Abstract
L1 retrotransposons are pervasive in the human genome. Approximately 25% of recent L1 insertions in the genome are inverted and truncated at the 5′ end of the element, but the mechanism of L1 inversion has been a complete mystery. We analyzed recent L1 inversions from the genomic database and discovered several findings that suggested a mechanism for the creation of L1 inversions, which we call twin priming. Twin priming is a consequence of target primed reverse transcription (TPRT), a coupled reverse transcription/integration reaction that L1 elements are thought to use during their retrotransposition. In TPRT, the L1 endonuclease cleaves DNA at its target site to produce a double-strand break with two single-strand overhangs. During twin priming, one of the overhangs anneals to the poly(A) tail of the L1 RNA, and the other overhang anneals internally on the RNA. The overhangs then serve as primers for reverse transcription. The data further indicate that a process identical to microhomology-driven single-strand annealing resolves L1 inversion intermediates.
The human genome contains >500,000 L1 sequences (International Human Genome Sequencing Consortium 2001), yet only 3000–5000 are full-length elements. One reason for the relative paucity of full-length elements is that older L1 elements have become fragmented by insertion of other retrotransposons or by genomic rearrangements. Another reason is that full-length elements are potentially more detrimental to the genome than shorter elements and may be eliminated by negative selection (Boissinot et al. 2001). However, the main reason that L1 sequences are usually not full-length is a consequence of the mechanism of L1 retrotransposition.
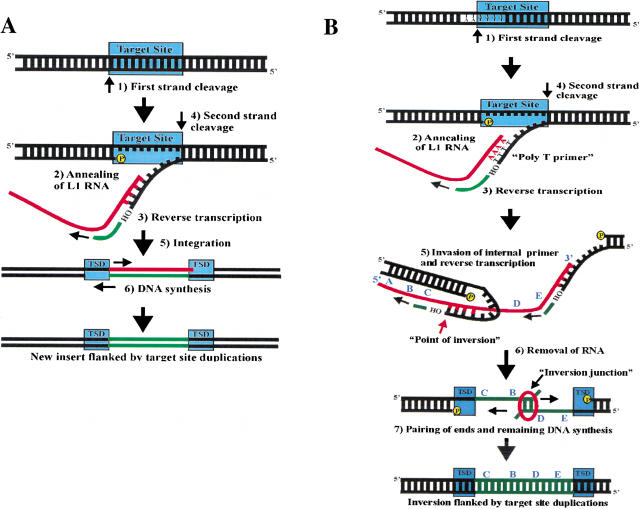
L1 elements are members of the non-long terminal repeat (nonLTR) retrotransposon family. NonLTR retrotransposons encode a protein with both endonuclease and reverse transcriptase (RT) activity (Malik et al. 1999), and are thought to integrate by a coupled reverse transcription/integration process called target primed reverse transcription (TPRT) (Fig. 1A; Luan et al. 1993). During TPRT, the endonuclease cleaves one strand of DNA at its target site, producing a free 3′-hydroxyl at the DNA nick. After the retrotransposon RNA anneals at the break, the RT uses the RNA as a template and the 3′-hydroxyl as a primer to perform reverse transcription. The remaining steps of TPRT include cleavage of the second DNA strand, integration of the cDNA, and completion of DNA synthesis. Upon the completion of TPRT, a copy of the original retrotransposon is integrated at a new genomic location and is flanked by target site duplications (TSDs).
Figure 1.
Target primed reverse transcription and twin priming. (A) This is a schematic of target primed reverse transcription (TPRT), based on in vitro studies of the R2 element from Bombyx mori (Luan et al. 1993). TPRT involves the following steps: (1) Cleavage of first DNA strand at the target site by the retrotransposon endonuclease (EN). (2) Annealing of retrotransposon RNA at the nick. (3) Reverse transcription from the free 3′-hydroxyl by the retrotransposon reverse transcriptase (RT). (4) Cleavage of second DNA strand. (5) Integration at the double-strand break. (6) Removal of RNA and completion of DNA synthesis. The TPRT process produces target site duplications (TSDs) at the flanks of the newly integrated retrotransposon. (B) Twin priming is a modification of the TPRT reaction with the following steps: (1) The L1 EN cleaves one strand of its DNA target site, producing the poly T primer. (2) The poly(A) tail of the L1 RNA anneals on the poly T primer. (3) L1 RT uses the L1 RNA as a template and the poly T primer to initiate reverse transcription. (4) The L1 EN cleaves the second DNA strand before reverse transcription has been completed, producing the internal primer. (5) The internal primer invades the L1 RNA and primes reverse transcription. (6) The RNA is removed from the RNA/cDNA structure. (7) The single-stranded cDNAs pair at a region of limited complementarity, and the remaining DNA synthesis is completed. The entire process results in an L1 inversion flanked by perfect target site duplications. The L1 RNA sequence is represented by 5′-A-B-C-D-E-3′. After the inversion, the insertion sequence is 5′-C-B-D-E-3′.
The 5′ ends of most L1 elements in the genome are either truncated or both inverted and truncated. Truncation has been hypothesized to occur because of low processivity by the L1 reverse transcriptase. If the RT disassociates from the RNA template before the completion of reverse transcription, then the resultant insertion will be truncated at the 5′ end. More difficult to explain are the inversions, which always involve both truncation and inversion of the 5′ end of the molecule. For example, if the sequence of the L1 RNA is 5′-A-B-C-D-E-3′, then the sequence of the insertion may be 5′-C-B-D-E-3′. The point of inversion may contain a deletion, duplication, or neither. A model of L1 inversion has been proposed previously, but it does not account for all of the L1 inversion structures found in the genome and it is not consistent with the data found in the present study of L1 inversion (Hutchison et al. 1989). We performed an analysis of inversion-containing L1 insertions in the human genome database, and from this analysis we created a model of L1 inversion. We propose that inversion is a consequence of the L1 TPRT process, and suggest a mechanism, twin priming, that creates L1 inversions.
RESULTS AND DISCUSSION
L1 Inversion Occurs Frequently
In a previous study of L1 elements in the genome, we characterized 66 L1 insertions from the Ta and pre-Ta subfamilies (Goodier et al. 2000). These were selected by performing a BLAST search of the human database using the last 100 bp of the 3′ untranslated region (3′UTR) from the Ta subfamily consensus sequence. The Ta subfamily is characterized by the nucleotides ACA 89–91 bp upstream of the L1 polyadenylation [poly(A)] signal (Skowronski and Singer 1986), whereas pre-Ta members have ACG at these positions and older L1s usually have GAG. We chose to analyze L1 inversions from this dataset for several reasons. First, these subfamilies are evolutionarily young and are easy to distinguish from older L1s. The young age of the Ta and pre-Ta subfamily members means that genomic copies of these elements represent relatively recent insertion events, within the last four million years in the case of Ta (Boissinot et al. 2000). Analyzing recent insertions is important for this study because they are unlikely to have undergone significant mutations since their insertion. Second, all of the L1s from the subfamilies studied are >99% identical to each other. One can therefore predict the sequence of the RNA that led to the insertion with relative certainty. Third, all of the insertions are flanked by identifiable TSDs, and therefore, likely inserted by TPRT.
Of the 66 insertions, 24 were full-length (36%) and the remaining 42 were truncated, or inverted and truncated (64%) (Goodier et al. 2000). These percentages are in agreement with a subsequent study of L1 elements from the Ta subfamily, which revealed that 34% were full-length (Boissinot et al. 2000). Sixteen of the 66 insertions were inverted and truncated (24%). Therefore, inversion of L1 elements is a frequent occurrence.
Twin Priming: A Novel Mechanism for Inversion
We analyzed 15 of the 16 L1 inversions. The 16th was discarded because it contained an ambiguous nucleotide within the TSD. We also analyzed one additional pre-Ta inversion found in the database, and one additional Ta inversion, which was previously described as a de novo insertion into the dystrophin gene (Table 1; Holmes et al. 1994). From our analysis, we discovered that L1 inversions share some interesting features: (1) The last four nucleotides at the end of the 5′ target site duplication are often complementary to the nucleotides predicted to be just proximal to the inversion point on the preintegration RNA; (2) the inversion points are highly clustered; and (3) there are usually 1–4 nucleotides at the inversion junction that, in theory, could have originated from either the noninverted or the inverted L1 sequence. These features suggest a model for the mechanism of L1 inversion.
Table 1.
L1 Inversions Analyzed
| Accession # | Subfamily | % Identitya | Target Site Duplication (TSD) | TSD length |
|---|---|---|---|---|
| AC007486 | Ta | 99.3 | aatt/AAAAAATTTTGG/gcgc | 12 |
| AC006131 | Pre-Ta | 99.3 | ggtc/ACAAAAGATACACTCCTTT/tgac | 19 |
| AC002122 | Ta | 99.7 | cttt/AAAATTTTTTAATG/aatc | 14 |
| aL022153 | Pre-Ta | 99.5 | cttt/AAAAAATACCGATTCC/tgag | 16 |
| AC004883 | Ta | 99.9 | attt/GATAATATGTT/gcat | 11 |
| AC004491 | Ta | 99.2 | ctct/AAAGAAGATATAT/acaa | 13 |
| AL031117 | Pre-Ta | 99.5 | cttc/AAAATGTTAAGGGTC/atct | 15 |
| AL0390998 | Ta | 99.0 | atat/AAAAGATCGGTGA/aaaa | 13 |
| Z84814 | Ta | 99.6 | catt/AAAAACAGCTATAGTTT/atca | 17 |
| Z95325 | Ta | 99.5 | tctc/AAAAACAAAACAA/aaca | 13 |
| AC004053 | Ta | 99.3 | gtat/AAGAATGCTTGTGATTTTTG/taca | 20 |
| AL034425 | Pre-Ta | 99.3 | actc/AAAACTTGGCTGTC/gagg | 14 |
| Z70758 | Pre-Ta | 99.1 | actt/AGAAGTCCATGAATCCA/tgct | 17 |
| AC004220 | Ta | 99.4 | agtt/AAGAAGGAGGGGA/gact | 13 |
| AL023284 | Ta | 99.1 | catt/AAAAAGCATATAGTAT/acac | 16 |
| AF)36938 | Pre-Ta | 99.0 | aaag/AAAAAAATGTTTCTAATTC/aaga | 19 |
| U09115 | Ta | 99.4 | agtt/AAATCATCTGCTGCT/gtgg | 15 |
Comparison with a consensus of active L1 elements.
A variation of the TPRT reaction, called twin priming, could lead to the formation of inverted L1 insertions (Fig. 1B). First, the L1 endonuclease cleaves one strand of its double-stranded DNA target site. The L1 endonuclease cleaves DNA at the loose consensus of 3′-AA|TTTT-5′ (Feng et al. 1996; Jurka 1997; Cost and Boeke 1998). Cleavage produces a nick in the DNA consisting of a free 3′hydroxyl and a T-rich stretch of DNA that can be used as a primer for reverse transcription. We refer to this primer as the poly(T) primer. Second, the poly(A) tail of the L1 RNA invades the DNA nick and anneals to the poly T primer. Third, the L1 reverse transcriptase uses the L1 RNA as a template and the poly T primer to initiate reverse transcription. Fourth, the L1 endonuclease cleaves the second DNA strand before reverse transcription has been completed, producing an additional 3′ hydroxyl and a stretch of single-stranded DNA. We refer to this additional potential primer as the internal primer. Fifth, the internal primer anneals to the L1 RNA internally and primes reverse transcription at a site distinct from the reverse transcription occurring at the 3′ end of the L1 RNA (primed by the poly T primer). Therefore, two different primers at two different locations are used on the L1 RNA template. Sixth, the RNA is removed from the RNA/cDNA structure. Seventh, the single-stranded cDNAs pair at a region of limited complementarity. Lastly, the remaining DNA synthesis is completed. The entire process results in an L1 inversion flanked by perfect target site duplications. Depending on the extent of reverse transcription from the poly T primer, the point of inversion may contain a deletion, duplication, or neither. This model predicts all of the typical L1 inversion structures that are found in the genome database and does not predict structures that are not found (such as internal inversions).
For this model to be correct, cleavage of the second DNA strand must occur before reverse transcription has been completed. During in vitro experiments on the R2 TPRT process, the cleavage of the second DNA strand occurs after reverse transcription (Luan et al. 1993). Therefore, according to our model one would not expect L1-like inversions to occur during R2 retrotransposition. Indeed, R2 inversions have not been observed (T.H. Eickbush, pers. comm.).
This model predicts that the nucleotides at the 3′ end of the internal primer will complement the nucleotides on the L1 RNA template that are just proximal to the point of inversion (the nucleotides at the red arrow in Fig. 1B). This prediction is strongly supported by our analysis of L1 inversions (Fig. 2). From each inversion, we checked the first six nucleotides at the 3′ end of the TSD (which would serve as the internal primer) to determine whether they were complementary to the predicted nucleotides just proximal to the inversion point on the RNA which likely created the insertion. We determined the likely RNA nucleotides from a consensus sequence of active L1 elements. One would predict a one -in four likelihood of a complementary match at each position by chance. However, the number of complementary matches occurring at the first four positions is far greater than expected (Fig. 2). When considering the first four nucleotides of each potential primer, one would expect only one complementary match by chance. The average number of matches in the first four nucleotides is 3.1 per primer. Four primers have a perfect four -of four complementary nucleotides, while eleven have three of four, and two have two of four. The frequency of complementarity decreases with distance from the 3′-hydroxyl. This finding would be expected if these nucleotides were used as a primer, since complementarity of the nucleotides nearest the 3′-hydroxyl is more important in determining the efficiency of the primer than the complementarity of the nucleotides positioned more 5′.
Figure 2.
Complementarity of the primers. (A) The internal primer and the poly T primer were analyzed for complementarity to their predicted binding sites on the L1 RNA. The first six nucleotides, numbered from the 3′-hydroxyl, are listed. Nucleotides are highlighted in yellow if they are complementary to the corresponding nucleotide on the L1 RNA. The last row lists the number of complementary nucleotides at each position, out of a possible total of seventeen. (B) The number of matches (r) at each position and the corresponding P-values, representing the likelihood of obtaining r matches or greater by chance alone.
How does this level of complementarity compare with that of the poly T primer? If the TPRT model is correct, then the T-rich stretch of the poly T primer anneals to the poly(A) tail of the L1 RNA. We checked the first six nucleotides at the end of the poly T primer from each inversion to determine how often they were complementary to the poly(A) tail. As predicted, these nucleotides were also complementary much more often than expected by chance. Interestingly, the frequency of a complementary match over the first four nucleotides was nearly identical to the frequencies found for the internal primer (Fig. 2). The main difference between the two potential primers was that significant complementarity extends to the fifth position for the poly T primer, while ending at the fourth position for the internal primer.
These data indicate that very limited complementarity is required for primer binding and reverse transcription, an average of three of the first four nucleotides and as few as two of the first four nucleotides. Surprisingly, there are a few examples for both the internal primer and the poly T primer where the nucleotide closest to the 3′-hydroxyl is not predicted to be complementary to the RNA template. It is possible that these represent cases where either the nucleotides of the insertion have mutated since the time of integration or the predicted sequence of the preintegration RNA is incorrect. However, the choice of insertions that are >99% identical to the consensus used to predict the preintegration RNA sequence greatly limits the likelihood of these possibilities. Interestingly, other reverse transcriptases can initiate reverse transcription when the terminal 3′ nucleotide of the primer is not complementary to the primer binding site (Perrino et al. 1989; Yu and Goodman 1992; Pulsinelli and Temin 1994; Kulpa et al. 1997).
A second interesting finding is that the points of inversion, and therefore the predicted invasion sites of the internal primers, are highly clustered (Fig. 3). Although the L1 RNA is 6022 nucleotides long from the 5′ end to the poly(A) signal, all of the inversion points occur between nucleotides 4328 and 5780. It is possible that many of the inversions occur near the 3′ end of the L1 RNA, because this end of the molecule is annealed to the poly T primer and may also be in close proximity to the potential internal primer. However, even within this region, eight of the inversion points occur in a small 269 bp region ranging from nucleotides 5030 to 5298. Perhaps the secondary structure of the L1 RNA is important for determining sites that are amenable to invasion by the internal primer.
Figure 3.
Clustering of the inversion points. The L1 RNA is represented by a black line ending in a poly(A) tail (A(n)). The position on the RNA is numbered from the first nucleotide (1) to the end of the AATAAA poly(A) signal (6022). The region of the L1 RNA from nucleotides 4000–6000 is expanded on the line above. A black arrow represents the inversion point of each element. Each inversion is labeled with its accession number and the position of its inversion point.
Microhomology-Driven Single-Strand Annealing: A Mechanism for Resolution of L1 Inversion Intermediates
Analysis of the inversion junctions (red circle in Fig. 1B) reveals that in most cases, there are 1–4 nucleotides that, in theory, could have arisen from either the noninverted L1 sequence or from the inverted sequence. One way of interpreting this finding is that the nucleotides came from both the noninverted sequence and the inverted sequence. If the cDNA/RNA structure that has undergone twin priming resolves by pairing at small regions of complementarity, then the nucleotides left at the inversion junction would appear as if they could have come from either the noninverted DNA (cDNA from the poly T primer) or the inverted DNA (cDNA from the internal primer). Such a mechanism of resolution is identical to microhomology-driven single-strand annealing (SSA), a form of nonhomologous end joining (NHEJ) (Thacker et al. 1992; Gottlich et al. 1998). Microhomology-driven SSA can resolve double-strand breaks when the extent of complementarity is limited to a single nucleotide match (Pfeiffer et al. 1994). One expects by chance alone that one of four inversions would contain inversion junctions with evidence of a single complementary nucleotide, 1 of 16 inversions would contain evidence of two complementary nucleotides, and so on. However, we found evidence of complementary nucleotides in 14 of the 17 inversions (Fig. 4). Two inversions had four complementary nucleotides, one inversion had three nucleotides, eight had two nucleotides, and three had one nucleotide. Of the three cases without evidence of complementary nucleotides, two inversion junctions had additional nucleotides added at the junction that are apparently nontemplated (Z95325 and AC006131) and one had neither complementary nucleotides, nor nontemplated nucleotides (AF036938). Interestingly, in experiments using 3′ single-strand overhangs without homology as the substrate for NHEJ, nontemplated nucleotides are sometimes found at the junction (Pfeiffer et al. 1994). Therefore, a resolution mechanism based upon microhomology-driven SSA can explain all of the types of inversion junctions seen in our dataset. Alternatively, some reverse transcriptases are able to add nontemplated nucleotides to the ends of cDNAs (Gabriel and Mules 1999).
Figure 4.
The structure of each inversion. A full-length L1 element is represented by the hashed line, numbered from the first nucleotide (1) to nucleotide 6000 near the end of the sequence. The structure of each inversion is represented below the schematic L1. A left-facing arrow represents noninverted sequence with the length denoted above (distance from the AATAAA poly(A) signal to the end of the noninverted sequence). A right-facing arrow represents inverted sequence with the length denoted above (distance from the inversion point to the target site duplication). For example, the sequence at the tail of the inverted sequence from Z95325 is represented on the L1 consensus sequence near nucleotide 5450, and the sequence at the head of the arrow is represented near nucleotide 1380. Each inversion has nucleotides from the consensus sequence either deleted (del) or duplicated (dup) at the end of the noninverted sequence (head of left-facing arrow). The size of the deletion or duplication is indicated above its corresponding location relative to the L1 consensus sequence. Note that in the case of a duplication, the duplicated sequence is located both in its usual univerted position at the end of the noninverted sequence (head of the left-facing arrow) and also is inverted at the beginning of the inverted sequence (tail of right-facing arrow). Most inversions have evidence of complementary nucleotides at the inversion junction that could be associated with either the end of the noninverted sequence (head of left-facing arrow) or the end of the inverted sequence (head of right-facing arrow). The number of complementary nucleotides is indicated in brackets under the column labeled “Overlap.” In the case of complementary nucleotides, the sizes of the inverted sequence, the noninverted sequence, and the deletion or duplication become variable, as indicated by the numbering. In one case there are no complementary nucleotides [0], and in two cases there are a number of nontemplated nucleotides. A negative number in brackets indicates the number of nontemplated nucleotides, and the nontemplated nucleotides are listed in parentheses.
The fact that the great majority of the junctions contain evidence of complementary nucleotides suggests that either cDNAs with complementary ends are the only ones which are able to resolve, or that pairing at limited complementarity can occur at internal nucleotides. Pairing at very small regions of internal complementarity is common in microhomology-mediated SSA, but requires a 3′-5′ exonuclease or endonuclease activity to remove the excess single-strand DNA flaps that are created (Fig. 1B). An in vitro study of microhomology-driven SSA suggested that 3′-5′ exonuclease activity could be provided by DNA polymerase ɛ (Gottlich et al. 1998), but a specific 3′ flap endonuclease activity, analogous to that of the RAG1/RAG2 complex (Santagata et al. 1999), is another possibility. DNA ligase III also appears to be important in the microhomology-driven SSA process (Gottlich et al. 1998). Other proteins required in this process are unknown, but the Ku70/80 proteins, which are essential for cohesive-end and blunt-end NHEJ, appear to be unimportant (Gottlich et al. 1998; Feldmann et al. 2000).
Other Insights Into the Mechanism of L1 Inversion
All of the inversion structures have nucleotides at the point of inversion that are either deleted or duplicated (Fig. 4). The inversions can be divided into three categories: Three have large deletions (>50 nucleotides), nine have small deletions (1–50 nucleotides), and five have small duplications (1–50 nucleotides).
The different inversion structures are easily explainable by variable disassociation of the RT from the L1 RNA template, the mechanism thought to produce 5′ truncation of noninverted L1s. L1 elements from the Ta and pre-Ta subfamilies display a full spectrum of variably 5′ truncated insertions (Boissinot et al. 2000). Accordingly, one would expect the inversions from our dataset to display variability in the length of the noninverted L1 sequence. Although this sequence is variable in length, the extent of reverse transcription from the poly T primer seems to be limited by the point of invasion of the internal primer. The invasion points are highly clustered towards the 3′ end of the L1 (Fig. 3), and therefore, the length of the noninverted L1 sequence tends to be on the short side. We interpret this to mean that invasion of the internal primer onto the L1 RNA has already occurred either before reverse transcription has begun at the poly T primer, or shortly thereafter. The L1 RT progresses from the poly T primer until it either disassociates on its own accord before it reaches the internal primer, thereby producing a large deletion, or disassociates because it has reached the impeding internal primer. The fact that 14 of 17 inversions contain small deletions or duplications suggests that the latter possibility is more common. Perhaps the L1 RT often reaches the impeding internal primer and then stalls and disassociates, producing a small deletion, or progresses a short distance while displacing the primer and associated downstream cDNA before disassociating from the RNA template. Providing that reverse transcription has already proceeded from the internal primer, the latter possibility would produce a small duplication of the inversion point sequence.
The extent of reverse transcription from the internal primer should not be limited in the same manner as that from the poly T primer, because there is no possibility of encountering a downstream primer. Accordingly, the lengths of the inverted sequences are more variable than the lengths of the noninverted sequence and include three instances of sequences >2 kb. On the other hand, 35% of insertions from these subfamilies are full-length (Boissinot et al. 2000; Goodier et al. 2000), suggesting that, if left unimpeded, the L1 RT can often reach the end of the L1 RNA. Therefore, one might expect reverse transcription from the internal primer to occasionally reach the 5′ end of the L1 RNA, but there is no evidence of this occurring in any of the inversions from our dataset. We provide three possible explanations for this discrepancy. First, the complex cDNA/RNA structure formed during twin priming may be antagonistic to prolonged reverse transcription from the internal primer by an unknown mechanism. Second, the RT working from the internal primer may occasionally proceed to the end of the L1 RNA, but these structures may be difficult to resolve and therefore observed less frequently. We favor a third possibility. Reverse transcription may occasionally proceed to the end of the L1 RNA and then microhomology-driven SSA resolves the structure using internal complementarity.
One question regarding the twin priming mechanism is whether priming occurs simultaneously, using two molecules of the L1 RT, or involves only one molecule that terminates and reinitiates. In either case, there is also the possibility of the use of either one or two L1 RNA molecules as template. The structures of the L1 inversions offer insight into the answers to these questions. In the case of two RNA molecules, with poly T priming on one molecule and internal priming on the other, there would be no internal primer to limit the extent of reverse transcription from the poly T primer. Therefore, it would be difficult to explain the finding that 14 of 17 inversions have small duplications or deletions at the inversion point. Furthermore, one would expect duplications of all sizes at the point of inversion, potentially up to several kilobases in length, yet all five inversions that we analyzed with duplications at the inversion point contained duplications of just 2–28 nucleotides.
The exclusion of two molecules of RNA from the model leaves three mechanistic possibilities. (1) One molecule of the L1 RT completes reverse transcription from the poly T primer, terminates, and reinitiates at the internal primer. (2) One molecule of RT completes reverse transcription from the internal primer, terminates, and reinitiates at the poly T primer. (3) Two molecules of RT perform simultaneous reverse transcription.
The first possibility cannot be ruled out but is not supported by the data. Large deletions could be created by this mechanism if the RT disassociates from the RNA and then jumps ahead to the internal primer. Small deletions also are not problematic; the RT progresses to the internal primer, stalls, disassociates, and then reinitiates at the internal primer. However, the presence of small duplications and the exclusion of large duplications are hard to explain by this mechanism. To create a duplication, the L1 RT would have to progress past the point of invasion of the internal primer, then the RNA/cDNA complex would need to be displaced by the internal primer, and finally the RT would have to dissociate and jump back to the internal primer. If this unlikely sequence of events could occur, one might also expect to see larger duplications created by the same process.
Our findings do not contradict the second possibility, in which the L1 RT completes reverse transcription from the internal primer, disassociates, and then reinitiates reverse transcription from the poly T primer. However, it is somewhat counterintuitive that the RT of a retrotransposon that has evolved to use the poly T primer would instead favor initiation at an internal primer.
The third possibility, in which twin priming occurs simultaneously with two molecules of RT, is a simple and attractive model. As discussed, this model easily explains all of the inversion types. One possible argument against two RT molecules is the fact that the L1 machinery tends to retrotranspose the RNA which encoded it (cis preference) (Wei et al. 2001). However, cis preference does not preclude a single L1 RNA synthesizing two RT molecules, which then interact preferentially with the RNA which encoded them.
Consequences of L1 Inversion
The fact that L1 elements are pervasive throughout the human genome taken together with the high occurrence of L1 inversion suggests that the L1 inversion process likely has important consequences towards determining the structure and function of the genome. Indeed, the inversion of an L1 element which inserted into the CYBB gene of a chronic granulomatous patient created a new splice site and branch site which resulted in heterogeneous splicing of the gene (Meischl et al. 2000). As a general mechanism, twin priming may limit the expansion of L1 retrotransposons by limiting the number of full-length insertions capable of subsequent retrotransposition. Structurally similar inversions occur in the mouse (AC073296 and AC005403) and dog (AB012217) (Choi et al. 1999), suggesting that L1 inversion and its consequences are not limited to humans.
METHODS
The L1 insertions were selected as described previously (Goodier et al. 2000). Briefly, we performed a BLAST search (Altschul et al. 1990) of the nr (nonredundant) human sequences in GenBank (Benson et al. 2000) using the last 100 nucleotides of the consensus sequence of active L1 elements. We determined the precise structure of each inversion by comparison with the consensus sequence of active L1 elements. We analyzed the complementarity of the nucleotides from the internal primer and poly T primer (the 3′ ends of the TSD) with their annealing sites by comparing the first six nucleotides of each potential primer with their predicted priming sites on a consensus sequence of active L1 elements. The nucleotides of each internal primer were compared to their predicted annealing site at the inversion point. Thymidine residues were considered a match if they occurred at any position of the poly T primer, which presumably binds to the L1 poly(A) tail. If each of the 17 inversions are analyzed in this manner, then X is a binomial random variable representing the number of complementary matches at any position, where X ∼ B (17, .25). The expected mean and variance of X are E(X) = 17*.25 = 4.25 and Var(X) = 17*.25*.75V= 3.19, respectively. The number of actual complementary matches at each position is represented by r. The p-value is the probability of finding as many or more complementary matches at each position by chance alone; P-value = P(X ≥ ≥ |H0), where H0 : p = .25 and HA : p gt; .25. We used the binomial probability distribution for probability calculations.
Acknowledgments
We thank J.L. Goodier for helpful comments in the preparation of this manuscript. E.M.O. is supported by a Howard Hughes Predoctoral Fellowship, and H.H.K. Jr. is supported by grants from the NIH.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL kazazian@mail.med.upenn.edu; FAX (215) 573-7760.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.205701.
REFERENCES
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Rapp BA, Wheeler DL. GenBank. Nucleic Acids Res. 2000;28:15–18. doi: 10.1093/nar/28.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boissinot S, Entezam A, Furano AV. Selection against deleterious LINE-1-containing loci in the human lineage. Mol Biol Evol. 2001;18:926–935. doi: 10.1093/oxfordjournals.molbev.a003893. [DOI] [PubMed] [Google Scholar]
- Choi Y, Ishiguro N, Shinagawa M, Kim CJ, Okamoto Y, Minami S, Ogihara K. Molecular structure of canine LINE-1 elements in canine transmissible venereal tumor. Anim Genet. 1999;30:51–53. doi: 10.1046/j.1365-2052.1999.00400.x. [DOI] [PubMed] [Google Scholar]
- Cost GJ, Boeke JD. Targeting of human retrotransposon integration is directed by the specificity of the L1 endonuclease for regions of unusual DNA structure. Biochemistry. 1998;37:18081–18093. doi: 10.1021/bi981858s. [DOI] [PubMed] [Google Scholar]
- Feldmann E, Schmiemann V, Goedecke W, Reichenberger S, Pfeiffer P. DNA double-strand break repair in cell-free extracts from Ku80-deficient cells: Implications for Ku serving as an alignment factor in non-homologous DNA end joining. Nucleic Acids Res. 2000;28:2585–2596. doi: 10.1093/nar/28.13.2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Q, Moran JV, Kazazian HH, Boeke JD. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell. 1996;87:905–916. doi: 10.1016/s0092-8674(00)81997-2. [DOI] [PubMed] [Google Scholar]
- Gabriel A, Mules EH. Fidelity of retrotransposon replication. Ann NY Acad Sci. 1999;870:108–118. doi: 10.1111/j.1749-6632.1999.tb08871.x. [DOI] [PubMed] [Google Scholar]
- Goodier JL, Ostertag EM, Kazazian HH., Jr Transduction of 3′-flanking sequences is common in L1 retrotransposition. Hum Mol Genet. 2000;9:653–657. doi: 10.1093/hmg/9.4.653. [DOI] [PubMed] [Google Scholar]
- Gottlich B, Reichenberger S, Feldmann E, Pfeiffer P. Rejoining of DNA double-strand breaks in vitro by single-strand annealing. Eu J Biochem. 1998;258:387–395. doi: 10.1046/j.1432-1327.1998.2580387.x. [DOI] [PubMed] [Google Scholar]
- Holmes SE, Dombroski BA, Krebs CM, Boehm CD, Kazazian HH. A new retrotransposable human L1 element from the LRE2 locus on chromosome 1q produces a chimaeric insertion. Nat Genet. 1994;7:143–148. doi: 10.1038/ng0694-143. [DOI] [PubMed] [Google Scholar]
- Hutchison CA, Hardies SC, Loeb DD, Shehee WR, Edgell MH. LINES and related retroposons: Long interspersed sequences in the eucaryotic genome. In: Berg DE, Howe MM, editors. Mobile DNA. Washington, D. C.: ASM Press; 1989. [Google Scholar]
- International Human Genome Sequencing Consortium. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Jurka J. Sequence patterns indicate an enzymatic involvement in integration of mammalian retroposons. Proc Natl Acad Sci. 1997;94:1872–1877. doi: 10.1073/pnas.94.5.1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulpa D, Topping R, Telesnitsky A. Determination of the site of first strand transfer during Moloney murine leukemia virus reverse transcription and identification of strand transfer-associated reverse transcriptase errors. EMBO J. 1997;16:856–865. doi: 10.1093/emboj/16.4.856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan DD, Korman MH, Jakubczak JL, Eickbush TH. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: A mechanism for non-LTR retrotransposition. Cell. 1993;72:595–605. doi: 10.1016/0092-8674(93)90078-5. [DOI] [PubMed] [Google Scholar]
- Malik HS, Burke WD, Eickbush TH. The age and evolution of non-LTR retrotransposable elements. Mol Biol Evol. 1999;16:793–805. doi: 10.1093/oxfordjournals.molbev.a026164. [DOI] [PubMed] [Google Scholar]
- Meischl C, de Boer M, Ahlin A, Roos D. A new exon created by intronic insertion of a rearranged LINE-1 element as the cause of chronic granulomatous disease. Eur J Hum Genet. 2000;8:697–703. doi: 10.1038/sj.ejhg.5200523. [DOI] [PubMed] [Google Scholar]
- Perrino FW, Preston BD, Sandell LL, Loeb LA. Extension of mismatched 3′ termini of DNA is a major determinant of the infidelity of human immunodeficiency virus type 1 reverse transcriptase. Proc Natl Acad Sci. 1989;86:8343–8347. doi: 10.1073/pnas.86.21.8343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer P, Thode S, Hancke J, Vielmetter W. Mechanisms of overlap formation in nonhomologous DNA end joining. Mol Cell Biol. 1994;14:888–895. doi: 10.1128/mcb.14.2.888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulsinelli GA, Temin HM. High rate of mismatch extension during reverse transcription in a single round of retrovirus replication. Proc Natl Acad Sci. 1994;91:9490–9494. doi: 10.1073/pnas.91.20.9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santagata S, Besmer E, Villa A, Bozzi F, Allingham JS, Sobacchi C, Haniford DB, Vezzoni P, Nussenzweig MC, Pan ZQ, et al. The RAG1/RAG2 complex constitutes a 3′ flap endonuclease: implications for junctional diversity in V(D)J and transpositional recombination. Mol Cell. 1999;4:935–947. doi: 10.1016/s1097-2765(00)80223-3. [DOI] [PubMed] [Google Scholar]
- Skowronski J, Singer MF. The abundant LINE-1 family of repeated DNA sequences in mammals: Genes and pseudogenes. Cold Spring Harb Symp Quant Biol. 1986;51 Pt 1:457–464. doi: 10.1101/sqb.1986.051.01.055. [DOI] [PubMed] [Google Scholar]
- Thacker J, Chalk J, Ganesh A, North P. A mechanism for deletion formation in DNA by human cell extracts: The involvement of short sequence repeats. Nucleic Acids Res. 1992;20:6183–6188. doi: 10.1093/nar/20.23.6183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W, Gilbert N, Ooi SL, Lawler JF, Ostertag EM, Kazazian HH, Boeke JD, Moran JV. Human L1 retrotransposition: Cis preference versus trans complementation. Mol Cell Biol. 2001;21:1429–1439. doi: 10.1128/MCB.21.4.1429-1439.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Goodman MF. Comparison of HIV-1 and avian myeloblastosis virus reverse transcriptase fidelity on RNA and DNA templates. J BiolChem. 1992;267:10888–10896. [PubMed] [Google Scholar]