Abstract
To increase the density of a gene map of the zebrafish, Danio rerio, we have placed 3119 expressed sequence tags (ESTs) and cDNA sequences on the LN54 radiation hybrid (RH) panel. The ESTs and genes mapped here join 748 SSLp markers and 459 previously mapped genes and ESTs, bringing the total number of markers on the LN54 RH panel to 4226. Addition of these new markers brings the total LN54 map size to 14,372 cR, with 118 kb/cR. The distribution of ESTs according to linkage groups shows relatively little variation (minimum, 73; maximum, 201). This observation, combined with a relatively uniform size for zebrafish chromosomes, as previously indicated by karyotyping, indicates that there are no especially gene-rich or gene-poor chromosomes in this species. We developed an algorithm to provide a semiautomatic method for the selection of additional framework markers for the LN54 map. This algorithm increased the total number of framework markers to 1150 and permitted the mapping of a high percentage of sequences that could not be placed on a previous version of the LN54 map. The increased concentration of expressed sequences on the LN54 map of the zebrafish genome will facilitate the molecular characterization of mutations in this species.
The zebrafish (Danio rerio) has emerged as an excellent model organism to study vertebrate biology and human diseases, largely because of the availability of a large number of mutations affecting a wide range of developmental pathways and physiological systems (Driever et al. 1996; Haffter et al. 1996; Dooley and Zon 2000). Many of the mutant phenotypes in zebrafish resemble human clinical disorders. The candidate gene approach and chromosomal walks have allowed the molecular characterization of a few dozen mutations in the zebrafish thanks, in part, to the availability of meitoic and radiation hybrid (RH) maps for this species (Knapik et al. 1998; Postlethwait et al. 1998; Gates et al. 1999; Geisler et al. 1999; Hukriede et al. 1999; Woods et al. 2000). For example, comparative genomics and RH mapping allowed the identification of β-spectrin as the gene affected in riesling mutants that suffer from hemolytic anemia (Liao et al. 2000).
Efforts to systematically identify genes in the zebrafish before the complete sequencing of its genome are under way. An expressed sequence tags (EST) project (http://www.genetics.wustl.edu/fish_lab/frank/cgi-bin/fish/) is being performed using fingerprint-normalized cDNA libraries (Clark et al. 2001). The fingerprinted libraries were made from mRNA either from embryos of various stages of development or from adult tissues such as brain, liver, kidney, retina, olfactory epithelium, fin, and fin regenerates.
In the past few years, RH mapping has been established as the most efficient method for generating large-scale genomic maps of both mammalian and nonmammalian species. The two zebrafish RH panels, LN54 and T51 (Geisler et al. 1999; Hukriede et al. 1999), constitute valuable resources to rapidly increase density of the zebrafish gene map through the placing of zebrafish ESTs. In an effort to provide a powerful and reliable tool for the molecular identification of zebrafish mutations by use of a candidate gene approach and to facilitate establishment of gene orthology relationships through conserved synteny analysis, we report an EST map of the zebrafish genome with ∼3600 ESTs, all but 459 previously uncharacterized. This brings the total number of markers on LN54 RH panel to 4226. We have also increased the number of framework markers on the LN54 map from 703 to 1150. The increase in the density of framework markers was shown to further increase the efficiency of placing markers on LN54.
RESULTS
Placement of ESTs on the LN54 RH Map
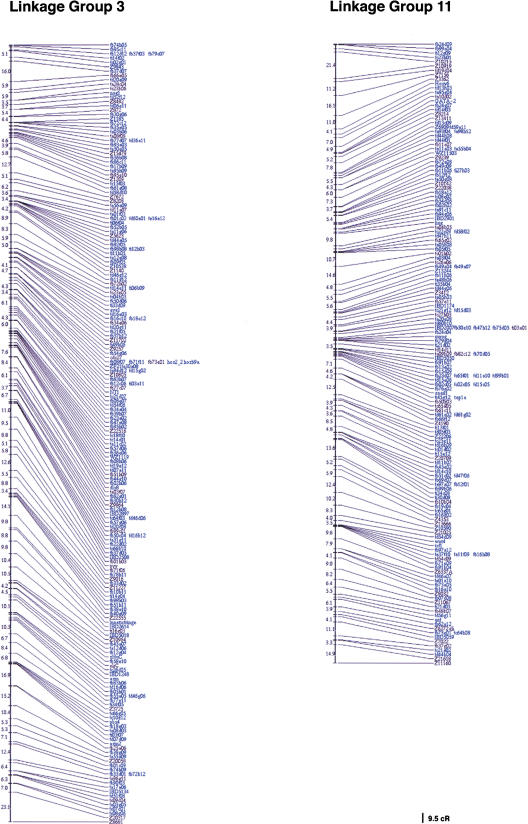
We have obtained RH mapping data for 3119 previously unmapped genes and EST sequences, bringing the total of genes and ESTs mapped on the LN54 RH panel to 3578. With a haploid genome size of 1700 Mbp, this represents 2.1 ESTs per Mb. For the mapping of ESTs, oligonucleotide primer sets for RH mapping were chosen using 3′ clusters. Information on the origin of the cDNA clones mapped in this study can be obtained by accessing the Washington University EST database (http://www.genetics.wustl.edu/fish_lab/frank/cgi-bin/fish/) or the Web site of Igor Dawid's laboratory's in situ screen (http://zf.nichd.nih.gov/pubzf; Kudoh et al. 2001). A map of two of the LGs (linkage groups) of the LN54 RH map is shown in Figure 1. The remaining LGs can be seen at http://dir.nichd.nih.gov/nichdlmg/lmgdevb.htm. The distribution of mapped genes ESTs per linkage group is shown in Table 1. The number of ESTs per linkage group varied between 73 for LG9 and 201 for LG2.
Figure 1.
Two linkage groups of the LN54 radiation hybrid (RH) map. Framework markers are shown in black; placement markers, in blue.
Table 1.
Distribution of Mapped ESTs/Genes per Linkage Group
| Linkage group | EST/genes mapped | Linkage group | EST/genes mapped |
|---|---|---|---|
| LG1 | 129 | LG14 | 116 |
| LG2 | 201 | LG15 | 115 |
| LG3 | 174 | LG16 | 160 |
| LG4 | 109 | LG17 | 127 |
| LG5 | 191 | LG18 | 105 |
| LG6 | 172 | LG19 | 157 |
| LG7 | 174 | LG20 | 86 |
| LG8 | 180 | LG21 | 139 |
| LG9 | 73 | LG22 | 142 |
| LG10 | 93 | LG23 | 194 |
| LG11 | 143 | LG24 | 116 |
| LG12 | 139 | LG25 | 99 |
| LG13 | 152 |
After integration of the above EST retention data to the overall LN54 set of RH vectors, the overall retention of LN54 is 21.7%, with a discordance of 1.4%. Thus, retention remained identical to that of the original LN54 map (22% for 1055 markers; Hukriede et al. 1999). Retention values per linkage group are given in Table 2.
Table 2.
Quantitative Characteristics of the LN54 Radiation Hybrid Maps of the 25 Zebrafish Linkage Groups
| Linkage group | % Retention | Framework markers | Length | cR/cM | |||
|---|---|---|---|---|---|---|---|
| Total | Previous | % Increase | cR | % Increase | |||
| 1 | 18.0 | 46 | 24 | 92 | 566 | 36 | 5.1 |
| 2 | 20.7 | 51 | 33 | 55 | 567 | 20 | 3.4 |
| 3 | 18.6 | 44 | 26 | 69 | 611 | 7 | 4.7 |
| 4 | 22.9 | 38 | 25 | 52 | 456 | 44 | 3.8 |
| 5 | 14.6 | 53 | 28 | 89 | 466 | 52 | 2.9 |
| 6 | 25.4 | 70 | 32 | 119 | 784 | 53 | 4.9 |
| 7 | 18.8 | 46 | 28 | 64 | 452 | 3 | 2.4 |
| 8 | 20.1 | 60 | 33 | 81 | 833 | 36 | 5.8 |
| 9 | 18.6 | 40 | 26 | 54 | 671 | 57 | 5.2 |
| 10 | 21.4 | 44 | 27 | 63 | 674 | 31 | 7.3 |
| 11 | 19.9 | 43 | 24 | 79 | 596 | 24 | 5.4 |
| 12 | 10.9 | 46 | 25 | 84 | 409 | 65 | 3.7 |
| 13 | 20.9 | 38 | 23 | 65 | 510 | 43 | 3.6 |
| 14 | 36.9 | 47 | 32 | 47 | 746 | 16 | 4.6 |
| 15 | 18.3 | 40 | 24 | 67 | 459 | 55 | 3.2 |
| 16 | 23.8 | 52 | 24 | 117 | 640 | 39 | 4.6 |
| 17 | 16.5 | 54 | 32 | 69 | 549 | 48 | 4.2 |
| 18 | 21.6 | 40 | 27 | 48 | 579 | 19 | 4.1 |
| 19 | 20.3 | 54 | 32 | 69 | 660 | 53 | 4.9 |
| 20 | 41.9 | 34 | 28 | 21 | 736 | 0 | 4.6 |
| 21 | 20.6 | 35 | 24 | 46 | 367 | 26 | 2.5 |
| 22 | 15.4 | 38 | 23 | 65 | 315 | 22 | 4.3 |
| 23 | 26.9 | 51 | 27 | 89 | 551 | 41 | 4.2 |
| 24 | 25.5 | 40 | 27 | 48 | 536 | 12 | 4.6 |
| 25 | 27.4 | 46 | 30 | 53 | 639 | 21 | 6.9 |
We compared the efficiency and reproducibility of mapping EST markers when these were assayed in only one polymerase chain reaction (PCR) assay or in duplicate PCR assays. We sampled ∼300 ESTs at random and found that for 92% of them, analysis of a single RH vector placed the EST to the same linkage group as the consensus RH vector obtained from duplicate PCR assays. Furthermore, 87% of the above markers placed to the same position. Most of those ESTs for which a single RH vector did not give the same LG placement as the consensus RH vector were cases in which one of the RH vectors produced multiple linkages. Only one EST in our sample produced placement to two distinct LGs when the individual RH vectors were compared. Duplicate mapping markedly increases the cost and diminishes the throughput of RH mapping. We found that duplicate mapping can help resolve uncertainties for those markers for which the initial RH vector yielded ambiguous results. However, mapping of single RH vectors can, in most instances, be sufficient. Similar observations were made during the building of a high-density EST map of the rat genome by RH mapping (Scheetz et al. 2001).
Selection of Additional Framework Markers for the LN54 RH Map
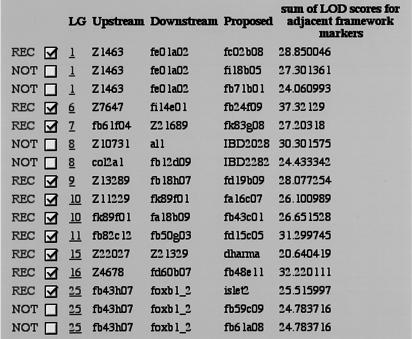
One of the challenges of growing RH maps is the need to carefully convert some placement markers into framework markers, thereby improving map resolution while maintaining its quality. Framework markers are important as they are used to establish placement of newly tested genes and sequences. Thus, the higher the density of framework markers, the more likely it will be possible to place new genes and markers on the RH map. Furthermore, a dense framework will sometimes enable the placement of genes or markers that are slightly mis-scored, for example, those caused by false negatives. Because of the volume of marker data, we have developed a software that uses RHMAPPER in a subsidiary fashion and provides a semiautomatic method of selecting candidate markers for this conversion (Hudson et al. 1995). The algorithm considers each pair of adjacent framework markers as A and B. A placement marker C is a candidate for conversion to a framework marker between A and B if it is closer to both A and B than A and B are to each other. More specifically, lod (A, C) > lod (A, B) + Δ and lod (B, C) > lod (A, B) + Δ, where in practice we choose Δ = 1, and typical good lod scores are ≥8. For each round of automated framework analysis, the software recommends against converting more than one candidate marker between an existing pair of framework markers. The software also recommends that confidential markers be excluded from the framework. Figure 2 shows a sample of the machine-generated recommendations, which are implemented automatically after user consultation.
Figure 2.
Sample output of the program for selection of framework markers. Only those candidate markers that satisfy the selection criteria (see text) are shown in the output. When more than one candidate in a given interval between two known framework markers is acceptable (e.g., LG1), the one with the highest sum of lod scores for adjacent framework markers is chosen (REC). Markers that have been submitted to the LN54 Web site in confidence (e.g., LG8) are temporarily rejected, even if they satisfy selection criteria. The program was re-run with the new frameworks in place until all possible candidates have been tested.
The number of framework markers at the time of publication of the original LN54 RH map was 684, most of them simple sequence-length polymorphic markers (Hukriede et al. 1999). We generated an additional 466 framework markers, using the software described above, bringing the total number of framework markers to 1150. The repartition of framework markers per linkage group is shown in Table 2 and varies from 34 for LG20 to 70 for LG6.
To determine how the increase in the number of framework markers improved our ability to map genes and ESTs with LN54, we tested a set of markers that had been previously submitted to the LN54 mapping Web site but had failed to map with the previous framework. We limited our analysis to markers with retention between 10% and 60%. Of 113 such markers, we find that 56 of them (49.6%) can now be placed on the map. Thus, the increase in framework markers improved the ability to map with LN54, at least for those sequences that do not show exceptionally high or low retention.
The total length of the LN54 map, following addition of the new framework markers, is 14,372 cR, for a 118 kb/cR correspondence. Thus, the addition of ∼3000 additional markers increased the total map length by 25%. There were wide variations in the percentage increase in the length of each linkage group. The length of some, like LG7 and LG20, did not change much, whereas the length of others, like LG5 and LG6, increased by ∼50% (Table 2). This variation in the length of the LGs was not related to the abundance or paucity of markers in the previous version of the map and correlated only weakly with the percentage increase in new framework markers (r = 0.62, excluding data for LG20).
To compare the RH map presented here to existing meiotic maps for the zebrafish, we calculated the cR/cM values using data from the meiotic map reported by Woods and collaborators (2000). We obtained a value of 4.43 cR/cM averaged across all linkage groups. Values ranged from 2.43 for LG7 to 7.33 for LG10, with most values (18 of 25) between 3.4 and 5.4 (Table 2).
DISCUSSION
Because a large number of ESTs from diverse sources were placed on the 25 linkage groups of the LN54 map (Table 1), we get a first indication of the gene density of each of the 25 chromosomes in zebrafish. The range of values in Table 1 (73 to 201) is small compared with the ratio observed in the human chromosome, for which there are up to eightfold differences (excluding chromosome Y) between chromosomes (Venter et al 2001). The zebrafish karyotype indicates a relatively uniform size for most of the zebrafish chromosomes (Piknacker and Ferwerda 1995; Daga et al. 1996; Gornung et al. 1997; Amores and Postlethwiat 1998). Although the 25 linkage groups of the RH and genetic maps of the zebrafish genome have yet to be assigned to specific chromosomes, the gene distribution presented here seems to indicate the zebrafish does not have chromosomes that are especially poor in genes.
Mapping of ESTs and genes has also allowed us to increase the density of framework markers on the LN54 RH map. This had a positive impact on our ability to map zebrafish genes, ESTs, and other markers with the LN54 panel. We have tried to determine whether or not we were approaching saturation in the number of framework markers. Plotting the number of framework markers selected with the algorithm presented here as a function of the total markers placed on LN54 indicates that we have not yet reached saturation in framework markers, although the curve is starting to plateau (data not shown). Therefore, further mapping of ESTs and other zebrafish sequences with the LN54 panel should lead to additional increases in the density of the framework map, thus increasing the ability to place markers on the map and increasing the confidence of marker order.
To be placed on the LN54 RH map, ESTs, cloned genes, and other markers had to map with a lod score >5 with respect to one of the framework markers, and a lod difference >3 had to be obtained between the best and the second best placement if the latter is found on a different linkage group. Using these criteria, 46 of a set of 50 cDNAs, for which the mapping was attempted recently on LN54, mapped successfully (N. Hukriede and M. Tsang, unpubl.). This 92% success rate is slightly higher than our initial value of 88%, which we achieved when the initial framework map was built (Hukriede et al. 1999). Thus, the increased density of the framework map may have resulted in a modest increase in the ability to map with the LN54 panel. When performing high-throughput mapping of ESTs, 84% of primer pairs showing retention on the LN54 panel were mapped successfully. This somewhat lower rate can be attributed to the fact that EST mapping was attempted a maximum of two times in this study, whereas the successful mapping of a specific gene or marker may require a larger number of PCR assays and adjustment of the PCR conditions.
The RH mapping of 3119 new ESTs on the LN54 panel complements the recently reported gene map based on one of the meiotic mapping panels and comprising 1503 genes and ESTs (Woods et al. 2000). Correspondence between the linkage groups of the RH and meiotic maps of the zebrafish genome has been performed (see http://zfin.org/cgi-bin/ZFIN_jump?record=JUMPTOREFCROSS). The previous version of the LN54 showed a high percentage of concordance with meiotic maps (Hukriede et al. 1999). A detailed comparison of the new EST mapping data with other gene maps of the zebrafish could not be performed because few of the ESTs mapped in the current study were mapped on the meiotic panels (Gates et al. 1999; Woods et al. 2000).
Comparative genomics using zebrafish and human gene maps has shown the existence of several blocks of synteny that existed in the common ancestor of these two species, ∼450 million years ago (Postlethwait et al. 1998, 2000; Gates et al. 1999; Barbazuk et al. 2000). Comparisons of gene sequence, function, and regulation have revealed a high degree of conservation across vertebrate species. The RH map presented here can be useful in the establishment of gene orthology relationships based on these conserved syntenies. For example, conserved synteny between a region of zebrafish LG12 and human chromosome 10p11.2 supported the presumed orthology between the recently identified zebrafish nma, which encodes a protein involved in attenuation of BMP signaling during development, and human NMA (Tsang et al. 2000).
The expansion of gene families in the genome of the zebrafish and other euteleosts compared with that of mammals (Robinson-Rechavi et al. 2001a) has led investigators to suggest the occurrence of a whole genome duplication event shortly after the divergence of teleost from lobe-finned fish (Amores et al. 1998), although the model of evolution by genome duplication versus local duplications is still debated (Hughes et al. 2001; Robinson-Rechavi et al. 2001b). As the number of genes mapped in zebrafish increases and the analysis of conserved syntenies with other vertebrate genomes is expanded, one should be able to bring support to one of the two models, although the presence of multiple local duplication events in addition to a whole genome duplication will be difficult to exclude.
Plans have been made to sequence the zebrafish genome and should be completed within the coming years. Meanwhile dense maps of the zebrafish, such as the one presented here, will be instrumental in the cloning of mutant loci by chromosomal walking or by the candidate gene approach. The availability of the RH mapping tools is particularly valuable for the analysis of data sets obtained from gene expression screens such as that of Kudoh and collaborators (2001). In this in situ hybridization-based screen, cDNAs are selected for analysis according to their embryonic expression pattern. The ability to routinely obtain map positions for many of these cDNAs adds greatly to their potential utility as markers and as candidate genes for mutations and thus constitutes an important application of the LN54 RH mapping panel.
METHODS
Selection of ESTs, Primer Design, and RH Mapping
ESTs from several zebrafish libraries (see Table 1 legend) were selected for RH mapping. EST 3′ clusters were mainly used for primer selection. In some cases, we attempted to map singleton 5′ ESTs with no corresponding 3′ EST when these were less likely to be redundant with 3′ clusters. Primer pairs were selected with the OSP program (Hillier and Green 1991). Parameters were set so primer length was between 19 and 22 nucleotides, primer-annealing temperatures were between 55°C and 65°C, and PCR products had a predicted size between 120 and 400 bp. In the event OSP failed to indicate a primer set for a particular 3′ cluster, we ran the sequence of a second EST from the same cluster through the program. When OSP designed too many pairs for a given cluster, we increased the requested annealing temperature to between 60°C and 65°C or asked for a narrower range of PCR product sizes. Using these approaches, primers could be designed for nearly every 3′ cluster. Primer pairs were used in PCR reactions as previously described (Hukriede et al. 1999). A primer set had to generate a clear amplicon from zebrafish genomic DNA but no band with genomic DNA from the mouse, the recipient species used to make the LN54 RH panel (Hukriede et al. 1999).
RH mapping was performed by PCR as previously described (Hukriede et al. 1999), and data were analyzed with RHMAPPER using the same parameters and modifications as previously (Hukriede et al. 1999).
Placement Map Construction and Selection of New Framework Markers
For placement map construction, RH vectors for ESTs are first compared with the whole genome map. If a lod placement value with ≥5 is found and a lod difference of ≥3 is obtained between the best placement and the second best placement on a different LG, the EST is placed on the LG with the best placement. Then, if the distance between the EST and the nearest framework marker is within 50 cR, the EST is added to the placement map. We find distances >50 cR allow the marker to cause an inappropriate expansion of the map and affect the correct order of markers on an LG. For each placement marker, the position on the map is based on the most likely interval compared with less likely intervals. The most likely interval is determined by whether the proximal or distal framework marker has a higher lod score with respect to the EST. To ensure that location of placement markers is as accurate as possible, we randomly performed multipoint map evaluations to confirm the positions for the placement markers. Furthermore, we compared positions of ESTs on the LN54 and the T51 RH maps for markers assessed on both RH panels.
A computer program was written to allow for selection of additional framework markers for the LN54 RH map. This program was briefly described in Results, and a more detailed description of how the data are being manipulated can be found on two flowcharts posted at http://dir.nichd.nih.gov/lmg/lmgdevb.htm. These flowcharts not only provide details of the novel approach but also show how placement maps are created, which is the step before the novel approach. The way the program is designed, a two-point analysis is used to assign a marker to a proper LG and a subsequent multipoint analysis is used to (1) assign the position on the LG, (2) evaluate the order of the marker with adjacent markers, and (3) assign the best marker in the interval between two existing framework markers to become a new framework marker.
Acknowledgments
We thank Thomas J. Hudson, the staff of the Montreal Genome Center, K. Vaillancourt, and D. Brez for technical assistance. This work was supported by grant GOP-12781 from the Canadian Institutes of Health Research (M.E. and M.C.) and by National Institutes of Health grants RO1 DK55379 (S.L.J.) and RO1 DK55381 (L.Z.). M.E. is an investigator of the Canadian Institutes of Health Research, and M.C is a “chercheur-boursier” of the Fonds de Recherches en Santé du Québec.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL mekker@ohri.ca; FAX (613) 761-5036.
Article published on-line before print: Genome Res., 10.1101/gr.210601.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.210601.
REFERENCES
- Amores A, Postlethwiat JH. Banded chromosomes and the zebrafish karyotype. In: Detrich WH, et al., editors. The zebrafish: Genetics and genomics. Vol. 60. San Diego: Academic Press; 1998. pp. 323–338. [DOI] [PubMed] [Google Scholar]
- Amores A, Force A, Yan Y-L, Joly L, Amemiya C, Fritz A, Ho RK, Langeland J, Prince V, Wang Y-L, et al. Zebrafish hox clusters and vertebrate genome evolution. Science. 1998;282:1711–1714. doi: 10.1126/science.282.5394.1711. [DOI] [PubMed] [Google Scholar]
- Barbazuk WB, Korf I, Kadavi C, Heyen J, Tate S, Wun E, Bedell JA, McPherson JD, Johnson SL. The syntenic relationship of the zebrafish and human genomes. Genome Res. 2000;10:1351–1358. doi: 10.1101/gr.144700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark MD, Henning S, Herwig S, Clifton SW, Marra MA, Lehrach H, Johnson SL. An oligonucleotide fingerprint normalized and EST characterized zebrafish cDNA library. Genome Res. 2001;11:1594–1602. doi: 10.1101/gr.186901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daga RR, Thode G, Amores A. Chromosome complement, C-banding, Ag-NOR and replication banding in the zebrafish Danio rerio. Chromosome Res. 1996;4:29–32. doi: 10.1007/BF02254941. [DOI] [PubMed] [Google Scholar]
- Dooley K, Zon L I. Zebrafish: A model system for the study of human disease. Curr Opin Genet Dev. 2000;10:252–256. doi: 10.1016/s0959-437x(00)00074-5. [DOI] [PubMed] [Google Scholar]
- Driever W, Solnica-Krezel L, Schier AF, Neuhauss SCF, Malicki J, Stemple DL, Stainier DYR, Zwartkruis F, Abdelilah S, Rangini Z, et al. A genetic screen for mutations affecting embryogenesis in zebrafish. Development. 1996;123:37–46. doi: 10.1242/dev.123.1.37. [DOI] [PubMed] [Google Scholar]
- Gates MA, Kim L, Egan ES, Cardozo T, Sirotkin HI, Dougan ST, Lashkari D, Abagyan R, Schier AF, Talbot WS. A genetic linkage map for zebrafish: Comparative analysis and localization of genes and expressed sequences. Genome Res. 1999;9:334–347. [PubMed] [Google Scholar]
- Geisler R, Rauch G-J, Baier H, van Bebber FV, Bro L, Dekens MPS, Finger K, Fricke C, Gates MA, Geiger H, et al. A radiation hybrid map of the zebrafish genome. Nat Genet. 1999;23:86–89. doi: 10.1038/12692. [DOI] [PubMed] [Google Scholar]
- Gornung E, Gabrielli I, Cataudella S, Sola L. CMA3-banding pattern and fluorescence in situ hybridization with 18S rRNA genes in zebrafish chromosomes. Chromosome Res. 1997;5:40–46. doi: 10.1023/a:1018441402370. [DOI] [PubMed] [Google Scholar]
- Haffter P, Granato M, Brand M, Mullins MC, Hammerschmidt M, Kane DA, Odenthal J, van Eeden FJM, Jianmg Y-J, Heisenberg C-P, et al. The identification of genes with unique and essential functions in the development of the zebrafish, Danio rerio. Development. 1996;123:1–36. doi: 10.1242/dev.123.1.1. [DOI] [PubMed] [Google Scholar]
- Hillier L, Green P. OSP: A computer program for choosing PCR and DNA sequencing primers. PCR Methods Appl. 1991;1:124–128. doi: 10.1101/gr.1.2.124. [DOI] [PubMed] [Google Scholar]
- Hudson TJ, Stein LD, Gerety SS, Ma J, Castle AB, Silva J, Slonim DK, Baptista R, Kruglyak L, Xu SH, et al. An STS-based map of the human genome. Science. 1995;270:1945–1954. doi: 10.1126/science.270.5244.1945. [DOI] [PubMed] [Google Scholar]
- Hughes AL, da Silva J, Friedman R. Ancient genome duplications did not structure the human Hox-bearing chromosomes. Genome Res. 2001;11:771–780. doi: 10.1101/gr.160001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hukriede NA, Joly L, Tsang M, Miles J, Tellis P, Epstein JA, Barbazuk WB, Li FN, Paw B, Postlethwiat JH, et al. Radiation hybrid mapping of the zebrafish genome. Proc Natl Acad Sci. 1999;96:9745–9750. doi: 10.1073/pnas.96.17.9745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapik EW, Goodman A, Ekker M, Chevrette M, Delgado J, Neuhaus S, Shimoda N, Driever W, Fishman MC, Jacob HJ. A microsatellite genetic linkage map for the zebrafish (Danio rerio) Nat Genet. 1998;18:338–343. doi: 10.1038/ng0498-338. [DOI] [PubMed] [Google Scholar]
- Kudoh T, Tsang M, Hukriede NA, Chen J, Dedekian M, Clarker CJ, Kiang A, Schultz S, Epstein JA, Toyama R, et al. A gene expression screen in zebrafish embryogenesis. Genome Res. 2001;11:1979–1987. doi: 10.1101/gr.209601. [DOI] [PubMed] [Google Scholar]
- Liao EC, Paw BH, Peters LL, Zapata A, Pratt SJ, Do CP, Lieschke G, Zon LI. Hereditary spherocytosis in zebrafish riesling illustrates evolution of erythroid β-spectrin structure, and function in red cell morphogenesis and membrane stability. Development. 2000;127:5123–5132. doi: 10.1242/dev.127.23.5123. [DOI] [PubMed] [Google Scholar]
- Piknacker LP, Ferwerda MA. Zebrafish chromosome banding. Genome. 1995;38:1052–1055. doi: 10.1139/g95-140. [DOI] [PubMed] [Google Scholar]
- Postlethwait JH, Yan Y-L, Gates MA, Horne S, Ekker M, Amores A, Brownlie A, Donovan A, Egan ES, Force A, et al. Vertebrate genome evolution and the zebrafish gene map. Nat Genet. 1998;18:345–349. doi: 10.1038/ng0498-345. [DOI] [PubMed] [Google Scholar]
- Postlethwait JH, Woods IG, Ngo-Hazelett P, Yan Y-L, Kelly PD, Chu F, Huang H, Hill-Force A, Talbot WS. Zebrafish comparative genomics and the origins of vertebrate chromosomes. Genome Res. 2000;10:1890–1902. doi: 10.1101/gr.164800. [DOI] [PubMed] [Google Scholar]
- Robinson-Rechavi M, Marchand O, Escriva H, Bardet P-L, Zelus D, Hughes S, Laudet V. Euteleost fish genomes are characterized by expansion of gene families. Genome Res. 2001a;11:781–788. doi: 10.1101/gr.165601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson-Rechavi M, Marchand O, Escriva H, Laudet V. An ancestral whole-genome duplication may not have been responsible for the abundance of duplicated fish genes. Curr Biol. 2001b;11:R458–R459. doi: 10.1016/s0960-9822(01)00280-9. [DOI] [PubMed] [Google Scholar]
- Scheetz TE, Raymond MR, Nishimura DY, McClain A, Roberts C, Birkett C, Gardiner J, Zhang J, Butters N, Sun C, et al. Generation of a high-density rat EST map. Genome Res. 2001;11:497–502. doi: 10.1101/gr.151601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang M, Kim R, de Caestecker MP, Kudoh T, Robert AB, Dawid IG. Zebrafish nma is inbolved in TGFb family signaling. Genesis. 2000;28:47–57. doi: 10.1002/1526-968x(200010)28:2<47::aid-gene20>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- Woods IG, Kelly PD, Chu F, Ngo-Hazelett P, Yan Y-L, Huang H, Postlethwait JH, Talbot WS. A comparative gene map of the zebrafish genome. Genome Res. 2000;10:1903–1914. doi: 10.1101/gr.10.12.1903. [DOI] [PMC free article] [PubMed] [Google Scholar]