerase I inhibitors that can act on Gram-negative bacteria remain to be discovered. Such new inhibitors could be very useful in the development of antibacterial therapy against drug-resistant Gram-negatives.
In addition to inhibiting the relaxation of negative DNA supercoils generated during transcription, topoisomerase I inhibitors also have the potential of acting in the ‘poison’ mechanism and initiate cell-killing pathways from the accumulation of the cleaved covalent complex. Bacterial cell killing induced by the stabilized topoisomerase I cleavage complex in E. coli was found to be enhanced by the stress challenges from trimethoprim treatment or acidified nitrite in this study. The increased binding of topoisomerase I to the single-stranded region of DNA due to the transcription-driven hypernegative supercoils formed during the SOS and stress response may increase the level of DNA cleavage complex. It is also possible that the formation of ROS in the oxidative cell death pathway initiated by topoisomerase I cleavage complex accumulation14 could be further boosted by nitrosative stress. Poison inhibitors that can stabilize the topoisomerase I cleavage complex could provide highly effective bactericidal therapy when combined with other bactericidal or bacteriostatic antibiotics in the host defence environment.
Funding
This work was supported by Public Health Service grant R01 AI069313 (to Y.-C. T.-D.) from the National Institutes of Health.
Transparency declarations
Y.-C. T.-D. has ongoing collaboration with AstraZeneca India. All other authors: none to declare.
Acknowledgements
We thank Tina Van Dyk for providing plasmid pDEW237 clone.
References
- 1.Schoeffler AJ, Berger JM. DNA topoisomerases: harnessing and constraining energy to govern chromosome topology. Q Rev Biophys. 2008;41:41–101. doi: 10.1017/S003358350800468X. [DOI] [PubMed] [Google Scholar]
- 2.Wang JC. Cellular roles of DNA topoisomerases: a molecular perspective. Nat Rev Mol Cell Biol. 2002;3:430–40. doi: 10.1038/nrm831. doi:10.1038/nrm831. [DOI] [PubMed] [Google Scholar]
- 3.Mitscher LA. Bacterial topoisomerase inhibitors: quinolone and pyridone antibacterial agents. Chem Rev. 2005;105:559–92. doi: 10.1021/cr030101q. doi:10.1021/cr030101q. [DOI] [PubMed] [Google Scholar]
- 4.Tse-Dinh YC. Exploring DNA topoisomerases as targets of novel therapeutic agents in the treatment of infectious diseases. Infect Disord Drug Targets. 2007;7:3–9. doi: 10.2174/187152607780090748. doi:10.2174/187152607780090748. [DOI] [PubMed] [Google Scholar]
- 5.Nitiss JL. DNA topoisomerases in cancer chemotherapy: using enzymes to generate selective DNA damage. Curr Opin Investig Drugs. 2002;3:1512–6. [PubMed] [Google Scholar]
- 6.Pommier Y, Leo E, Zhang H, et al. DNA topoisomerases and their poisoning by anticancer and antibacterial drugs. Chem Biol. 2010;17:421–33. doi: 10.1016/j.chembiol.2010.04.012. doi:10.1016/j.chembiol.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drlica K, Malik M, Kerns RJ, et al. Quinolone-mediated bacterial death. Antimicrob Agents Chemother. 2008;52:385–92. doi: 10.1128/AAC.01617-06. doi:10.1128/AAC.01617-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hooper DC. Quinolone mode of action. Drugs. 1995;49(Suppl 2):10–5. doi: 10.2165/00003495-199500492-00004. doi:10.2165/00003495-199500492-00004. [DOI] [PubMed] [Google Scholar]
- 9.Dwyer DJ, Kohanski MA, Hayete B, et al. Gyrase inhibitors induce an oxidative damage cellular death pathway in Escherichia coli. Mol Syst Biol. 2007;3:91. doi: 10.1038/msb4100135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kohanski MA, Dwyer DJ, Hayete B, et al. A common mechanism of cellular death induced by bactericidal antibiotics. Cell. 2007;130:797–810. doi: 10.1016/j.cell.2007.06.049. doi:10.1016/j.cell.2007.06.049. [DOI] [PubMed] [Google Scholar]
- 11.Forterre P, Gadelle D. Phylogenomics of DNA topoisomerases: their origin and putative roles in the emergence of modern organisms. Nucleic Acids Res. 2009;37:679–92. doi: 10.1093/nar/gkp032. doi:10.1093/nar/gkp032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tse-Dinh YC. Bacterial topoisomerase I as a target for discovery of antibacterial compounds. Nucleic Acids Res. 2009;37:731–7. doi: 10.1093/nar/gkn936. doi:10.1093/nar/gkn936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheng B, Shukla S, Vasunilashorn S, et al. Bacterial cell killing mediated by topoisomerase I DNA cleavage activity. J Biol Chem. 2005;280:38489–95. doi: 10.1074/jbc.M509722200. doi:10.1074/jbc.M509722200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu IF, Annamalai T, Sutherland JH, et al. Hydroxyl radicals are involved in cell killing by the bacterial topoisomerase I cleavage complex. J Bacteriol. 2009;191:5315–9. doi: 10.1128/JB.00559-09. doi:10.1128/JB.00559-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sutherland JH, Cheng B, Liu IF, et al. SOS induction by stabilized topoisomerase IA cleavage complex occurs via the RecBCD pathway. J Bacteriol. 2008;190:3399–403. doi: 10.1128/JB.01674-07. doi:10.1128/JB.01674-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garcia MT, Blazquez MA, Ferrandiz MJ, et al. New alkaloid antibiotics that target the DNA topoisomerase I of Streptococcus pneumoniae. J Biol Chem. 2011;286:6402–13. doi: 10.1074/jbc.M110.148148. doi:10.1074/jbc.M110.148148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheng B, Liu I, Tse-Dinh YC. Compounds with antibacterial activity that enhance DNA cleavage by bacterial DNA topoisomerase I. J Antimicrob Chemother. 2007;59:640–5. doi: 10.1093/jac/dkl556. doi:10.1093/jac/dkl556. [DOI] [PubMed] [Google Scholar]
- 18.Drolet M. Growth inhibition mediated by excess negative supercoiling: the interplay between transcription elongation, R-loop formation and DNA topology. Mol Microbiol. 2006;59:723–30. doi: 10.1111/j.1365-2958.2005.05006.x. doi:10.1111/j.1365-2958.2005.05006.x. [DOI] [PubMed] [Google Scholar]
- 19.Blot N, Mavathur R, Geertz M, et al. Homeostatic regulation of supercoiling sensitivity coordinates transcription of the bacterial genome. EMBO Rep. 2006;7:710–5. doi: 10.1038/sj.embor.7400729. doi:10.1038/sj.embor.7400729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rui S, Tse-Dinh YC. Topoisomerase function during bacterial responses to environmental challenge. Front Biosci. 2003;8:d256–63. doi: 10.2741/984. doi:10.2741/984. [DOI] [PubMed] [Google Scholar]
- 21.Cheung KJ, Badarinarayana V, Selinger DW, et al. A microarray-based antibiotic screen identifies a regulatory role for supercoiling in the osmotic stress response of Escherichia coli. Genome Res. 2003;13:206–15. doi: 10.1101/gr.401003. doi:10.1101/gr.401003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lesley SA, Jovanovich SB, Tse-Dinh YC, et al. Identification of a heat shock promoter in the topA gene of Escherichia coli. J Bacteriol. 1990;172:6871–4. doi: 10.1128/jb.172.12.6871-6874.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qi H, Menzel R, Tse-Dinh YC. Regulation of Escherichia coli topA gene transcription: involvement of a σS-dependent promoter. J Mol Biol. 1997;267:481–9. doi: 10.1006/jmbi.1997.0901. doi:10.1006/jmbi.1997.0901. [DOI] [PubMed] [Google Scholar]
- 24.Liu LF, Wang JC. Supercoiling of the DNA template during transcription. Proc Natl Acad Sci USA. 1987;84:7024–7. doi: 10.1073/pnas.84.20.7024. doi:10.1073/pnas.84.20.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheng B, Rui S, Ji C, et al. RNase H overproduction allows the expression of stress-induced genes in the absence of topoisomerase I. FEMS Microbiol Lett. 2003;221:237–42. doi: 10.1016/S0378-1097(03)00209-X. doi:10.1016/S0378-1097(03)00209-X. [DOI] [PubMed] [Google Scholar]
- 26.Drolet M, Phoenix P, Menzel R, et al. Overexpression of RNase H partially complements the growth defect of an Escherichia coli ΔtopA mutant: R-loop formation is a major problem in the absence of DNA topoisomerase I. Proc Natl Acad Sci USA. 1995;92:3526–30. doi: 10.1073/pnas.92.8.3526. doi:10.1073/pnas.92.8.3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cheng B, Zhu CX, Ji C, et al. Direct interaction between Escherichia coli RNA polymerase and the zinc ribbon domains of DNA topoisomerase I. J Biol Chem. 2003;278:30705–10. doi: 10.1074/jbc.M303403200. doi:10.1074/jbc.M303403200. [DOI] [PubMed] [Google Scholar]
- 28.Qi H, Menzel R, Tse-Dinh YC. Increased thermosensitivity associated with topoisomerase I deletion and promoter mutations in Escherichia coli. FEMS Microbiol Lett. 1999;178:141–6. doi: 10.1111/j.1574-6968.1999.tb13770.x. doi:10.1111/j.1574-6968.1999.tb13770.x. [DOI] [PubMed] [Google Scholar]
- 29.Qi H, Menzel R, Tse-Dinh YC. Effect of the deletion of the sigma 32-dependent promoter (P1) of the Escherichia coli topoisomerase I gene on thermotolerance. Mol Microbiol. 1996;21:703–11. doi: 10.1046/j.1365-2958.1996.241390.x. doi:10.1046/j.1365-2958.1996.241390.x. [DOI] [PubMed] [Google Scholar]
- 30.Tse-Dinh YC. Increased sensitivity to oxidative challenges associated with topA deletion in Escherichia coli. J Bacteriol. 2000;182:829–32. doi: 10.1128/jb.182.3.829-832.2000. doi:10.1128/JB.182.3.829-832.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stewart N, Feng J, Liu X, et al. Loss of topoisomerase I function affects the RpoS-dependent and GAD systems of acid resistance in Escherichia coli. Microbiology. 2005;151:2783–91. doi: 10.1099/mic.0.28022-0. doi:10.1099/mic.0.28022-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dwyer DJ, Kohanski MA, Collins JJ. Role of reactive oxygen species in antibiotic action and resistance. Curr Opin Microbiol. 2009;12:482–9. doi: 10.1016/j.mib.2009.06.018. doi:10.1016/j.mib.2009.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Da Re S, Garnier F, Guerin E, et al. The SOS response promotes qnrB quinolone-resistance determinant expression. EMBO Rep. 2009;10:929–33. doi: 10.1038/embor.2009.99. doi:10.1038/embor.2009.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beaber JW, Hochhut B, Waldor MK. SOS response promotes horizontal dissemination of antibiotic resistance genes. Nature. 2004;427:72–4. doi: 10.1038/nature02241. doi:10.1038/nature02241. [DOI] [PubMed] [Google Scholar]
- 35.Singh R, Ledesma KR, Chang KT, et al. Impact of recA on levofloxacin exposure-related resistance development. Antimicrob Agents Chemother. 2010;54:4262–8. doi: 10.1128/AAC.00168-10. doi:10.1128/AAC.00168-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cheng B, Annamalai T, Sorokin E, et al. Asp-to-Asn substitution at the first position of the DxD TOPRIM motif of recombinant bacterial topoisomerase I is extremely lethal to E. coli. J Mol Biol. 2009;385:558–67. doi: 10.1016/j.jmb.2008.10.073. doi:10.1016/j.jmb.2008.10.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Van Dyk TK, Wei Y, Hanafey MK, et al. A genomic approach to gene fusion technology. Proc Natl Acad Sci USA. 2001;98:2555–60. doi: 10.1073/pnas.041620498. doi:10.1073/pnas.041620498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O'Reilly EK, Kreuzer KN. Isolation of SOS constitutive mutants of Escherichia coli. J Bacteriol. 2004;186:7149–60. doi: 10.1128/JB.186.21.7149-7160.2004. doi:10.1128/JB.186.21.7149-7160.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tomasz M, Palom Y. The mitomycin bioreductive antitumor agents: cross-linking and alkylation of DNA as the molecular basis of their activity. Pharmacol Ther. 1997;76:73–87. doi: 10.1016/s0163-7258(97)00088-0. doi:10.1016/S0163-7258(97)00088-0. [DOI] [PubMed] [Google Scholar]
- 40.Amyes SG, Smith JT. Trimethoprim action and its analogy with thymine starvation. Antimicrob Agents Chemother. 1974;5:169–78. doi: 10.1128/aac.5.2.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weinstein-Fischer D, Elgrably-Weiss M, Altuvia S. Escherichia coli response to hydrogen peroxide: a role for DNA supercoiling, topoisomerase I and Fis. Mol Microbiol. 2000;35:1413–20. doi: 10.1046/j.1365-2958.2000.01805.x. doi:10.1046/j.1365-2958.2000.01805.x. [DOI] [PubMed] [Google Scholar]
- 42.Fang FC. Antimicrobial reactive oxygen and nitrogen species: concepts and controversies. Nat Rev Microbiol. 2004;2:820–32. doi: 10.1038/nrmicro1004. doi:10.1038/nrmicro1004. [DOI] [PubMed] [Google Scholar]
- 43.Bower JM, Mulvey MA. Polyamine-mediated resistance of uropathogenic Escherichia coli to nitrosative stress. J Bacteriol. 2006;188:928–33. doi: 10.1128/JB.188.3.928-933.2006. doi:10.1128/JB.188.3.928-933.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pruss GJ, Manes SH, Drlica K. Escherichia coli DNA topoisomerase I mutants: increased supercoiling is corrected by mutations near gyrase genes. Cell. 1982;31:35–42. doi: 10.1016/0092-8674(82)90402-0. doi:10.1016/0092-8674(82)90402-0. [DOI] [PubMed] [Google Scholar]
- 45.DiNardo S, Voelkel KA, Sternglanz R, et al. Escherichia coli DNA topoisomerase I mutants have compensatory mutations in DNA gyrase genes. Cell. 1982;31:43–51. doi: 10.1016/0092-8674(82)90403-2. doi:10.1016/0092-8674(82)90403-2. [DOI] [PubMed] [Google Scholar]
- 46.Dorr T, Lewis K, Vulic M. SOS response induces persistence to fluoroquinolones in Escherichia coli. PLoS Genet. 2009;5:e1000760. doi: 10.1371/journal.pgen.1000760. doi:10.1371/journal.pgen.1000760. [DOI] [PMC free article] [PubMed] [Google Scholar]