Abstract
Genotype-phenotype studies in pharmacogenomics promise to identify the genetic factors that contribute substantially to variation in individual drug response. While most genetic association studies have failed to deliver the promise, several recent examples serve as a reminder that these associations do exist and can be identified when investigated using well-designed studies. Here, we describe the path taken to identify the association between common VKORC1 genetic variation and warfarin dosing in patients. We also describe the key elements that led the way, such as definition of the phenotype, confirmation of a genetic component, determination of biological plausibility, and selection of genetic polymorphisms. We also describe several avenues yet-to-be explored for the specific VKORC1 warfarin example that can also be generalized as the future directions for many genetic association studies in pharmacogenomics. These future avenues will best be explored using diverse approaches encompassing clinical, statistical, and genomic methods currently being developed for genotype-phenotype studies in human populations.
Keywords: warfarin, VKORC1, genetic association studies, pharmacogenomics, SNPs
Introduction
Since DNA's first molecular description by Watson and Crick [1] followed by the first draft of the human genome nearly fifty years later [2, 3], there has been an explosion in the knowledge of genetic variation contained within human populations. One field of scientific inquiry that has capitalized on the recent deluge of genetic variation information is pharmacogenomics, a field of study where genetics explains individual differences in drug responses [4]. Despite the present-day technologies available to study pharmacogenomics, such as the development of whole-genome genotyping methods [5] and the promise of low-cost whole genome re-sequencing [6–8], many challenges still remain that impede the ability to identify specific genetic variations associated with variable drug response in human populations. In this report, we briefly summarize a few of the challenges faced in performing genetic association studies that, in general, are also applicable to studies in the field of pharmacogenomics. We then outline several key elements contained within a well-designed association study using the well-established and recent example of the association between the candidate gene vitamin K epoxide reductase complex subunit 1 (VKORC1) and warfarin dose. The functional role of VKORC1 as one of the rate controlling enzymes in the vitamin K cycle, leads to the production of vitamin K dependent, gamma carboxylated proteins, which are composed primarily of clotting factors II, VII, IX, X, protein C, S, and Z and have broad effects on the coagulation cascade. Finally, we outline several burgeoning areas of research in pharmacogenomics requiring the exploration of different methodologies and techniques to fully mine the datasets of interest. As more data are produced, it is hoped that diverse approaches encompassing elegant clinical, statistical, and genomic methods will expand and lead to additional high-impact discoveries in the field of pharmacogenomics.
Challenges in genetic association studies
The Achilles heel of genetic association studies has been the issue of replication. It is well-known that most initial, published genetic associations do not replicate in subsequent studies [9–11]. The causes of non-replication in genetic association studies are many and complex. Some of these initial association findings may be due to improper matching of cases and controls [12, 13] or cryptic population subdivision [14–16], spurious false-positives from studies with small sample sizes [17], and over-interpretation of the results and/or biological implausibility of the candidate gene [13, 18]. Also, technical issues related to genotype accuracy and efficiency as well as SNP selection using linkage disequilibrium can play a role in the failure to replicate the initial association [13, 19]. Even in well-designed association studies, failure to replicate in subsequent studies is expected due to genetic and allelic heterogeneity (encouraged by poor phenotype definitions), differences in prevalence and effect size across populations [14], and differences in environmental exposures across study populations [20].
Given these numerous factors, it seems almost all genetic association studies are doomed to non-replication. Despite the dismal outlook, there are a few examples in the field of pharmacogenomics where the initial genetic association finding was and continues to be replicated in subsequent studies. These examples include the association between genetic variants of VKORC1 and CYP2C9 and the anticoagulant warfarin, ADRB1 and β-blockers, and UGT1A1 and irinotecan, to name a few [21–23]. The last example, the association between UGT1A1 variants and the development of neutropenia in individuals that take irinotecan, is an example where the Federal Drug Administration (FDA) in the United States has approved re-labeling of this medication to mention that the gene and its variants are associated with adverse event in individuals prescribed the mediation [24]. More recently, the FDA's Clinical Pharmacology Subcommittee has suggested re-labeling warfarin to incorporate information about VKORC1 and CYP2C9 variants and their ability to predict dosing. Thus, despite the negative findings so well documented in the literature, it is certain that true and strong genetic associations exist and, when detected, can be extremely useful in the prevention or treatment of disease.
VKORC1 and warfarin: dissection of a successful genetic association study
The phenotype
In retrospect, there are several key characteristics of the VKORC1/warfarin association study that enhanced its ability to be detected using present-day technologies and methodologies. The first and arguably most important characteristic of this successful association is the phenotype of interest: warfarin dosing. While warfarin dosing can vary widely from individual to individual, it is tailored to each individual and strictly monitored by physicians using the international normalized ratio (INR). INR is a standardized ratio of the patient's prothrombin time (the time it takes for the plasma to clot) to the control individual's prothrombin time. For patients taking anticoagulants (warfarin), the therapeutic INR generally ranges from 2.0 to 3.0. INR levels falling above or below this therapeutic window leads to increased risk of adverse bleeding events or subtherapeutic anticoagulation levels that put a patient at risk for another clotting event. The narrow therapeutic range and the standardization of prothrombin time make INR a homogenous phenotype, which presumably decreases the amount of trait heterogeneity in the study and subsequently decreases the amount of locus heterogeneity. INR can be used directly as a phenotype measure to determine time within or outside the therapeutic range and the risk for adverse events. Furthermore, stabilized warfarin dose is determined as a function of the time spent consistently within the INR therapeutic range, and therefore serves a proxy for a patient's anticoagulation state. Although it is these clean, accurate phenotype definitions that give hope for success to other association studies, a caveat to this hope is the fact that many complex diseases do not have such an easily defined phenotype for study.
Evidence for a Genetic Component and Biological Plausibility
The second and third characteristics of this successful candidate gene association study are evidence that the phenotype has a tangible genetic component and there is biological plausibility of the candidate gene chosen for the study. Heritability estimates from twin and family studies can give clues to whether or not there is a measurable genetic component to a phenotype [25]. In the case of warfarin sensitivity, there are no heritability estimates per se for this specific phenotype; however, several decades before present, it was noted that warfarin resistance was heritable in rats [26]. In humans, the more extreme and rare phenotype of familial multiple coagulation factor deficiency was observed in two families with autosomal recessive deficiency of all vitamin K dependent coagulation factors [27]. These two families, along with linkage studies in rats and mice for warfarin resistance [28], provided the opportunity to localize the VKORC1 locus to chromosome 16 using traditional linkage analysis [29].
In 2004, simultaneous reports were published describing different strategies and methods that lead to the identification of VKORC1. In one report, Li and colleagues used short interfering RNA (siRNA) pools against candidate genes in the region localized by previous mapping efforts to identify the gene that was responsible for VKOR activity in human cells [30]. In the other report, Rost and colleagues identified VKORC1 by sequencing the exons contained within the 4.0 Mb region of chromosome 16 linked to warfarin resistance in a small sample of patients with either warfarin resistance or with combined deficiency of vitamin-K-dependent clotting factors type 2 from the original linkage studies [31]. In this mutational screen, VKORC1 missense mutations were identified in all warfarin resistant patients screened, and, in the case of the patients from the linkage analysis, the patients and their affected siblings were homozygous for the mutations while their unaffected parents were heterozygous for the mutations [31]. While these studies were designed to elucidate the function of VKORC1, further examination of the coding region in ‘control’ individuals did not reveal any commonly occurring protein altering polymorphisms which would be hypothesized to affect VKOR activity in a broader spectrum of individuals [30, 31]. Subsequent reports have described additional VKORC1 mutations which alter protein structure and cause warfarin resistance [32–35], establishing that this candidate gene is at least important in the rare, extreme phenotype of resistance, and invoking the idea that it may be important in warfarin dose variability commonly observed in patient populations on normal dosing regimens.
Common genetic variation and linkage disequilibrium
Given the plausibility that the candidate gene VKORC1 could be important in warfarin dosing among patients responding to warfarin, interest grew in examining the possible association between common genetic variation and the dosing phenotype. Public resources such as the International HapMap Project [36] and the SeattleSNPs Program for Genomic Applications [37] provide information on the genetic variations known within VKORC1, and they also provide information about linkage disequilibrium (LD) for this candidate gene in several human populations. Knowledge of LD provides the investigator with an opportunity to rationally select a small set of common genetic variations to genotype in their study so that each SNP would represent or "tag" the other SNPs not genotyped in the study. This "tagSNP" approach to a genetic association study is often referred to as an "indirect" association study design because a significant association between the tagSNP and the phenotype implies that either the tagSNP confers risk or, more likely, a SNP in LD with the tagSNP confers risk [19].
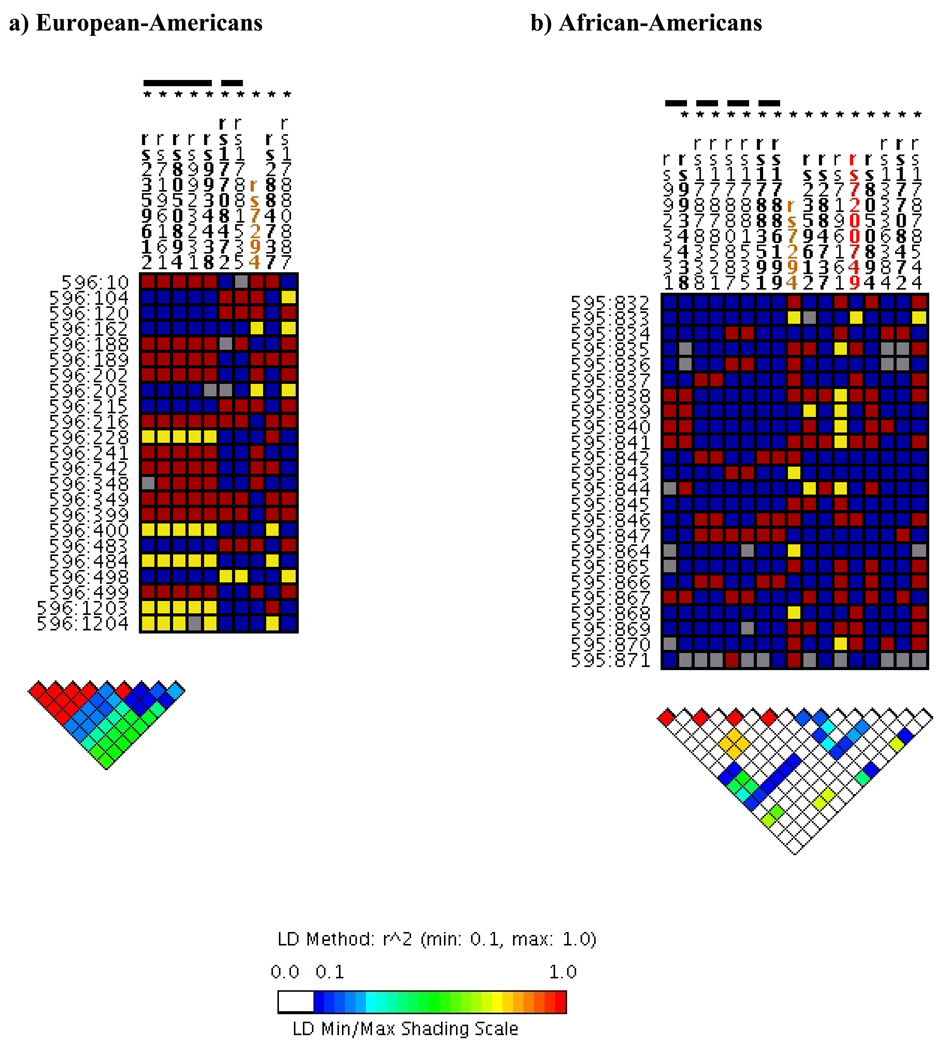
Knowledge of LD in the candidate gene and surrounding genomic regions can also help the investigator interpret the result suggesting that several SNPs are associated with the phenotype of interest. As an example, the Figure (a) displays ten common VKORC1 SNPs (minor allele frequency >5%) recently described in a European-descent patient population [38]. From these ten SNPs, seven were significantly associated with warfarin dose after adjustment for covariates at p<0.001, yet five of these seven significant SNPs (rs7196161, rs9923231, rs9934438, rs8050894, and rs2359612) were in LD with one another [Figure (a); 38]. Had tagSNPs been chosen prior to genotyping, only five of the ten SNPs would have been genotyped (Figure a), and, of those, only three tagSNPs would have been significantly associated with warfarin dosing, representing the seven common SNPs originally described as significant in the association study.
Figure. Linkage disequilibrium and tagSNP selection for common VKORC1 genetic variation in two populations.
VKORC1 was re-sequenced in 23 Euorepan-Americans and 24 African-Americans as previously described [38]. Linkage disequilibrium was calculated using r2 by the Genome Variation Server (http://gvs.gs.washington.edu/GVS/) for SNPs with a minor allele frequency (MAF) >5% for each population sample separately. TagSNPs were also determined using the Genome Variation Server for common VKORC1 SNPs (MAF>5%) at default settings (r2>0.80). SNPs are labeled using rs numbers and are color coded so that red represent nonsynonymous SNPs, orange represents SNPs in the untranslated region, bolded represents SNPs in unique sequence regions, and unbolded represents SNPs in repeat sequence regions. DNA samples used in re-sequencing are labeled to the left of the figure using dbSNP nomenclature (population ID:individual ID). SNP genotypes (squares) are color coded so that blue represents homozygosity for the major allele, red represents heterozygosity, and yellow represents homozygosity for the minor allele. Gray squares represent missing data. The bar above the SNPs represents a group or “bin” of SNPs that can be represented by a single tagSNP (denoted by an asterisk above the rs number) from the bin. The triangle plot represents the pair-wise linkage disequilibrium statistics (r2), and the results are color coded so that red represents the highest and blue represents lowest linkage disequilibrium statistics, respectively.
Finding the "functional" SNP
Given that there are several SNPs associated with warfarin dosing, the next step in possibly understanding the underlying biology or functional mechanism behind the phenotype is to identify the risk conferring SNP from among the SNPs represented by the tagSNPs. In one report [38], a stepwise regression was performed to determine which of the seven originally associated SNPs were most predictive of warfarin dosing. Of the original seven, the five SNPs in LD (rs7196161, rs9923231, rs9934438, rs8050894, and rs2359612) were most predictive of warfarin dosing (i.e., explained the largest portion of the dose variability), suggesting that one of the five SNPs is the putative "functional" SNP. None of the five SNPs were located in any of the exons of VKORC1: two were located 5' upstream of VKORC1 (rs7196161 and rs9923231), and three were located in introns (rs9934438, rs8050894, and rs2359612). To help narrow the list of functional or risk conferring SNPs even further, the human SNPs were mapped to other species (mouse, rat, and dog) in hopes of identifying conserved non-coding regions that contained the SNPs associated with warfarin dosing. The sequences containing two of the five SNPs (rs9934438 and rs8050894) were conserved across species, strongly implicating that these SNPs are the putative functional SNPs responsible for the association observed between VKORC1 and warfarin dosing [38]. However, other SNPs in the upstream promoter region (such as −1639 G>A; rs9923231) clearly would be functional candidates simply by virtue of their location independent of sequence conservation.
Replication and other supportive data
Despite the strength of the initial descriptions of the association between VKORC1 common SNPs and warfarin dosing [38, 39], the association is considered tenuous without replication in an independent population sample In the report described above [38], replication was achieved in a second, larger study of European-descent patients using a subset of the original 10 SNPs described in VKORC1, and subsequent studies performed by other investigators have reported similar findings for these SNPs in their patient populations [40]. Supportive data came from experimental data generated to define a molecular mechanism for the observed genotype-dose association. For example, VKORC1 SNPs associated with warfarin dosing are also associated with VKORC1 mRNA levels in human liver tissue [38]. Further supportive data have shown that significant alterations in promoter expression, due to a promoter SNP (−1639 G>A; rs9923231) among the five putative functional SNPs in LD, gives further evidence for transcriptional regulation of VKORC1 and may provide a more mechanistic explanation for the phenotypic effects observed in patients [41].
Effect size
The final characteristic of this association that led to its identification is the effect size of the genetic component. Not accounting for complicating factors such as linkage disequilibrium and allele frequency of the risk conferring SNP and the genotyped SNP [42], genetic variants with larger effect sizes require smaller sample sizes to detect a statistical association compared with smaller effect sizes. All studies except for one [39] estimate that ∼15–30% of the variability in warfarin dosing in European-descent populations could be explained by common VKORC1 genetic variants [38, 43–48]. In comparison to this relatively large effect size, many studies suggest that two genetic variants within CYP2C9 (known as CYP2C9*2 and CYP2C9*3 or rs1799853 and rs1057910, respectively) each account for only approximately 10% in the variability of warfarin dosing [38, 46, 47]. For the data available in other populations (mostly of Asian-descent), it seems that VKORC1 genetic variants also explain a large proportion of the variance observed in warfarin dosing in these populations [49–54]. Thus, the effect size of these VKORC1 genetic variants is relatively large for all populations studied to date. Unfortunately, it is difficult to predict a priori whether or not the genetic variants being assayed in a study will have a large effect on the phenotype of interest.
Outlook
The success of the replicable VKORC1 association with warfarin dosing has catapulted this candidate gene into clinical trials to establish whether or not knowledge of the genotypes associated with low dose or high dose is more effective in the determining the starting dose of warfarin compared with standard care (www.clinicaltrials.gov). While much excitement surrounds this prospect of clinical utility, many avenues of research have yet-to-be explored that can further our understanding of the genetic factors that influence warfarin dosing. These avenues include studying the VKORC1/warfarin association in populations of African-descent, searching for other genes and genomic regions that are associated with warfarin dosing, and exploring gene-gene and gene-environment interactions.
VKORC1/warfarin and African-descent populations
On average, African-American patients require a higher dose of warfarin compared with European-descent patients [55]. Interestingly, SNPs associated with higher warfarin dosing in other populations are also found at a much higher frequency in African-Americans [38]. It is possible that either novel or established VKORC1 associations could be identified in this population that would explain the difference across populations observed in the clinic. Most previous studies have included small sample sizes of African-descent patients, and these studies were inconclusive both because of the sample size and because of the study design [44, 45, 54]. However a recent report testing for an association between VKORC1 SNP 1173 C>T (rs9934438) and warfarin dosing in a large African-American patient population (n=162) suggests that while the minor allele of this SNP is associated with lower dosing in the African-American patient population compared with the carriers of the CC genotype, the SNP accounts for less of the variability in warfarin dosing compared with the European-descent population [56]. Moreover, the T allele of SNP 1173 C>T (rs9934438) is associated with lower odds of under- and over-anticoagulation (INRs <2 and >3, respectively) in the European-descent patient population, but this same allele is not associated with similar lower odds in the African-American patient population [56].
Schellemen and colleagues [56] offer several explanations for the differences they observed between the European- and African-descent patient populations. One explanation offered is the observation that European- and African-descent populations have different VKORC1 polymorphisms and patterns of linkage disequilibrium [38, 57], and it is possible that other VKORC1 SNPs confer stronger effects on warfarin dosing for the African-American patient population compared with those described originally in European-descent patients. To properly test for an association between VKORC1 and warfarin dosing in an African-descent patient population, tagSNPs or SNPs in general should be chosen specifically for this population rather than genotyping SNPs identified and characterized in a European-descent population (Figure b). This strategy could be used to both identify novel associations as well as further narrow the putative functional VKORC1 SNP (rs9934438 versus rs8050894) given the reduced linkage disequilibrium characteristic of an African-descent population.
If a novel VKORC1 association were identified in African-descent patients, it would be interesting to note if inclusion of these new data would alter the decision model for warfarin use, which currently takes into account the patient's race/ethnicity [58]. Presumably, knowledge of the patient's VKORC1 genotype prior to warfarin use will help guide clinicians in selecting a specific warfarin dose at the beginning of treatment rather than the standard dose that requires constant monitoring and adjustment. Clinical trials are now underway to test whether the pharmacogenetic-guided dosing method is superior to the standard dosing method (for example, PRospective Evaluation Comparing Initiation of Warfarin StrategiEs (PRECISE): Pharmacogenetic-Guided Versus Usual Care described at www.clinicaltrials.gov). Although using self-identified race in both research [59, 60] and treatment [61–63] is highly controversial, it cannot be denied that data from a variety of populations, however defined, is necessary to increase the chance that a treatment or preventive measure will benefit as many individuals as possible [64]. Ideally, identification of a definitive functional SNP or a consistent SNP-dose association across several racial groups (i.e. European, Asian and African-descent) could lead to the elimination of qualitative race categorization and treatment with replacement by quantitative and more accurate data based on genotype-dose associations.
Other genes and whole genome association studies
Currently, VKORC1 genetic variants combined with CYP2C9 genetic variants, body mass index, age, gender, and drug interactions can account for as much as 60% of the variance in warfarin dosing in European-descent populations [65, 66]. Conversely, up to 40% of the variance in warfarin dosing is unexplained. Recent efforts to identify associations between other candidate genes from the warfarin interactive pathways and warfarin dosing have not identified novel associations that contribute substantially to its variability as did VKORC1 [47].
It is still possible that other genes or genomic regions contribute substantially to the variability in warfarin dosing. Recently developed genotyping technologies now make it feasible to perform a whole genome association study in unrelated cases and controls to identify the previously unknown candidate genes or genomic regions associated with the phenotype of interest [5]. While is it now possible both technologically and economically to genotype 500,000 to 1 million markers across the genome, the techniques to fully analyze and interpret the data are still taking shape [67]. Much like the candidate gene association study, whole genome association studies are subject to false positives and require replication in several populations [68–73]. Despite these problems, early successes such as the association between CFH and age-related macular degeneration [74] and IL23R and Crohn's disease [75] provide the hope that novel associations with measurable impacts on the phenotype can be identified through this approach.
Exploring gene-gene and gene-environment interactions
Up to this point, the focus of this special report has been the association of a single genetic polymorphism with the phenotype of interest. The genetic architecture of any phenotype is, of course, more complicated, involving genes, the environment, and interactions between these entities. In the case of warfarin dosing, among the candidate genes associated with the phenotype, no significant gene-gene interactions have been reported. For our purposes here, gene-gene interaction (also known as epistasis) is defined as the combined effect of two or more genes on a phenotype that could not be predicted from their individual genotypes [76]. In the same vein, no significant gene-environment interactions have been reported despite the fact that dietary intake of vitamin K [77] and concurrent medication [78] can affect warfarin dosing in individuals. Gene-environment interaction is defined as the combined effect of genetic factors with environmental factors, whereby the resultant phenotype could not be predicted from either factor individually. As more data are collected, particularly for prospective studies [20] such as clinical trials, it will be possible to mine these datasets for interactions using traditional methodologies such as those based on regression [79] as well as newly developed methodologies such as multifactor dimensionality reduction or MDR [80, 81] and neural networks [82, 83]. MDR is a data reduction method designed to detect gene-gene and gene-environment interactions in the presence or absence of main effects in case-controls studies. MDR pools high-risk and low-risk multi-locus genotype combinations to identify predictive multi-locus models of association [80, 81]. Neural networks (NN), on the other hand, are a class of pattern recognition methods that can also be used for association studies. NN consider data in parallel, much like the human brain, rather than sequentially like most computers [82, 83]. A huge challenge for these analyses will be their interpretation as a single statistical interaction could be explained by many biological processes and pathways [84]. Yet another challenge will be collecting the large sample sizes that may be required to detect these interactions [20].
Summary
In this special report, we use the recently described association between common genetic variants in the candidate gene VKORC1 and warfarin dosing as a successful model for candidate gene association studies for pharmacogenomics. This successful genetic association study has several essential elements including a well-defined phenotype, biological plausibility of the candidate gene, large effect size of the genetic component, and good knowledge of the LD structure prior to the study (see Table). While these elements ensured rapid identification and replication of the association, several avenues of study remain including identifying the true VKORC1 risk allele(s) among the many SNPs in strong linkage disequilibrium with one another in the European-descent population, determining the function of the risk SNP(s), including other populations in the association study, identifying other relevant candidate genes or genomic regions, and searching for relevant gene-gene and gene-environment interactions. To accomplish these goals, no single approach is sufficient, thus necessitating and perpetuating the need for diverse approaches to identifying the genotype behind the phenotype of human complex disease.
Table.
Key elements of a successful candidate gene association study through the example VKORC1 and warfarin dosing.
| Study Element | Brief Description | References |
|---|---|---|
| Phenotype | Warfarin has a narrow therapeutic range |
|
| Warfarin dosage has a large inter- individual variability |
||
| Warfarin dosage monitored by homogenous measure (INR) |
||
| Substantial genetic Component |
No human heritabilility estimates available for warfarin sensitivity |
|
| Warfarin resistance heritable in rats |
[26] | |
| The rare phenotype of familial multiple coagulation factor deficiency was observed in two families with autosomal recessive deficiency of all vitamin K dependent coagulation factors |
[27] | |
| Biological plausibility |
Human and rodent linkage studies localize warfarin resistance locus to chromosome 16 |
[27–29] |
|
VKORC1 localized using short interfering RNA (siRNA) pools against candidate genes in the region localized by linkage |
[30] | |
| Mutations identified in VKORC1 exons of warfarin resistant patients and linkage study families |
[31] | |
| Knowledge of polymorphisms and linkage disequilibrium |
VKORC1 re- sequenced by SeattleSNPs |
[38] |
|
VKORC1 SNPs and genotypes are available in public databases and can be used to calculate linkage disequilibrium for tagSNP selection |
pga.gs.washington.edu www.ncbi.nlm.nih.gov/projects/SNP/ www.hapmap.org |
|
| Effect size |
VKORC1 in most populations accounts for 15–30% of the variability in warfarin dosing |
[38, 43–48] |
|
VKORC1 variants and covariates account for up to 60% of the variability in warfarin dosing |
[65, 66] | |
Acknowledgements
This work was supported by National Institutes of Health grants NS053646 (M.J.R), AG20135 (M.D.R.), and HL65962 as part of the Pharmacogenomics of Arrhythmia Therapy U01 site of the Pharmacogenetics Research Network (M.D.R).
Reference List
- 1.Watson JD, Crick FH. Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature. 1953;17:737–738. doi: 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- 2.Venter JC, Adams MD, Myers EW, Li PW, Mural RJ. The sequence of the human genome. Science. 2001;291:1304. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 3.Initial sequencing and analysis of the human genome. Nature. 2001;409:806. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 4.Motulsky A. Drug reactions, enzymes and biochemical genetics. JAMA. 1957;165:835–837. doi: 10.1001/jama.1957.72980250010016. [DOI] [PubMed] [Google Scholar]
- 5.Fan JB, Chee MS, Gunderson KL. Highly parallel genomic assays. Nat Rev Genet. 2006;7:632–644. doi: 10.1038/nrg1901. [DOI] [PubMed] [Google Scholar]
- 6.Bennett ST, Barnes C, Cox A, Davies L, Brown C. Toward the $1000 human genome. Pharmacogenomics. 2005;6:373–382. doi: 10.1517/14622416.6.4.373. [DOI] [PubMed] [Google Scholar]
- 7.Gibbs R. Deeper into the genome. Nature. 2005;437:1233–1234. doi: 10.1038/4371233a. [DOI] [PubMed] [Google Scholar]
- 8.Bentley DR. Whole-genome re-sequencing. Current Opinion in Genetics & Development. 2006;16:545–552. doi: 10.1016/j.gde.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 9.Lohmueller KE, Pearce CL, Pike M, Lander ES, Hirschhorn JN. Meta-analysis of genetic association studies supports a contribution of common variants to susceptibility to common disease. Nat Genet. 2003;33:177–182. doi: 10.1038/ng1071. [DOI] [PubMed] [Google Scholar]
- 10.Ioannidis JPA, Ntzani EE, Trikalinos TA, Contopoulos-Ioannidis DG. Replication validity of genetic association studies. Nat Genet. 2001;29:306–309. doi: 10.1038/ng749. [DOI] [PubMed] [Google Scholar]
- 11.Hirschhorn JN, Lohmueller K, Byrne E, Hirschhorn K. A comprehensive review of genetic association studies. Genet Med. 2002;4:45–61. doi: 10.1097/00125817-200203000-00002. [DOI] [PubMed] [Google Scholar]
- 12.Zondervan KT, Cardon LR, Kennedy SH. What makes a good case-control study? Design issues for complex traits such as endometriosis. Hum Reprod. 2002;17:1415. doi: 10.1093/humrep/17.6.1415. [DOI] [PubMed] [Google Scholar]
- 13.Hattersley AT, McCarthy MI. What makes a good genetic association study? The Lancet. 366:1315–1323. doi: 10.1016/S0140-6736(05)67531-9. [DOI] [PubMed] [Google Scholar]
- 14.Ardlie KG, Lunetta KL, Seielstad M. Testing for population subdivision and association in four case-control studies. Am J Hum Genet. 2002;71:304–311. doi: 10.1086/341719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cardon LR, Palmer LJ. Population stratification and spurious allelic association. Lancet. 2003;361:598. doi: 10.1016/S0140-6736(03)12520-2. [DOI] [PubMed] [Google Scholar]
- 16.Freedman ML, Reich D, Penney KL, McDonald GJ, Mignault AA, Patterson N, et al. Assessing the impact of population stratification on genetic association studies. Nat Genet. 2004;36:388–393. doi: 10.1038/ng1333. [DOI] [PubMed] [Google Scholar]
- 17.Ioannidis JP, Trikalinos TA, Ntzani EE, Contopoulos-Ioannidis DG. Genetic associations in large versus small studies: an empirical assessment. The Lancet. 2003;361:567–571. doi: 10.1016/S0140-6736(03)12516-0. [DOI] [PubMed] [Google Scholar]
- 18.Cardon LR, Bell JI. Association study designs for complex diseases. Nat Rev Genet. 2001;2:91. doi: 10.1038/35052543. [DOI] [PubMed] [Google Scholar]
- 19.Palmer LJ, Cardon LR. Shaking the tree: mapping complex disease genes with linkage disequilibrium. The Lancet. 2005;366:1223–1234. doi: 10.1016/S0140-6736(05)67485-5. [DOI] [PubMed] [Google Scholar]
- 20.Manolio TA, Bailey-Wilson JE, Collins FS. Genes, environment and the value of prospective cohort studies. Nat Rev Genet. 2006;7:812–820. doi: 10.1038/nrg1919. [DOI] [PubMed] [Google Scholar]
- 21.Goldstein DB, Tate SK, Sisodiya SM. Pharmacogenetics goes genomic. Nat Rev Genet. 2003;4:937. doi: 10.1038/nrg1229. [DOI] [PubMed] [Google Scholar]
- 22.Need AC, Motulsky AG, Goldstein DB. Priorities and standards in pharmacogenetic research. Nat Genet. 2005;37:671–681. doi: 10.1038/ng1593. [DOI] [PubMed] [Google Scholar]
- 23.Roden DM, Altman RB, Benowitz NL, Flockhart DA, Giacomini KM, Johnson JA, et al. Pharmacogenomics: Challenges and Opportunities. Ann Intern Med. 2006;145:749–757. doi: 10.7326/0003-4819-145-10-200611210-00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marsh S, McLeod HL. Pharmacogenomics: from bedside to clinical practice. Hum Mol Genet. 2006;15:R89–R93. doi: 10.1093/hmg/ddl087. [DOI] [PubMed] [Google Scholar]
- 25.Vitzthum VJ. A number no greater than the sum of its parts: the use and abuse of heritability. Hum Biol. 2003;75:539. doi: 10.1353/hub.2003.0064. [DOI] [PubMed] [Google Scholar]
- 26.Greavses JH, Ayres P. Heritable resistance to warfarin in rats. Nature. 1967;215:877–878. doi: 10.1038/215877a0. [DOI] [PubMed] [Google Scholar]
- 27.Oldenburg J, von Brederlow B, Fregin A, Rost S, Wolz W, Eberl W, et al. Congenital deficiency of vitamin K dependent coagulation factors in two families presents as a genetic defect of the vitamin K-epoixide reductase-complex. Thromb Haemost. 2000;84:937–941. [PubMed] [Google Scholar]
- 28.Kohn MH, Pelz HJ. A gene-anchored map position of the rat warfarin-resistance locus, Rw, and its orthologs in mice and humans. Blood. 2000;96:1996–1998. [PubMed] [Google Scholar]
- 29.Fregin A, Rost S, Wolz W, Krebsova A, Muller CR, Oldenburg J. Homozygosity mapping of a second gene locus for hereditary combined deficiency of vitamin K-dependent clotting factors to the centromeric region of chromosome 16. Blood. 2002;100:3229–3232. doi: 10.1182/blood-2002-03-0698. [DOI] [PubMed] [Google Scholar]
- 30.Li T, Chang CY, Jin DY, Lin PJ, Khvorova A, Stafford DW. Identification of the gene for vitamin K epoxide reductase. Nature. 2004;427:541–544. doi: 10.1038/nature02254. [DOI] [PubMed] [Google Scholar]
- 31.Rost S, Fregin A, Ivaskevicius V, Conzelmann E, Hortnagel K, Pelz HJ, et al. Mutations in VKORC1 cause warfarin resistance and multiple coagulation factor deficiency type 2. Nature. 2004;427:537–541. doi: 10.1038/nature02214. [DOI] [PubMed] [Google Scholar]
- 32.Loebstein R, Dvoskin I, Halkin H, Vecsler M, Lubetsky A, Rechavi G, et al. A coding VKORC1 Asp36Tyr polymorphism predisposes to warfarin resistance. Blood. 2006 doi: 10.1182/blood-2006-08-038984. blood-2006. [DOI] [PubMed] [Google Scholar]
- 33.D'AMBROSIO RL, D'ANDREA G, CAFOLLA A, FAILLACE F, MARGAGLIONE M. A new vitamin K epoxide reductase complex subunit-1 (VKORC1) mutation in a patient with decreased stability of CYP2C9 enzyme. Journal of Thrombosis and Haemostasis. 2007;5:191–193. doi: 10.1111/j.1538-7836.2006.02261.x. [DOI] [PubMed] [Google Scholar]
- 34.BODIN L, HORELLOU MH, FLAUJAC C, LORIOT MA, SAMAMA MM. A vitamin K epoxide reductase complex subunit-1 (VKORC1) mutation in a patient with vitamin K antagonist resistance. Journal of Thrombosis and Haemostasis. 2005;3:1533–1535. doi: 10.1111/j.1538-7836.2005.01449.x. [DOI] [PubMed] [Google Scholar]
- 35.Harrington DJ, Underwood S, Morse C, Shearer MJ, Tuddenham EG, Mumford AD. Pharmacodynamic resistance to warfarin associated with a Val66Met substitution in vitamin K epoxide reductase complex subunit 1. Thromb Haemost. 2005;93:23–26. doi: 10.1160/TH04-08-0540. [DOI] [PubMed] [Google Scholar]
- 36.The International HapMap Consortium. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Crawford DC, Akey DT, Nickerson DA. The patterns of natural variation in human genes. Annual Review of Genomics and Human Genetics. 2005;6:287–312. doi: 10.1146/annurev.genom.6.080604.162309. [DOI] [PubMed] [Google Scholar]
- 38.Rieder MJ, Reiner AP, Gage BF, Nickerson DA, Eby CS, McLeod HL, et al. Effect of VKORC1 haplotypes on transcriptional regulation and warfarin dose. N Engl J Med. 2005;352:2285–2293. doi: 10.1056/NEJMoa044503. [DOI] [PubMed] [Google Scholar]
- 39.D'Andrea G, D'Ambrosio RL, Di Perna P, Chetta M, Santacroce R, Brancaccio V, et al. A polymorphism in the VKORC1 gene is associated with an interindividual variability in the dose-anticoagulant effect of warfarin. Blood. 2005;105:645–649. doi: 10.1182/blood-2004-06-2111. [DOI] [PubMed] [Google Scholar]
- 40.Wadelius M, Pirmohamed M. Pharmacogenetics of warfarin: current status and future challenges. Pharmacogenomics J. 2006 doi: 10.1038/sj.tpj.6500417. [DOI] [PubMed] [Google Scholar]
- 41.Yuan HY, Chen JJ, Lee MTM, Wung JC, Chen YF, Charng MJ, et al. A novel functional VKORC1 promoter polymorphism is associated with inter-individual and inter-ethnic differences in warfarin sensitivity. Human Molecular Genetics. 2005;14:1745–1751. doi: 10.1093/hmg/ddi180. [DOI] [PubMed] [Google Scholar]
- 42.Zondervan KT, Cardon LR. The complex interplay among factors that influence allelic association. Nat Rev Genet. 2004;5:89. doi: 10.1038/nrg1270. [DOI] [PubMed] [Google Scholar]
- 43.Carlquist JF, Horne BD, Muhlestein JB, Lappe DL, Whiting BM, Kolek MJ, et al. Genotypes of the cytochrome p450 isoform, CYP2C9, and the vitamin K epoxide reductase complex subunit 1 conjointly determine stable warfarin dose: a prospective study. Journal of Thrombosis and Thrombolysis. 2006;22:191–197. doi: 10.1007/s11239-006-9030-7. [DOI] [PubMed] [Google Scholar]
- 44.Li T, Lange LA, Li X, Susswein L, Bryant B, Malone R, et al. Polymorphisms in the VKORC1 gene are strongly associated with warfarin dosage requirements in patients receiving anticoagulation. J Med Genet. 2006;43:740–744. doi: 10.1136/jmg.2005.040410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aquilante CL, Langaee TY, Lopez LM, Yarandi HN, Tromberg JS, Mohuczy D, et al. Influence of coagulation factor, vitamin K epoxide reducatse complex subunit 1, and cytochrome P450 2C9 gene polymorphisms on warfarin dose requirements. Clinical Pharmacology & Therapeutics. 2006;79:291–302. doi: 10.1016/j.clpt.2005.11.011. [DOI] [PubMed] [Google Scholar]
- 46.Wadelius M, Chen LY, Downes K, Ghori J, Hunt S, Eriksson N, et al. Common VKORC1 and GGCX polymorphisms associated with warfarin dose. Pharmacogenomics J. 2005;5:262–270. doi: 10.1038/sj.tpj.6500313. [DOI] [PubMed] [Google Scholar]
- 47.Wadelius M, Chen LY, Eriksson N, Bumpstead S, Ghori J, Wadelius C, et al. Association of warfarin dose with genes involved in its action and metabolism. Hum Genet. 2006 doi: 10.1007/s00439-006-0260-8. Ref Type: In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sconce EA, Khan TI, Wynne HA, Avery P, Monkhouse L, King BP, et al. The impact of CYP2C9 and VKORC1 genetic polymorphism and patient characteristics upon warfarin dose requirements: proposal for a new dosing regimen. Blood. 2005;106:2329–2333. doi: 10.1182/blood-2005-03-1108. [DOI] [PubMed] [Google Scholar]
- 49.Lee SC, Ng SS, Oldenburg J, Chong PY, Rost S, Guo JY, et al. Interethnic variability of warfarin maintenance requirement is explained by VKORC1 genotype in an Asian population. Clin Pharmacol Ther. 2006;79:197–205. doi: 10.1016/j.clpt.2005.11.006. [DOI] [PubMed] [Google Scholar]
- 50.Obayashi K, Nakamura K, Kawana J, Ogata H, Hanada K, Kurabayashi M, et al. VKORC1 gene variations are the major contributors of variation in warfarin dose in Japanese patients. Clin Pharmacol Ther. 2006;80:169–178. doi: 10.1016/j.clpt.2006.04.010. [DOI] [PubMed] [Google Scholar]
- 51.Kimura Rina, Miyashita Kotaro, Kokubo Yoshihiro, Akaiwa Yasuhisa, Otsubo Ryoichi, Nagatsuka Kazuyuki, et al. Genotypes of vitamin K epoxide reductase, [gamma]-glutamyl carboxylase, and cytochrome P450 2C9 as determinants of daily warfarin dose in Japanese patients. Thrombosis Research. 2007 doi: 10.1016/j.thromres.2006.09.007. Ref Type: In Press. [DOI] [PubMed] [Google Scholar]
- 52.Veenstra DL, You JH, Rieder MJ, Farin FM, Wilkerson HW, Blough DK, et al. Association of vitamin K epoxide reductase complex 1 (VKORC1) variants with warfarin dose in a Hong Kong Chinese population. Pharmacogenetics and Genomics. 2005;15:687–691. doi: 10.1097/01.fpc.0000174789.77614.68. [DOI] [PubMed] [Google Scholar]
- 53.Vecsler M, Loebstein R, Almog S, Kurnik D, Goldman B, Halkin H, et al. Combined genetic profiles of components and regulators of the vitamin K-dependent gamma-carboxylation system affect individual sensitivity to warfarin. Thromb Haemost. 2006;95:205–211. doi: 10.1160/TH05-06-0446. [DOI] [PubMed] [Google Scholar]
- 54.Takahashi H, Wilkinson GR, Nutescu EA, Morita T, Ritchie MD, Scordo MG, et al. Different contributions of polymorphisms in VKORC1 and CYP2C9 to intra- and inter-population differences in maintenance dose of warfarin in Japanese, Caucasians and African-Americans. Pharmacogenetics and Genomics. 2006;16:101–110. doi: 10.1097/01.fpc.0000184955.08453.a8. [DOI] [PubMed] [Google Scholar]
- 55.Absher RK, Moore ME, Parker MH. Patient-specific factors predictive of warfarin dosage requirements. Ann Pharmacother. 2002;36:1512–1517. doi: 10.1345/aph.1C025. [DOI] [PubMed] [Google Scholar]
- 56.Schelleman H, Chen Z, Kealey C, Whitehead AS, Christie J, Price M, et al. Warfarin response and vitamin K epoxide reductase complex 1 in African Americans and Caucasians. Clin Pharmacol Ther. 2007 doi: 10.1038/sj.clpt.6100144. Ref Type: In Press. [DOI] [PubMed] [Google Scholar]
- 57.Geisen C, Watzka M, Sittinger K, Steffens M, Daugela L, Seifried E, et al. VKORC1 haplotypes and their impact on the inter-individual and inter-ethnical variability of oral anticoagulation. Thromb Haemost. 2005;94:773–779. doi: 10.1160/TH05-04-0290. [DOI] [PubMed] [Google Scholar]
- 58.Gage BF, Eby C, Milligan PE, Banet GA, Duncan JR, McLeod HL. Use of pharmacogenetics and clinical factors to predict the maintenance dose of warfarin. Thromb Haemost. 2004;91:87–94. doi: 10.1160/TH03-06-0379. [DOI] [PubMed] [Google Scholar]
- 59.Burchard EG, Ziv E, Coyle N, Gomez SL, Tang H, Karter AJ, et al. The Importance of Race and Ethnic Background in Biomedical Research and Clinical Practice. N Engl J Med. 2003;348:1170–1175. doi: 10.1056/NEJMsb025007. [DOI] [PubMed] [Google Scholar]
- 60.Cooper RS, Kaufman JS, Ward R. Race and Genomics. N Engl J Med. 2003;348:1166–1170. doi: 10.1056/NEJMsb022863. [DOI] [PubMed] [Google Scholar]
- 61.Kahn J. Misreading race and genomics after BiDil. Nat Genet. 2005;37:655–656. doi: 10.1038/ng0705-655. [DOI] [PubMed] [Google Scholar]
- 62.Temple R, Stockbridge NL. BiDil for Heart Failure in Black Patients: The U.S. Food and Drug Administration Perspective. Ann Intern Med. 2007;146:57–62. doi: 10.7326/0003-4819-146-1-200701020-00010. [DOI] [PubMed] [Google Scholar]
- 63.Bloche MG. Race-Based Therapeutics. N Engl J Med. 2004;351:2035–2037. doi: 10.1056/NEJMp048271. [DOI] [PubMed] [Google Scholar]
- 64.Tate SK, Goldstein DB. Will tomorrow's medicines work for everyone? Nat Genet. 2004 doi: 10.1038/ng1437. [DOI] [PubMed] [Google Scholar]
- 65.Yin T, Miyata T. Warfarin dose and the parmacogenomics of CYP2C9 and VKORC1--Rationale and perspectives. Thromb Res. 2007 doi: 10.1016/j.thromres.2006.10.021. Ref Type: In Press. [DOI] [PubMed] [Google Scholar]
- 66.Kimmel SE, Christie J, Kealey C, Chen Z, Price M, Thorn CF, et al. Apolipoprotein E genotype and warfarin dosing among Caucasians and African Americans. Pharmacogenomics J. 2007 doi: 10.1038/sj.tpj.6500445. [DOI] [PubMed] [Google Scholar]
- 67.Thomas DC, Haile RW, Duggan D. Recent Developments in Genomewide Association Scans: A Workshop Summary and Review. Am J Hum Genet. 2005;77:337–345. doi: 10.1086/432962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dina C, Meyre D, Samson C, Tichet J, Marre M, Jouret B, et al. Comment on "A Common Genetic Variant Is Associated with Adult and Childhood Obesity". Science. 2007;315 doi: 10.1126/science.1129402. 187b. [DOI] [PubMed] [Google Scholar]
- 69.Ozaki K, Ohnishi Y, Iida A, Sekine A, Yamada R, Tsunoda T, et al. Functional SNPs in the lymphotoxin-[alpha] gene that are associated with susceptibility to myocardial infarction. Nat Genet. 2002;32:650–654. doi: 10.1038/ng1047. [DOI] [PubMed] [Google Scholar]
- 70.Clarke R, Xu P, Bennett D, Lewington S, Zondervan K, Parish S, et al. Lymphotoxin-alpha gene and risk of myocardial infarction in 6,928 cases and 2,712 controls in the ISIS case-control study. PLoS Genet. 2006;2:107. doi: 10.1371/journal.pgen.0020107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Herbert A, Gerry NP, McQueen MB, Heid IM, Pfeufer A, Illig T, et al. A Common Genetic Variant Is Associated with Adult and Childhood Obesity. Science. 2006;312:279–283. doi: 10.1126/science.1124779. [DOI] [PubMed] [Google Scholar]
- 72.Rosskopf D, Bornhorst A, Rimmbach C, Schwahn C, Kayser A, Kruger A, et al. Comment on "A Common Genetic Variant Is Associated with Adult and Childhood Obesity". Science. 2007;315 doi: 10.1126/science.1130571. 187d. [DOI] [PubMed] [Google Scholar]
- 73.Loos RJF, Barroso I, O'Rahilly S, Wareham NJ. Comment on "A Common Genetic Variant Is Associated with Adult and Childhood Obesity". Science. 2007;315 doi: 10.1126/science.1130012. 187c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Klein RJ, Zeiss C, Chew EY, Tsai JY, Sackler RS, Haynes C, et al. Complement factor H polymorphism in age-related macular degeneration. Science. 2005;308:385–389. doi: 10.1126/science.1109557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Duerr RH, Taylor KD, Brant SR, Rioux JD, Silverberg MS, Daly MJ, et al. A Genome-Wide Association Study Identifies IL23R as an Inflammatory Bowel Disease Gene. Science. 2006;314:1461–1463. doi: 10.1126/science.1135245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Moore JH. The ubiquitous nature of epistasis in determining susceptibility to common human diseases. Hum Hered. 2003;56:73–82. doi: 10.1159/000073735. [DOI] [PubMed] [Google Scholar]
- 77.Franco V, Polanczyk CA, Clausell N, Rohde LE. Role of dietary vitamin K intake in chronic oral anticoagulation: prospective evidence from observational and randomized protocols. The American Journal of Medicine. 2004;116:651–656. doi: 10.1016/j.amjmed.2003.12.036. [DOI] [PubMed] [Google Scholar]
- 78.Greenblatt DJ, von Moltke LL. Interaction of Warfarin With Drugs, Natural Substances, and Foods. J Clin Pharmacol. 2005;45:127–132. doi: 10.1177/0091270004271404. [DOI] [PubMed] [Google Scholar]
- 79.Musani SK, Shriner D, Liu N, Feng R, Coffey CS, Yi N, et al. Detection of gene x gene interactions in genome-wide association studies of human population data. Hum Hered. 2007;63:67–84. doi: 10.1159/000099179. [DOI] [PubMed] [Google Scholar]
- 80.Ritchie MD, Hahn LW, Roodi N, Bailey LR, Dupont WD, Parl FF, et al. Multi-factor dimensionality reduction reveals high-order interactions among estrogen-metabolism genes in sporadic breast cancer. Am J Hum Genet. 2001;69:138–147. doi: 10.1086/321276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bush WS, Dudek SM, Ritchie MD. Parallel multifactor dimensionality reduction: a tool for the large-scale analysis of gene-gene interactions. Bioinformatics. 2006;22:2173–2174. doi: 10.1093/bioinformatics/btl347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ritchie MD, White BC, Parker JS, Hahn LW, Moore JH. Optimization of neural network architecture using genetic programming improves detection and modeling of gene-gene interactions in studies of human diseases. BMC Bioinformatics. 2003;28:4. doi: 10.1186/1471-2105-4-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Motsinger AA, Lee SL, Mellick G, Ritchie MD. GPNN: power studies and applications of a neural network method for detecting gene-gene interactions in studies of human disease. BMC Bioinformatics. 2006;7:39–49. doi: 10.1186/1471-2105-7-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cordell HJ. Epistasis: what it means, what it doesn't mean, and statistical methods to detect it in humans. Human Molecular Genetics. 2002;11:2463–2468. doi: 10.1093/hmg/11.20.2463. [DOI] [PubMed] [Google Scholar]