Abstract
The Dlk1–Gtl2 domain on mouse chromosome 12 contains reciprocally imprinted genes with the potential to contribute to our understanding of common features involved in imprinting control. We have sequenced this conserved region in the mouse and sheep and included the human sequence in a three species comparison. This analysis resulted in a precise conservation map and identification of highly conserved sequence elements, some of which we have shown previously to be differentially methylated in the mouse. Additionally, this analysis facilitated identification of a CpG-rich tandem repeat array located ∼13–15 kb upstream of Gtl2. Furthermore, we have identified a third imprinted transcript that overlaps with the last Dlk1 exon in the mouse. This transcript lacks a conserved open reading frame and is probably generated by cleavage of extended Dlk1 transcripts. Because Dlk1 and Gtl2 share many of the imprinting properties of the well-characterized Igf2–H19 domain, it has been proposed that the two regions may be regulated in the same way. Comparative genomic examination of the two domains indicates that although there are similarities, other features are very different, including the location of conserved CTCF-binding sites, and the level of conservation at regulatory regions.
[The sequence data described in this paper have been submitted to the GenBank data library under accession no. AJ320506.]
To date, >40 imprinted genes, characterized by preferential expression from only one of their parental alleles, have been identified in human and mouse. Although regulation of imprinted gene expression is under intensive investigation, little is known about common elements that may be responsible for the parental origin-specific silencing of one allele. During development, the imprint that differentially marks the parental alleles is likely to be set in the germline and after fertilization is stably transmitted during somatic cell division. For that reason, the regulation of imprinting involves heritable epigenetic modifications that affect chromatin structure and the ability of the DNA to interact with regulatory factors. DNA methylation is one modification known to have a key role in the regulation of imprinted genes and differential modifications to chromatin-associated proteins may also be involved. So far, however, it is not known whether there are common genomic features that distinguish imprinted domains from the majority of other genes that are expressed from both parental alleles. One approach to address this issue is to look for genomic features common to imprinted domains within species and to conduct comparative genomic analysis of imprinted regions between species. This approach becomes more feasible as more imprinted domains are being cloned and characterized and more mammalian genomic sequence is being generated.
Two pairs of reciprocally imprinted genes in the mouse, Dlk1–Gtl2 on distal chromosome 12 (Schmidt et al. 2000; Takada et al. 2000) and Igf2–H19 on distal chromosome 7, share a number of intriguing features. Dlk1 and Igf2 are both paternally expressed, whereas Gtl2 and H19 are maternally expressed and appear to encode untranslated RNAs. Both pairs of genes are located ∼80 kb apart and share similar patterns of differential DNA methylation. In general, there is some evidence that Dlk1 and Gtl2 are co-expressed in the same tissues during development as are Igf2 and H19 (Takada et al. 2000). Furthermore, both pairs of genes exhibit the same reciprocal behavior in Dnmt1−/− mice (Schmidt et al. 2000).
There is compelling evidence that Dlk1–Gtl2 and Igf2–H19 are both involved in the regulation of prenatal growth. For Igf2–H19, this has been documented extensively (DeChiara et al. 1991; Ferguson-Smith et al. 1991; Leighton et al. 1995; Eggenschwiler et al. 1997; Sun et al. 1997). Evidence for the involvement of Dlk1–Gtl2 in growth regulation is derived from different types of imprinting anomalies in mouse, sheep, and human. Mouse embryos harboring maternal or paternal uniparental disomy for chromosome 12 have growth defects and are inviable. (Georgiades et al. 2000, 2001). In the sheep, the Callipyge phenotype is a muscular hypertrophy that is subject to a parent-of-origin effect. The phenotype is only present in individuals that have a single mutated allele of the Callipyge (Clpg) locus inherited from the father. The Clpg locus has been mapped to a 400-kb long interval that includes Dlk1 and Gtl2 on ovine chromosome 18 (Berghmans et al. 2000). The exact nature of the mutation is not yet known. Further evidence for a role of this locus in growth regulation is derived from transgenic mice that carry a lacZ insertion in the upstream region of Gtl2 (Schuster-Gossler et al. 1998). When the lacZ transgene is paternally inherited, the mice are growth retarded (Schuster-Gossler et al. 1996). The lacZ integration located in the intergenic Dlk1–Gtl2 region, indicates the presence either of a third growth-regulating gene or of an important regulatory element whose function is disturbed by the lacZ integration on the paternal allele. In the human, DLK1 and GTL2 are located on chromosome 14q and have also been shown to be imprinted (Wylie et al. 2000). In agreement with the observed deregulation of growth in the described animal models, patients with maternal uniparental disomy for chromosome 14q exhibit growth retardation (Georgiades et al. 1998; Sutton and Schaffer 2000).
Here we have sequenced 112 kb encompassing the mouse Dlk1–Gtl2 domain and conducted a three species comparison of the same regions in sheep and human with the aim of identifying conserved genomic features that may be functionally important in imprinting control. In addition, we used this information to compare the Dlk1–Gtl2 domain with the well characterized Igf2–H19 domain. This study indicates that although the two regions have similarities, there are also striking differences in their genomic properties. This analysis provides further insight into our understanding of the genomics of imprinting.
RESULTS
The Dlk1–Gtl2 Regions Are Highly Conserved in Human, Mouse, and Sheep
Using a Gtl2 cDNA probe, genomic clones were isolated from a genomic bacterial artificial chromosome (BAC) library derived from the mouse strain 129/SvJ. One of these clones, Bac 103N10, harbored Gtl2 as well as Dlk1 and was therefore chosen for sequence analysis (see Methods). The obtained sequence (GenBank accession no. AJ320506) is 111805 bp long and can be regarded as high-quality sequence; on average, each nucleotide is covered by nine sequence reads, and the error rate is estimated as <0.005%.
An initial analysis of the new mouse sequence revealed that the entire Dlk1 gene and also 6.5 kb of upstream region was covered by the genomic mouse sequence, whereas the Gtl2 gene was missing the last two 3′ exons (exon 9 and exon 10). Therefore, the comparison between the mouse and human sequences encompasses the entire Dlk1 gene and terminates in intron 8 of mouse Gtl2. Both genes are separated by an 80-kb-long intergenic region. The homologous human and sheep genomic sequences are 124 kb and 110 kb long (GenBank accession no. AL117190, AL132711, AF354168), see Methods. The organization of the studied regions in the three species is shown in Figure 1.
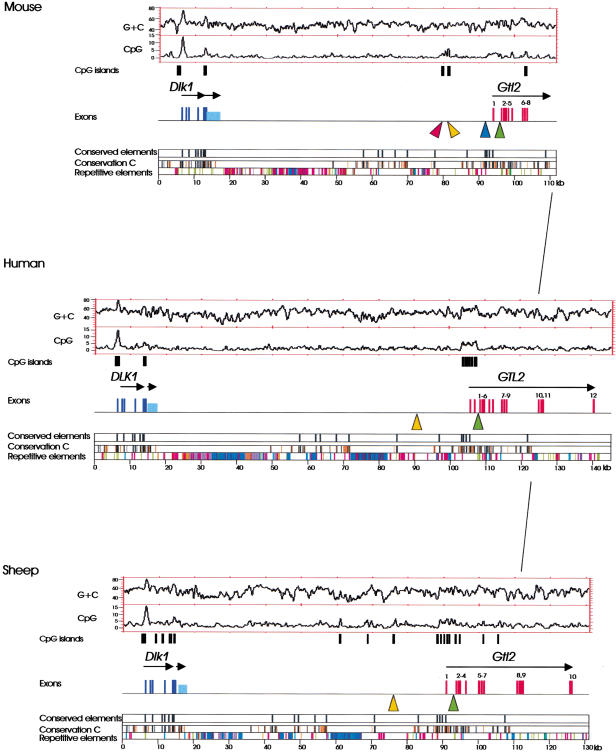
Figure 1.
Conserved organization of the Dlk1–Gtl2 regions in mouse, human, and sheep. Above colored panels that describe sequence conservation and distribution of repetitive elements in the analyzed sequences, the structures of Dlk1 (dark blue), the transcript downstream of Dlk1 (light blue), and Gtl2 (red) are indicated by bars that represent exons. Arrows above the depicted exons indicate the orientation of gene transcription. (Conservation C) Conserved elements that are present in the mouse–human, sheep–human, mouse–sheep alignments. Elements that are identical in at least 40% of all positions (>40 bp in length, aligned without gap) are shown in orange, elements that are identical in at least 70% of all positions are shown in black (>40 bp, aligned without gap). (Conserved elements) Elements that are aligned without gaps, identical in at least 70% of all positions, and at least 100 bp long. (Repetitive elements) LINE elements (blue); SINE elements (red); LTR elements (purple); DNA elements (orange); small RNAs (grey); satellites, simple repeats, low complexity DNA (green). (Orange triangles) Conserved CpG-rich repeats; (red triangle) nonconserved CpG-rich repeat; (blue triangle) lacZ integration site; (green triangles) conserved putative CTCF-binding sites. The overall G+C and CpG densities (window size 500 bp, values given as percentages) are presented as plots. CpG islands identified by the CpG plot software (http://www.ebi.ac.uk/index.html) are shown as black bars below the plots.
As suitable computer software for one multiple alignment that encompasses all three sequences is not freely available, we developed a new approach making use of the existing software. The genomic sequences were pairwise aligned using PipMaker software (Schwartz et al. 2000), generating three alignment pairs: Human–mouse, human–sheep and mouse–sheep. Interestingly, the closer phylogenetic relationship between mouse and human rather than human and sheep (Madsen et al. 2001; Murphy et al. 2001) is not reflected in the similarities of the sequences analyzed here. The average similarity in the local alignments is 58.9% identity in the human–sheep comparison, and 56.4% identity in the human–mouse comparison. Whereas 104,046 bp of the human sequence are spanned by local alignments in the human–sheep comparison, in the human–mouse alignment, the coverage is lower (65,685 bp). One reason for that might be a faster evolution of the mouse genome.
In all three sequences, the stretches of homologies were mainly interrupted by blocks of repetitive elements (Fig. 1), indicating that during evolution no significant insertions/deletions of unique sequences, for example of entire genes, have occurred. The content of repetitive elements is highest in the human and is the major cause for the expansion of the human genomic sequence. Furthermore, the positions of the two most prominent clusters of repetitive elements are conserved, located ∼5–8 kb downstream of Dlk1, and ∼20 kb upstream of Gtl2 (Fig. 1).
Conservation of the Dlk1 and Gtl2 Genes
As expected for a protein-encoding gene, similarities of the Dlk1 genomic sequences were most pronounced in the exons of this gene. The human and mouse cDNA sequences (GenBank accession no. U15979, U15980) are identical in 84.81% of all positions (84.9% of amino acids). The exons of the sheep Dlk1 gene are identical to the human cDNA in 87.11% of all positions, and to the mouse cDNA sequence in 81.47% of all positions (82.0% and 80.8%, respectively, of amino acids). In contrast, the Gtl2 genes show a general conservation in their physical organization (Fig. 1) but are less conserved at the sequence level. Taking the originally identified mouse cDNA sequence (GenBank accession no. Y13832) (Schuster-Gossler et al. 1998) as a reference sequence, the homologous human and sheep cDNA sequences are identical in 72.99% and 71.26%, respectively, of all positions. For mouse Gtl2, 10 exons have been identified (Miyoshi et al. 2000). The human and sheep Gtl2 genes encompass 12 and 10 exons, respectively (Charlier et al. 2001). Homologs for human exons 2 and 6 have not been identified in sheep and mouse. The human GTL2 cDNA sequence described previously by Miyoshi et al. (2000) commences in exon 1 and is conserved in the three species. The human exon 1 shows sequence similarity to the mouse and sheep exon 1 (74.47% and 81.25% identity, respectively). This exon 1 is also confirmed by various expressed sequence tags (ESTs). A previous report (Wylie et al. 2000) appears to misplace the start of transcription at human exon 4.
A Conserved Imprinted Transcript Downstream of Dlk1
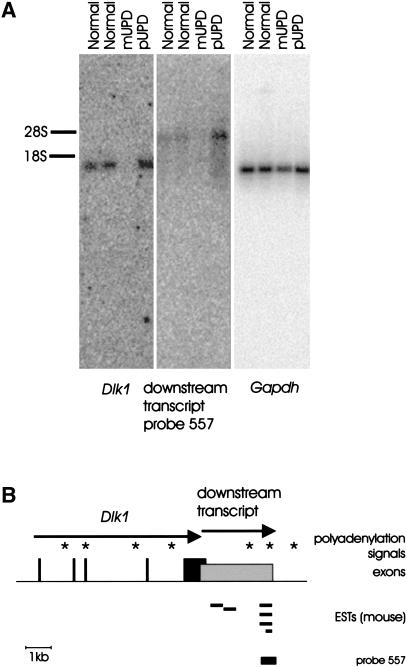
In an early approach for the isolation of probes that cover potential CpG islands, HpaII fragments derived from Bac103N10 DNA were randomly subcloned and sequenced. A 557-bp-long HpaII fragment showed homology to four mouse sequences (GenBank accession no. AA437756, AW60763, AI551552, AW120464) in the EST section of the GenBank database. The HpaII fragment was mapped to a position 2.2 kb downstream of mouse Dlk1. The presence of a polyadenylation signal 14 bp upstream of the poly A+ tail at the 3′ end of the ESTs indicated that these sequences were derived from mRNAs and that the orientation of transcription is the same as for Dlk1 and Gtl2. Northern blot hybridization to poly A+ RNA identified a transcript ∼2.5–3 kb in size (Fig. 2). This transcript was present in pUPD12 embryos and placentae but absent in mUPD12 mRNA, indicating expression solely from the paternal allele. Because the hybridization signal did not colocalize with signals obtained with probes that were specific for exon 1, 2, or 5 of Dlk1, we initially assumed that this transcript is not Dlk1.
Figure 2.
Imprinted expression and physical structure of a transcript downstream of Dlk1 in mouse. (A) Northern blot hybridization of enriched poly A+ RNA hybridized with probes specific for Dlk1, the transcript downstream of Dlk1, and Gapdh, respectively. The probe specific for Dlk1 spanned exons 1 and 2, the expression of the Dlk1 downstream transcript was tested using a genomic HpaII fragment overlapping with the 3– end of the transcript. Hybridization of Gapdh transcripts proved similar amounts of RNAs in all lanes. The positions of the 18S and 28S rRNA bands are indicated. (B) Dlk1 exons are shown as black bars. The Dlk1 downstream transcript overlaps with the last exon of Dlk and is shown as a grey box, starting at the most upstream 5′ end predicted by the 5′RACE experiment. Mouse ESTs that indicated the presence of a transcript downstream of Dlk1 are shown as horizontal bars. Asterisks represent polyadenylation signals. The position of the HpaII fragment used as a probe for the detection of transcripts downstream of Dlk1 is indicated. The schematic is drawn to scale.
To reconstruct the physical organization of the new transcript, RT-PCRs were performed. In 5' RACE experiments, three different-sized products were amplified. The longest product placed the assumed 5′ end of the transcript in the last exon of Dlk1, 217 bp upstream of the Dlk1 poly A+ tail (Fig. 2). From these analyses, we deduced a 2933-bp-long cDNA sequence (nucleotides 13452–16384, GenBank accession no. AJ320506) from the genomic sequence, consistent with the transcript size on Northern blots. The successful amplification of RT-PCR products using 5′ primers specific for all Dlk1 exons and 3′ primers specific for the downstream transcript, indicated the existence of spliced transcripts that cover both Dlk1 and the expressed region downstream of Dlk1. A probe for the entire Dlk1 exon 5, however, did not detect the downstream transcript on Northern blots, indicating that transcripts consisting of this small portion of Dlk1 extending into the downstream region may be less abundant. We suggest that the downstream transcript may be a cleavage product derived from extended Dlk1 transcripts. This may be similar to the post-transcriptional processing of IGF2 RNAs in human (Scheper et al. 1995).
Similar transcripts exist in the Dlk1 downstream regions of human and sheep (Charlier et al. 2001). Sequencing of the inserts of two human IMAGE cDNA clones (IMAGE ID 1753255, 4345285) enabled us to reconstruct a human 2892-bp-long cDNA sequence that starts 200 bp downstream of DLK1 (nucleotides 149945–152837, GenBank accession no. AL132711.4). We assume that the 5′ end of the deduced cDNA sequence is incomplete and that the 5′ end may be in the last exon of DLK1, similar to the situation in the mouse. Searches on potential protein-encoding open reading frames (ORFs) in the human and mouse cDNA sequences and also in the homologous genomic sheep sequence revealed the absence of a conserved ORF.
Apart from the Dlk1 downstream transcript, there is no strong evidence from sequence analyses for additional genes in the intergenic region between Dlk1 and Gtl2.
Identification of 20 Highly Conserved Elements Shared by Mouse, Human, and Sheep
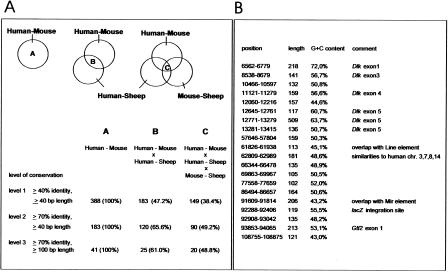
To identify conserved elements in this region, the alignments of the three sequence pairs, human–mouse, human–sheep, mouse–sheep were compared. Three hundred eighty-eight sequence elements that were aligned without any gaps, were at least 40 bp long and showing sequence conservation of at least 40% identity in the mouse–human alignment, were selected for further analysis. The developed scheme, shown in Figure 3, involves a progressive increase in the stringency of conservation, and conserved elements were identified that are present in all three alignments. The 149 elements that show at least 40% identity (>40 bp length) in all three alignments were used to generate a general picture of the sequence conservation in the Dlk1–Gtl2 region (Conservation C, Fig. 1).
Figure 3.
Identification of highly conserved elements. (A) The strategy for the identification of conserved elements is described by the scheme above the table. The table shows the numbers of conserved elements identified on different levels of sequence conservation. The inclusion of the human–sheep, and mouse–sheep alignments results in a reduction in numbers of identified elements. This reduction is described by the proportions of identified elements related to the values given for the human–mouse alignment only. (B) Positions of the 20 highly conserved elements (>100 bp, >70% identity) in the mouse sequence (GenBank accession no. AJ320506) that were identified in all three alignments. Additionally, the lengths, G+C content, and special features of the individual elements are shown.
Twenty identified elements of at least 100 bp in length were aligned without any gaps and were identical in at least 70% of all positions in all three alignments (Fig. 3B). The reliability of the alignment and selection procedure was proven using a different software (http://www-gsd.lbl.gov/vista/) for the alignments. This placed all but two elements (nucleotides 69863–69967 and 77558–77659, GenBank accession no. AJ320506) in regions that were highly conserved in all three species (data not shown). These two elements were present in two of the three species.
Six of these 20 elements represented exons of Dlk1, two overlapping with the differentially methylated region in intron 4/exon 5 (Takada et al. 2000) (Fig. 3B; conserved elements in Fig. 1). In contrast, among the Gtl2 exons, only exon 1, which is also embedded in a differentially methylated region, overlaps with a highly conserved element. Between the three species, the transcript downstream of Dlk1 exhibited similar lack of conservation as the Gtl2 exons downstream of exon 1. Ten highly conserved elements are present in the intergenic region between Dlk1 and Gtl2. Three of these elements are clustered in a region up to 2.5 kb upstream of the first Gtl2 exon, whereas highly conserved elements are not present immediately upstream of Dlk1. Precise localization of the 3′ sequence of the lacZ integration site described by Schuster-Gossler and colleagues (Schuster-Gossler et al. 1998) (see Introduction) localized the 3′ breakpoint of the integration within one of the conserved elements 1.7 kb upstream of the first Gtl2 exon. The consequences of this insertion for local gene regulation remain to be determined.
Two elements showed similarity with highly repetitive elements, one overlaps with a LINE element, the second with a Mir element. A third element appeared to be a slightly repetitive element, showing sequence homologies to genomic sequences on human chromosomes 3, 7, 8, and a second locus on human chromosome 14.
CpG-Rich Repeats Upstream of Gtl2
CpG islands that are important for the regulation of imprinted gene expression are expected to be conserved in mouse, human, and sheep. The G+C and CpG distributions in the analyzed sequences are shown in Figure 1. The overall regional G+C contents (49.37% in mouse, 51.37% in human, 53.70% in sheep) differ slightly and might reflect species-specific genome-wide differences in the G+C content (Gautier 2000).
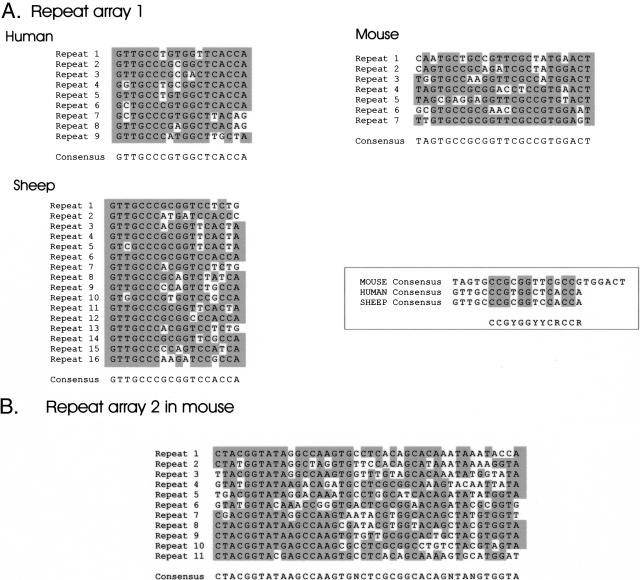
In this region, the average CpG content is 1.52% in mouse, 2.09% in human, and 2.81% in sheep. The average CpG/GC ratios are 0.28 in mouse, 0.33 in human, and 0.44 in sheep. These differences are also reflected in the number and distribution of CpG islands. Whereas the mouse sequence has five CpG islands (CpG/GC ratio > 0.6, length > 200 bp, G+C content > 50%) (http://www.ebi.ac.uk/index.html), the human sequence has eight, and the sheep sequence has 18 CpG islands (Fig. 1). All three species possess a strong CpG island in the promoter region of Dlk1 and a less pronounced CpG island in Dlk1 exon 5. CpG islands were identified in human and sheep at the transcriptional start site of Gtl2. In the mouse, this region can be regarded as a CpG-rich region, but is by definition not a CpG island. In the mouse, additional CpG islands were identified 12.3 and 14.1 kb upstream of Gtl2 exon 1 (nucleotides 81341–81686 and 79721–79937, GenBank accession no. AJ320506). A CpG island in a similar position is present in the sheep but is absent in the human. Absence of sequence homology in the alignment pairs showed that this region is not conserved in all three species. More detailed analysis of this region, however, revealed the presence of direct repeats in head to tail order in all three species in positions overlapping with the CpG island 12.3 kb upstream of Gtl2 in mouse and the CpG island in sheep (orange triangles in Fig. 1). In the mouse, the region between nucleotides 81291–81504 spans seven 24-bp-long repeated motifs (Figs. 1 and 4). In sheep and human, the repeat motifs are 18 bp long and are repeated 16 and nine times, respectively. The similarity of these motifs in human and sheep indicates that both arrays have the same phylogenetic origin (Fig. 4). In all three species, the repeats contain numerous CpG dinucleotides. The reduced length of this structure in the human compared with the sheep and the fact that in the human motif one CpG is replaced by a TpG are the reasons why pronounced CpG richness is not visible in the human CpG plot (Fig. 1). Interestingly, the central part of the mouse motif shows some similarities to the sheep and human motifs (Fig. 4), indicating that all three motifs may be derived from the same ancestral motif.
Figure 4.
Repeated sequence motifs in the Gtl2 upstream region in mouse, human, and sheep. (A) Repeat array 1 is located 12.5–15 kb upstream of Gtl2 in mouse, human, and sheep. Shown are the repeated motifs and the derived consensus sequences. Positions that fit the consensus sequences are shown on a grey background. The similarity of the mouse, human, and sheep consensus sequences is shown in the box. (B) Repeat array 2 in the mouse is located 13.3 kb upstream of Gtl2 in the mouse. Positions that are identical to the consensus sequence are shaded.
In the mouse, a second CpG-rich repeat array is present 590 bp upstream of the first repeat array at nucleotides 80151–80701 (GenBank accession no. AJ320506). This array encompasses 11 42-bp-long motifs (Fig. 4B). A similar array is not present in human and sheep.
Comparison of the Dlk1–Gtl2 and Igf2–H19 Loci
Dlk1–Gtl2 and Igf2–H19 share similarities in their reciprocal imprinting, aspects of their regulation, and their patterns of differential methylation. There has been speculation that the two domains may have common imprinting control elements (Schmidt et al. 2000; Takada et al. 2000; Wylie et al. 2000). Initial BLAST and FASTA searches for similarities to known regulatory elements in the Igf2–H19 region, such as the enhancer elements downstream of H19 and a muscle-specific repressor element 40 kb downstream of Igf2 (Ainscough et al. 2000), were unfruitful for the available sequence. Furthermore, searches using the sequences of the 20 conserved elements from the Dlk1–Gtl2 region did not reveal any similarities to the Igf2–H19 region. We then compared the Dlk1–Gtl2 and Igf2–H19 regions on the basis of features including the distribution of repetitive elements, G + C content, and the distribution of CpG islands. For this we selected the genomic sequences of the human and mouse Igf2 and H19 regions (Onyango et al. 2000), encompassing the entire Igf2 and H19 genes and 2 and 8 kb, respectively, of the Igf2 upstream regions, and in both cases, 11 kb of the H19 downstream regions. The analyzed human sequence is 138 kb long, the mouse sequence spans 101 kb.
For Igf2 and H19, the G + C content is 51.37% in the mouse, 59.50% in the human, and is higher than in the Dlk1–Gtl2 region (49.37% in the mouse, 51.37% in the human). Like the Dlk1-related CpG islands, the CpG islands at the Igf2 transcription start sites are the most pronounced.
Repetitive Elements in the Dlk1–Gtl2 and Igf2–H19 Regions
It has been proposed that LINE1 elements might have a role in X inactivation (Lyon 1998; Smit 1999; Bailey et al. 2000). To address whether this might be also the case for imprinted domains, we have analyzed the content of repetitive elements in the Dlk1–Gtl2 and also in the Igf2–H19 regions. In contrast to repetitive elements in mouse and human, little is known about the properties of these elements in the sheep genome. We therefore focused on the repetitive elements in the human and mouse sequences of both domains (Table 1).
Table 1.
Contents of Interspersed Repeats
| G+C content | Sine | Line | Line1 | LTR elements | DNA transposon-like elements | Unclassified elements | Total interspersed repeats | |
|---|---|---|---|---|---|---|---|---|
| mouse | ||||||||
| Dlk1-Gtl2 | 49.4% | 9.6% | 3.2% | 3.0% | 1.6% | 0.3% | 0.0% | 14.7% |
| lgf2-H19 | 51.4% | 1.8% | 0.9% | 0.7% | 2.0% | 0.1% | 0.0% | 4.8% |
| genome | 46–50% | 14.4% | n.d. | 3.4% | 9.9% | n.d. | n.d | 28.9% |
| 50–54% | 16.5% | n.d. | 1.4% | 3.6% | n.d. | n.d. | 20.3% | |
| human | ||||||||
| DLK1-GTL2 | 51.4% | 13.5% | 12.6% | 11.6% | 7.4% | 2.3% | 0.0% | 35.8% |
| lGF2-H19 | 59.5% | 3.6% | 4.4% | 3.4% | 3.1% | 1.2% | 0.0% | 12.3% |
| autosomes | 50–54% | 25.5%* | n.d. | 4.8% | 4.7% | 1.8% | n.d. | 41% |
| >54% | 20.1%* | n.d. | 3.0% | 2.9% | 1.2% | n.d. | 31% | |
| X chromosome | 50–54% | 24.9%* | n.d. | 6.1% | 2.7% | 1.1% | n.d. | 38% |
| >54% | 14.8%* | n.d. | 5.3% | 2.8% | 1.0% | n.d. | 26% | |
| sheep | ||||||||
| Dlk1-Gtl2 | 53.7% | 5.5% | 12.0% | 20.5% | 20.5% | 20.5% | 20.5% | 20.5% |
Shown are summaries for the interspersed repeats that were identified using RepeatMasker (http://ftp.genome.washington.edu/index.html) for the analyzed sequences. The average values for human autosomal and X chromosome linked sequences and mouse genome-wide sequences were taken from Smit (1999). Separate values for mouse autosomes and X chromosome were not available. Because the content of interspersed repeats is related to the G+C content, the values are given for sequences that show G+C contents similar to the imprinted regions analyzed here.
Sum of the values for Alu and Mir elements; n.d., not defined.
In general, the overall content of interspersed repeats (IR) is higher in the Dlk1–Gtl2 region than in the Igf2–H19 region in both species (Table 1). In both regions, however, the IR content is lower than the published average values for mouse and human sequences with similar G + C content (Smit 1999). A consistent enrichment of LINE1 elements in the analyzed imprinted domains compared with the published average values for autosomal sequences was not observed. In contrast to other subclasses of repetitive elements, a low proportion of SINE elements seems to be persistently related to the relatively low IR content in the Dlk1–Gtl2 and Igf2–H19 regions.
Conserved Putative CTCF-Binding Sites in the H19 and Gtl2 Regions Are Not at Corresponding Positions
CTCF-binding sites in the upstream region of H19 in mouse and human contribute to the function of this region as methylation-sensitive insulator elements by affecting interactions between Igf2 and the shared enhancers downstream of H19 on the maternal allele (Bell and Felsenfeld 2000; Hark et al. 2000; Kanduri et al. 2000; Szabo et al. 2000). We looked for conserved putative CTCF-binding sites in the Dlk1–Gtl2 regions in the three species. Among several different known motifs for CTCF-binding sites to date, only one motif (consensus sequence: CCGCNNGGNGNC; Wylie et al. 2000) is accessible to CpG methylation. A number of putative CTCF sites were identified in the Dlk1–Gtl2 regions in all three species (two in mouse, five in human, 12 in sheep), but only one of these was conserved in all three species (green triangles in Fig. 1, nucleotide 96071 in GenBank accession no. AJ320506, nucleotide 68347 in GenBank accession no. AL117190.4, nucleotide 140153 in GenBank accession no. AF354168). This putative CTCF-binding site is located in a homologous position in the first Gtl2 intron in mouse and sheep, and in the second intron in the human (Fig. 1).
DISCUSSION
Whereas previous sequence comparisons in imprinted regions were restricted to the comparison of the mouse and the human sequences (Engemann et al. 2000; Paulsen et al. 2000; Okamura et al. 2000; Onyango et al. 2000), we were able to include the sequence of third mammalian species, the genomic sheep sequence, in our analyses of the Dlk1–Gtl2 region on mouse chromosome 12. Compared with the human–mouse comparison alone, a three-species comparison can result in a more precise identification of conserved regions (Dubchak et al. 2000). As our chosen selection procedure excludes gaps in the alignment of the conserved elements, the 20 elements identified should be regarded as cores of highly conserved regions rather than as isolated conserved stretches of high homology. These elements were clustered in Dlk1 and upstream of Gtl2. The inclusion of the sheep sequence also facilitated the identification of short tandem repeats 13–15 kb upstream of Gtl2 in all three species, although this region is not well conserved at the level of the DNA sequence.
We were able to identify a third transcript in the Dlk1–Gtl2 region in mouse and human. This transcript resides in the downstream region of Dlk1 and is also present in the sheep (Charlier et al. 2001). Like Dlk1, this transcript is imprinted being silent on the maternal allele. The 5′ end of this transcript is in Dlk1 exon 5, and it is likely that it represents a cleavage product of Dlk1 transcripts. We cannot exclude, however, that expression of the transcript downstream of Dlk1 is independent from Dlk1 transcription and is initiated by a so-far-unknown promoter in the last Dlk1 exon. We have no further indications for additional genes in the intergenic Dlk1–Gtl2 region. This is in contrast to the Igf2–H19 region where an additional transcript has been described (Onyango et al. 2000).
As expected for an imprinted region, the DMRs in intron 4/exon 5 of Dlk1 and at the transcriptional start site of Gtl2 are highly conserved. In addition, we identified CpG-rich short direct repeats ∼12.5 –15 kb upstream of Gtl2. The similarity of the repeat cores in mouse, human, and sheep indicates that these repeats may be derived from the same ancestral motif. This indicates that either the repeat structure or the motif itself might be important for regulation in this domain. It has been hypothesized that short tandem repeat arrays might have a function in the regulation of imprinting (Neumann et al. 1995), however, the positions of such elements in imprinted regions are rarely conserved in mouse and human (Engemann et al. 2000; Paulsen et al. 2000). Nevertheless, CpG-rich tandem repeats have been identified upstream of Magel2 in human and mouse (Boccaccio et al. 1999). Interestingly, the imprinted Impact gene in the mouse possesses a CpG island that is characterized by tandem repeats, whereas in the nonimprinted human IMPACT gene such repeats are not present (Okamura et al. 2000). Furthermore, the CTCF-binding sites upstream of H19 are arranged in a repeated structure in both, mouse and human. In the mouse, however, the CTCF-binding sites are not short direct tandem repeats, therefore it is not very likely that they are functionally the same as the described repeats upstream of Gtl2. The G-rich short tandem repeats upstream of the mouse H19 gene may be similar, but their function is still unclear and an analog is absent upstream of human H19.
Because Dlk1–Gtl2 and Igf2–H19 share many imprinting properties, it has been suggested that imprinting in both regions may be regulated by common elements. Interestingly, the distribution and “shape” of CpG islands are similar in both regions: Igf2 and Dlk1 have pronounced unmethylated CpG islands in their promoter regions and additional CpG islands in their last exons that are differentially methylated in both genes (Sasaki et al. 1992; Feil et al. 1994; Takada et al. 2000). Conversely, the H19 and Gtl2 promoters are associated with “weaker” CpG islands (Sasaki et al. 1992; Ferguson-Smith et al. 1993; Takada et al. 2000; this study). Analysis of general features, however, revealed that both regions differ in the content of interspersed repeats and their G + C contents. We have identified a number of features of the Dlk1–Gtl2 region that do not have any sequence analogs in the Igf2–H19 region. This includes the different positions of conserved CTCF-binding sites, and a conserved CpG-rich repeat structure 13–15 kb upstream of Gtl2. This indicates that the regulation of imprinted gene expression may be different in both regions. Our findings do not exclude the possibility that some regulatory aspects, such as those that are required for reciprocal imprinting, are shared. It is also possible that common transcription factors are involved, but that their precise action may differ, as is indicated by the different positions of the (putative) CTCF-binding sites in H19 and Gtl2. Further analysis of the functional roles of these conserved and related features will contribute to our understanding of gene regulation at imprinted loci and the genomic evolution of imprinted domains.
METHODS
BAC Clone Isolation and DNA Sequencing
The BAC clone 103N10 was isolated from a genomic library (BAC ES (I), mouse strain 129/SvJ; Incyte Genomics Inc.) using a probe specific for exon 3 of Gtl2. Subsequently, the BAC DNA was sequenced at MWG Biotech (Milton Keynes). The assembled 213,094-bp-long sequence is covered in average by 9.06 sequence reads. The expected accuracy was estimated to be at least 99.995%. The first 100 kb of sequence belonged to a different locus indicating the BAC clone 103N10 was chimaeric. The breakpoint between both fragments was determined by sequencing a 14-kb-long BamHI fragment that contained Dlk1 and additional 7 kb of the true Dlk1 upstream region. This clone (kindly provided by Dr. J. Laborda, Universidad de Castilla-La Mancha, Albacete, Spain) was originally isolated in an independent screen from a cosmid library. The breakpoint in the chimaeric BAC sequence was localized 6576 bp upstream of the start site of transcription of Dlk1 and was chosen as the start site of the published sequence (GenBank accession no. AJ320506).
Sequences Taken from the GenBank Database
The human sequence was obtained by assembly of nucleotides 135001–184740 from GenBank accession no. AL132711.3 and nucleotides 10426–84685 from GenBank accession no. AL117190.4. Therefore, in Figure 1, nucleotide 1 in the human sequence is nucleotide 135001 (AL132711.3). The analyzed region in the sheep spans nucleotides 47001–157000 in the published genomic sequence from GenBank accession no. AF354168. Likewise, in Figure 1, nucleotide 1 in the sheep sequence is nucleotide 47001 (AF354168). The genomic mouse sequence of the Igf2 region was assembled using the genomic sequences of the Igf2 and H19 genes (nucleotides 1–27823 of GenBank accession no. U71085, nucleotides 1–19154 of GenBank accession no. AF049091) and an unfinished sequence for the intergenic region (Onyango et al. 2000) (reverse complement of nucleotides 57576–111598, downloaded from http://bio.cse.psu.edu/). The human genomic IGF2–H19 sequence was downloaded from http://bio.cse.psu.edu/ (Onyango et al. 2000) (reverse complement of nucleotides 39001–177000).
The human, mouse, and bovine Dlk1 cDNAs have the GenBank accession no. U15979, U15980, AB009278, and AF181462. The structure of the mouse Gtl2 gene was derived from alignment of the cDNA sequence (GenBank accession no. Y13182) to the genomic sequence. For the human Gtl2 gene, two different cDNA sequences have been characterized (GenBank accession no. AB032607, AF052114). The human Gtl2 exons 2, 6, 7, and 8 are represented by ESTs (GenBank accession no. AW163035, H58895, AV701976, W44755). The structure of sheep Gtl2 was established by alignment to bovine ESTs (GenBank accession no. AV594305, AV596262, BF076011, AV609668, BF601485).
Computational Characterization of the Genomic Sequences
Pairwise alignments were generated using the PipMaker software at Pennsylvania State University (Schwartz et al. 2000) (http://bio.cse.psu.edu/). The overall similarities of the sequence pairs were calculated using the obtained local alignments. The “concise” outputs contain lists of sequence matches in the analyzed sequence pairs. These lists were compared to identify conserved elements that were present in all three alignment pairs.
Interspersed repeats, small RNAs, satellites, simple repeats, and DNA elements of low complexity were detected using the RepeatMasker software at the University of Washington (http://ftp.genome.washington.edu/index.html). Additionally, tandem repeats were detected using the Compare (window size 21, stringency 14) and Dotplot programs of the Wisconsin package, version 10.0 (Genetics Computer Group).
CpG islands were identified using the CpG plot software at the European Bioinformatics Institute (http://www.ebi.ac.uk/index.html), choosing the following settings: Window size 200, step 1, Obs/Exp 0.6, MinPC 50, Length 200. CpG and G+C plots were generated using the window (window size 500, shift increment 50) and statplot programs of the Wisconsin package, version 10.0 (Genetics Computer Group). Putative CTCF-binding sites were identified using the “findpatterns” option of the Wisconsin package, version 10.0 (Genetics Computer Group).
Northern Blot Analysis
Total RNA was prepared from UPD12 embryos (eight) at 15.5 dpc according to standard protocols (Chomzcynski and Sacchi 1987). Poly A+ RNA was enriched using Oligo(dT)25 Dynabeads (Dynal Ltd.) according to the manufacturer's protocol. Separation by agarose gel electrophoresis and Northern blot transfer were performed according to standard protocols (Sambrook et al. 1989). The subsequent hybridizations were performed using the following probes. Dlk1 downstream transcript: genomic HpaII fragment 557 (nucleotides 15899–16456, GenBank accession no. AJ320506); Dlk1: 680-bp-long PstI fragment excised from IMAGE clone 604466; Gapdh: PCR product from genomic DNA (primers: 5′-ACAGTCCATGC CATCACTGCCACTC-3′, 5′-CCAGCCCCAGCATCAAAG GTGG-3′). These probes were radioactively labeled using the Megaprime DNA Labeling system (Amersham Pharmacia). The subsequent hybridization was performed according to Sambrook et al. (1989) with the following modifications: in 50% formamide, 5× SSPE, 0.5% SDS, 5% Bailey's Irish Cream Liquor, 50 μg/mL heat denatured salmon sperm DNA at 42°C overnight, the filters were subsequently washed to 65°C in 0.1× SSC, 0.1% SDS.
RT-PCRs and 5′RACE
RT-PCRs for the analysis of expression of the Dlk1 downstream transcript were performed using two different sets of primers. Set 1: 5′-GTAGTGGCTGTGTGCCAGGC-3′ and 5′-TGGCTAGGTGTTTGGGGATC-3′; set 2: 5′-CAGCCCCCAC CAAGGTTTGC -3′ and 5′-GGAAGCTAGAAAGAGCGCCC-3′ (1.5 mM MgCl2, 80 μM dNTPs, 0.03 U/μL BIOTAQTM DNA Polymerase (BioLine), 1× PCR buffer (BioLine), 60°C annealing temperature, 35 cycles). For the identification of expanded Dlk1 transcripts, the following primers were used: 5′-AACCCCCTGCGCCAACAATG-3′and 5′-GCTGGGTTAGG ACTAGGTCCCGAC-3′ (1.5 mM MgCl2, 80 μM dNTPs, 0.03 U/μL BIOTAQTM DNA Polymerase (BioLine), 1× PCR buffer (BioLine), 45 sec 95°C; 35 cycles: 30 sec 95°C, 30 sec 60°C, 3 min 72°C; 5 min 72°C). The 5′RACE PCR was performed on randomly primed cDNAs that had linkers ligated to their 5′ ends using the Marathon-Ready cDNA Kit (mouse 15.5 dpc) (Clontech) according to the manufacturer's protocol. For the nested PCR, the following specific primers were used: (1) Primer: 5′-GGTTGGAGGTGGGGGAATCTCGCC-3′; (2) Primer: 5′-GCTGGGTTAGGACTAGGTCCCG AC-3′.
Acknowledgments
We thank Maxine Tevendale and Dr. J. Laborda for providing information before publication; Helena Boixadera Espax and Takashi Sado for experimental contributions; and Prof. H. Winking for providing the homozygous breeders for the UPD12 mice. We gratefully acknowledge the sequencing team at MWG Biotech, in particular Gerald Nyakatura, for the careful subcloning and sequencing of the BAC clone, and for helpful discussions. This work was supported by the MRC.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL afsmith@mole.bio.cam.ac.uk; FAX 011-44-1223-333-786.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.206901.
REFERENCES
- Ainscough JFX, John RM, Barton SC, Surani MA. A skeletal muscle-specific mouse Igf2 repressor lies 40 kb downstream of the gene. Development. 2000;127:3923–3930. doi: 10.1242/dev.127.18.3923. [DOI] [PubMed] [Google Scholar]
- Bailey JA, Carrel L, Chakravarti A, Eichler EE. Molecular evidence for a relationship between LINE-1 elements and X chromosome inactivation: The Lyon repeat hypothesis. Proc Natl Acad Sci. 2000;97:6634–6639. doi: 10.1073/pnas.97.12.6634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell AC, Felsenfeld G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature. 2000;405:482–485. doi: 10.1038/35013100. [DOI] [PubMed] [Google Scholar]
- Berghmans S, Segers K, Shay T, Georges M, Cockett NE, Charlier C. Breakpoint mapping positions of the callipyge gene within a 450 kilobase chromosome segment containing the Gtl2 gene. Mamm Genome. 2000;12:183–185. doi: 10.1007/s003350010246. [DOI] [PubMed] [Google Scholar]
- Boccaccio I, Glatt-Deeley H, Watrin F, Roeckel N, Lalande M, Muscatelli F. The human MAGEL2 gene and its mouse homologue are paternally expressed and mapped to the Prader-Willi region. Hum Mol Genet. 1999;8:2497–2505. doi: 10.1093/hmg/8.13.2497. [DOI] [PubMed] [Google Scholar]
- Charlier C, Segers K, Karim L, Shay T, Gyapay G, Cockett N, Georges M. Human–ovine comparative sequencing of a 250 kilobase imprinted domain encompassing the clpg gene and identification of six imprinted transcripts: DLK1, GTL2, DAT, PERL, antiPERL, and MEGC. Genome Res. 2001;11:850–862. doi: 10.1101/gr.172701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. Single step method of RNA isolation by acid Guanidinium Thiocyanate-Phenol-Chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- DeChiara TM, Robertson EJ, Efstratiadis A. Parental imprinting of the mouse insulin-like growth factor II gene. Cell. 1991;64:849–859. doi: 10.1016/0092-8674(91)90513-x. [DOI] [PubMed] [Google Scholar]
- Dubchak I, Brudno M, Loots GG, Pachter L, Mayor C, Rubin EM, Frazer KA. Active conservation of noncoding sequences revealed by three-way species comparisons. Genome Res. 2000;10:1304–1306. doi: 10.1101/gr.142200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggenschwiler J, Ludwig T, Fisher P, Leighton PA, Tilghman SM, Efstratiadis A. Mouse mutant embryos overexpressing IGF-II exhibit phenotypic features of the Beckwith-Wiedemann and Simpson-Golabi-Behmel syndromes. Genes & Dev. 1997;11:3128–3142. doi: 10.1101/gad.11.23.3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engemann S, Strödicke M, Paulsen M, Franck O, Reinhardt R, Lane N, Reik W, Walter J. Sequence and functional comparison in the Beckwith-Wiedemann region: Implications for a novel imprinting centre and extended imprinting. Hum Mol Genet. 2000;9:2691–2706. doi: 10.1093/hmg/9.18.2691. [DOI] [PubMed] [Google Scholar]
- Feil R, Walter J, Allen ND, Reik W. Developmental control of allelic methylation in the imprinted mouse Igf2 and H19 genes. Development. 1994;120:2933–2943. doi: 10.1242/dev.120.10.2933. [DOI] [PubMed] [Google Scholar]
- Ferguson-Smith AC, Cattanach BM, Barton SC, Beechey CV, Surani MA. Embryological and molecular investigations of parental imprinting on mouse chromosome 7. Nature. 1991;351:667–670. doi: 10.1038/351667a0. [DOI] [PubMed] [Google Scholar]
- Ferguson-Smith AC, Sasaki H, Cattanach BM, Surani MA. Parental-origin-specific epigenetic modification of the mouse H19 gene. Nature. 1993;22:751–755. doi: 10.1038/362751a0. [DOI] [PubMed] [Google Scholar]
- Gautier C. Compositional bias in DNA. Curr Opin Genet Dev. 2000;10:656–661. doi: 10.1016/s0959-437x(00)00144-1. [DOI] [PubMed] [Google Scholar]
- Georgiades P, Chierakul C, Ferguson-Smith AC. Parental origin effects in human trisomy for chromosome 14q: Implications for genomic imprinting. J Med Genet. 1998;35:821–824. doi: 10.1136/jmg.35.10.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiades P, Watkins M, Surani MA, Ferguson-Smith AC. Parental origin-specific developmental defects in mice with uniparental disomy for chromosome 12. Development. 2000;127:4719–4728. doi: 10.1242/dev.127.21.4719. [DOI] [PubMed] [Google Scholar]
- Georgiades P, Watkins M, Burton G, Ferguson-Smith AC. Roles for genomic imprinting and the zygotic genome in placental development. Proc Natl Acad Sci. 2001;98:4522–4527. doi: 10.1073/pnas.081540898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hark AT, Schoenherr CJ, Katz DJ, Ingram RS, Levorse JM, Tilghman SM. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature. 2000;405:486–489. doi: 10.1038/35013106. [DOI] [PubMed] [Google Scholar]
- Kanduri C, Pant V, Loukinov D, Pugacheva E, Qi CF, Wolffe A, Ohlsson R, Lobanenkov VV. Functional association of CTCF with the insulator upstream of the H19 gene is parent of origin-specific and methylation-sensitive. Curr Biol. 2000;13:853–856. doi: 10.1016/s0960-9822(00)00597-2. [DOI] [PubMed] [Google Scholar]
- Leighton PA, Saam JR, Ingram RS, Stewart CL, Tilghman SM. An enhancer deletion affects both H19 and Igf2 expression. Genes & Dev. 1995;9:2079–2089. doi: 10.1101/gad.9.17.2079. [DOI] [PubMed] [Google Scholar]
- Lyon MF. X-chromosome inactivation: A repeat hypothesis. Cytogenet Cell Genet. 1998;80:133–137. doi: 10.1159/000014969. [DOI] [PubMed] [Google Scholar]
- Madsen O, Scally M, Douady CJ, Kao DJ, DeBry RW, Adkins R, Amrine HM, Stanhope MJ, de Jong WW, Springer MS. Parallel adaptive radiations in two major clades of placental mammals. Nature. 2001;409:610–614. doi: 10.1038/35054544. [DOI] [PubMed] [Google Scholar]
- Miyoshi N, Wagatsuma H, Wakana S, Shiroishi T, Nomura M, Aisaka K, Kohda T, Surani MA, Kaneko-Ishino T, Ishino F. Identification of an imprinted gene, Meg3/Gtl2 and its human homologue MEG3, first mapped on mouse distal chromosome 12 and human chromosome 14q. Genes Cells. 2000;5:211–220. doi: 10.1046/j.1365-2443.2000.00320.x. [DOI] [PubMed] [Google Scholar]
- Murphy WJ, Eizirik E, Johnson WE, Zhang YP, Ryder OA, O'Brien SJ. Molecular phylogenetics and the origins of placental mammals. Nature. 2001;409:614–618. doi: 10.1038/35054550. [DOI] [PubMed] [Google Scholar]
- Neumann B, Kubicka P, Barlow DP. Characteristics of imprinted genes. Nat Genet. 1995;9:12–13. doi: 10.1038/ng0195-12. [DOI] [PubMed] [Google Scholar]
- Okamura K, Hagiwara-Takeuchi Y, Li T, Vu TH, Hirai M, Hattori M, Sakaki Y, Hoffman AR, Ito T. Comparative genome analysis of the mouse imprinted gene Impact and its nonimprinted human homolog IMPACT: Toward the structural basis of species-specific imprinting. Genome Res. 2000;10:1878–1889. doi: 10.1101/gr.139200. [DOI] [PubMed] [Google Scholar]
- Onyango P, Miller W, Lehoczky J, Leung CT, Birren B, Wheelan S, Dewar K, Feinberg AP. Sequence and comparative analysis of the mouse 1-Megabase region orthologous to the human 11p15 imprinted domain. Genome Res. 2000;10:1697–1710. doi: 10.1101/gr.161800. [DOI] [PubMed] [Google Scholar]
- Paulsen M, El-Maari O, Engemann S, Strödicke M, Franck O, Davies K, Reinhardt R, Reik W, Walter J. Sequence conservation and variability of imprinting in the Beckwith-Wiedemann syndrome gene cluster in human and mouse. Hum Mol Genet. 2000;9:1829–1841. doi: 10.1093/hmg/9.12.1829. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch E, Maniatis T. Molecular cloning: A laboratory manual (second edition). Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sasaki H, Jones PA, Chaillet JR, Ferguson-Smith AC, Barton SC, Reik W, Surani MA. Parental imprinting: Potentially active chromatin of the repressed maternal allele of the mouse insulin-like growth factor II (Igf2) gene. Genes & Dev. 1992;6:1843–1856. doi: 10.1101/gad.6.10.1843. [DOI] [PubMed] [Google Scholar]
- Scheper W, Meinsma D, Holtzhuizen PE, Sussenbach JS. Long-range RNA interaction of two sequence elements required for endonucleolytic cleavage of human insulin-like growth factor II mRNAs. Mol Cell Biol. 1995;15:235–245. doi: 10.1128/mcb.15.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt JV, Matteson PG, Jones BK, Guan XJ, Tilghman SM. The Dlk1 and Gtl2 genes are linked and reciprocally imprinted. Genes & Dev. 2000;14:1997–2002. [PMC free article] [PubMed] [Google Scholar]
- Schuster-Gossler K, Simon D, Guénet J-L, Zachgo J, Gossler A. Gtl2lacZ an insertional mutation on mouse Chromosome 12 with parental origin-dependent phenotype. Mammalian Genome. 1996;7:20–24. doi: 10.1007/s003359900006. [DOI] [PubMed] [Google Scholar]
- Schuster-Gossler K, Bilinksi P, Sado T, Ferguson-Smith A, Gossler A. The mouse Gtl2 gene is differentially expressed during embryonic development, encodes multiple alternatively spliced transcripts, and may act as an RNA. Dev Dyn. 1998;212:214–228. doi: 10.1002/(SICI)1097-0177(199806)212:2<214::AID-AJA6>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Schwartz S, Zhang Z, Frazer KA, Smit A, Riemer C, Bouck J, Gibbs R, Hardison R, Miller W. PipMaker–A web server for aligning two genomic DNA sequences. Genome Res. 2000;10:577–586. doi: 10.1101/gr.10.4.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit AFA. Interspersed repeats and other mementos of transposable elements in mammalian genomes. Curr Opin Genet Dev. 1999;9:657–663. doi: 10.1016/s0959-437x(99)00031-3. [DOI] [PubMed] [Google Scholar]
- Sun FL, Dean WL, Kelsey G, Allen ND, Reik W. Transactivation of Igf2 in a mouse model of Beckwith-Wiedemann syndrome. Nature. 1997;389:809–815. doi: 10.1038/39797. [DOI] [PubMed] [Google Scholar]
- Sutton VR, Shaffer LG. Search for imprinted regions on chromosome 14: Comparison of maternal and paternal UPD cases with cases of chromosome 14 deletion. Am J Med Genet. 2000;93:381–387. doi: 10.1002/1096-8628(20000828)93:5<381::aid-ajmg7>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- Szabo P, Tang SH, Rentsendorj A, Pfeifer GP, Mann JR. Maternal-specific footprints at putative CTCF sites in the H19 imprinting control region give evidence for insulator function. Curr Biol. 2000;10:607–610. doi: 10.1016/s0960-9822(00)00489-9. [DOI] [PubMed] [Google Scholar]
- Takada S, Tevendale M, Baker J, Georgiades P, Campbell E, Freeman T, Johnson MH, Paulsen M, Ferguson-Smith AC. Dlk (Delta-like) and Gtl2 are closely linked reciprocally imprinted genes on mouse chromosome 12 and are paternally methylated and co-expressed during development. Curr Biol. 2000;10:1135–1138. doi: 10.1016/s0960-9822(00)00704-1. [DOI] [PubMed] [Google Scholar]
- Wylie AA, Murphy SK, Orton TC, Jirtle RL. Novel imprinted DLK1/GTL2 domain on human chromosome 14 contains motifs that mimic those implicated in Igf2/H19 regulation. Genome Res. 2000;10:1711–1718. doi: 10.1101/gr.161600. [DOI] [PMC free article] [PubMed] [Google Scholar]