Abstract
Caenorhabditis elegans sqv mutants are defective in vulval epithelial invagination and have a severe reduction in hermaphrodite fertility. The gene sqv-7 encodes a multitransmembrane hydrophobic protein resembling nucleotide sugar transporters of the Golgi membrane. A Golgi vesicle enriched fraction of Saccharomyces cerevisiae expressing SQV-7 transported UDP-glucuronic acid, UDP-N-acetylgalactosamine, and UDP-galactose (Gal) in a temperature-dependent and saturable manner. These nucleotide sugars are competitive, alternate, noncooperative substrates. The two mutant sqv-7 missense alleles resulted in a severe reduction of these three transport activities. SQV-7 did not transport CMP-sialic acid, GDP-fucose, UDP-N-acetylglucosamine, UDP-glucose, or GDP-mannose. SQV-7 is able to transport UDP-Gal in vivo, as shown by its ability to complement the phenotype of Madin-Darby canine kidney ricin resistant cells, a mammalian cell line deficient in UDP-Gal transport into the Golgi. These results demonstrate that unlike most nucleotide sugar transporters, SQV-7 can transport multiple distinct nucleotide sugars. We propose that SQV-7 translocates multiple nucleotide sugars into the Golgi lumen for the biosynthesis of glycoconjugates that play a pivotal role in development.
Most cell surface and secreted proteins and some lipids undergo covalent modifications by the addition of carbohydrates (1, 2). These macromolecules play essential roles in multicellular organisms by participating in normal embryonic development and cell–cell and cell–matrix interactions. The carbohydrate-deficient glycoprotein syndromes, a group of autosomal recessive multisystemic diseases characterized by defective glycosylation of N-glycans, and studies of null mutations of N-glycan biosynthetic enzymes in mice provide strong evidence that the glycan moieties of glycoproteins play essential roles in the normal development and physiology of mammals and probably of all multicellular organisms (3–5). Disorders affecting the assembly of the glycosaminoglycan moieties of proteoglycans suggest the importance of these macromolecules in connective tissue, cartilage, and bone development (3). Heparan sulfate glycosaminoglycans (GAGs) are critical components of Wingless and fibroblast growth factor signaling in Drosophila melanogaster, and defects in heparan sulfate GAG assembly are associated with effects on Drosophila embryonic development, such as abnormalities in segment-polarity cuticle patterns and in mesodermal and tracheal cell migrations (6–9). Mutations of the EXT genes, a tumor suppressor family that includes glycosyltransferases involved in polymerization of heparan sulfates, have been associated with hereditary multiple exostoses, a skeletal dysplasia characterized by multiple cartilage-capped skeletal tumors (10).
Nucleotide sugar transporters translocate nucleotide sugars from the cytosol, their site of synthesis, into the lumen of the Golgi apparatus, where they are used as sugar-donor substrates by glycosyltransferases (11). Yeast, Leishmania donovani, and mammalian cell line mutants impaired in the transport of specific nucleotide sugars into the Golgi lumen have severe deficiencies of the corresponding sugar in their macromolecules (11), demonstrating the essential role of the transport process in glycosylation (11–15). In addition, fibroblasts from a patient with a clinical phenotype resembling that of leukocyte adhesion deficiency II are defective in GDP-fucose (Fuc) transport into the Golgi (16).
A role of glycosylation during Caenorhabditis elegans development was revealed by the recent identification and analysis of the sqv (squashed vulva) genes. Mutation of any of eight sqv genes perturbs the process of vulval epithelial invagination without affecting cell lineage (17, 18). The molecular identities of the three sqv genes described to date suggest that these genes act in a glycosylation pathway. SQV-8 is similar to two vertebrate β1,3-glucuronyltransferases, and SQV-3 is similar to vertebrate β1,4-galactosyltransferases and a snail β1,4-N-acetylglucosaminyltransferase (18). The human counterpart of SQV-3 with the greatest similarity to SQV-3 was recently identified as galactosyltransferase I (19, 20), the enzyme that attaches the first galactose in the glycosaminoglycan-protein linkage region in proteoglycans. The SQV-7 protein is 20% identical to LPG2, a Leishmania donovani Golgi membrane GDP-mannose (Man) transporter, and the two proteins share a similar hydropathy profile (18).
C. elegans is the first multicellular organism for which a mutation in a putative nucleotide sugar transporter (SQV-7) and its resulting phenotype have been described. Because SQV-8 and SQV-3 are putative glucuronyl- and galactosyltransferases, respectively, Herman and Horvitz (18) proposed that SQV-7 transports UDP-glucuronic acid (GlcA) or UDP-galactose (Gal), which may be used as substrates by SQV-3 or SQV-8 (18).
Amino acid sequence similarity to nucleotide sugar transporters of known function is not a reliable indicator of the substrate specificity of putative nucleotide sugar transporters. For instance, the canine Madin-Darby canine kidney cell (MDCK) UDP-N-acetylglucosamine (GlcNAc) transporter is 53% identical to the human UDP-Gal transporter and 40% identical to the murine CMP-sialic acid (SA) transporter, yet each is highly substrate-specific (21). The UDP-GlcNAc transporters from MDCK and yeast are only 22% identical, yet the MDCK UDP-GlcNAc transporter can complement the mutant phenotype of the UDP-GlcNAc transporter-deficient yeast (21). Therefore, on the basis of sequence identity alone, we cannot confidently predict the substrates of SQV-7.
cDNAs encoding putative nucleotide sugar transporters have been functionally expressed in distantly related organisms lacking endogenous activity (21–23), suggesting that these cDNAs encode the transporter proteins rather than accessory proteins required for transporter function. Purified protein fractions containing a single protein as determined by radioiodination and reconstituted into proteoliposomes display nucleotide sugar transport activities with specificities and affinities comparable to those obtained in intact Golgi vesicles or crude extracts (24–27). This observation strongly suggests that translocation of each nucleotide sugar is mediated by a single protein transporter.
We have previously developed a heterologous expression system to determine the function of putative Golgi nucleotide sugar transporters (22). Saccharomyces cerevisiae Golgi and endoplasmic reticulum vesicles transport GDP-Man and UDP-glucose (Glu) but no other nucleotide sugar in vivo or in vitro, making these vesicles an excellent system with which to assay the functions of putative nucleotide sugar transporters. Expression of the murine Golgi CMP-SA transporter in S. cerevisiae, which does not contain SA, resulted in the transport of CMP-SA into Golgi-enriched vesicles (22). We have used this system to study SQV-7 and found it to be a novel multisubstrate nucleotide sugar transporter for UDP-GlcA, UDP-GalNAc, and UDP-Gal.
Materials and Methods
Molecular Biology.
Standard molecular biology protocols were used as described by Sambrook et al. (28) unless otherwise noted. UDP-[3H]Gal (60 Ci/mmol), UDP-[3H]GlcA (10 Ci/mmol), UDP-[3H]GlcNAc (60 Ci/mmol), UDP-[3H] GalNAc (10 Ci/mmol), and UDP-[3H]Glu (10 Ci/mmol) were purchased from ARC, St. Louis. CMP-[3H]N-acetylneuraminic acid (32.8 Ci/mmol), GDP-[3H]Fuc (16 Ci/mmol), GDP-[3H]Man (18.9 Ci/mmol), and Na-[3H]acetate (135 m Ci/mmol) were purchased from NEN-DuPont.
sqv-7 Transcript.
Genomic DNA sequence of the sqv-7 region was determined by the C. elegans Genome Sequencing Consortium, and primers used throughout this study were designed based on this sequence. A sqv-7 cDNA 990 bases long that consists of only the sqv-7 predicted ORF was obtained by reverse transcriptase–PCR using RNA isolated from a mixed-stage C. elegans population. To determine the 5′ and 3′ ends of the transcript, we performed 5′ and 3′ rapid amplification of cDNA ends (RACE) as described by the manufacturer (GIBCO/BRL) using mixed-stage RNA as the template. The 5′ RACE product contains a variant SL2 splice leader sequence, GGTTTTAACCCAGTTAACCAAG (29) and lacks a 5′ untranslated region. The 3′ RACE product contains 304 bases of 3′ untranslated region. We used the 990 bases corresponding to the sqv-7 ORF to probe a Northern blot containing mixed-stage RNA and detected a single transcript of approximately 1,700 nt (data not shown).
Plasmid Construction.
For expression in yeast, a NotI site was generated by PCR mutagenesis at the 3′ terminus of sqv-7 cDNA immediately upstream of the stop codon, and a 111-bp NotI fragment encoding a triple hemaglutinin (HA) tag was cloned into the NotI site. The resulting sqv-7-HA fragment was cloned into pPBO9, an expression plasmid that was generated by cloning the phosphoglycerokinase promoter and terminator sequences from E1 (21) into YEP352 (30). The HA-tagged versions of the sqv-7 mutant alleles n2844 (L151P) and n2839 (G95D) were generated by PCR mutagenesis using the QuickChange system (Stratagene. A 469-bp SalI fragment was removed from the previously described MluI–PstI minus SphI sqv-7 genomic rescuing fragment (18), and a NotI site was generated at the 3′ terminus of sqv-7 immediately upstream of the stop codon. The 111-bp NotI fragment encoding a triple HA tag was cloned into the new NotI restriction site to generate a construct that encodes a SQV-7-HA fusion protein in C. elegans. Wild-type sqv-7 cDNA was cloned into pCDNA3.1 + (Invitrogen) and expressed in MDCK Ricinus communis agglutinin (RCA) resistant cells.
Strains and Genetics.
S. cerevisiae strain PRY225 (ura3–52, lys2–801am, ade2–1020c, his3, leu2, trpl-1Δ1) was grown at 30°C in liquid yeast extract/peptone/dextrose or on solid yeast extract/peptone/dextrose media containing 2% Bacto-agar. Strains derived from PRY225 transformed with URA plasmids were grown at 30°C in synthetic complete medium lacking uracil (SC-URA) (31) prepared using SCM-URA (Bufferad, Lake Bluff, IL). C. elegans strains were cultured as described by Brenner (32). The sqv-7(n2844) allele that confers maternal-effect lethality was cis-marked with unc-4(e120) and balanced with the chromosomal rearrangement mnC1. Germ-line transformation was performed as described by Mello et al. (33). The sqv-7-HA rescuing genomic DNA construct (40–50 ng/μl) was coinjected with the dominant roller marker pRF4 (50–60 ng/μl), and lines of Rol transgenic animals were established. Rescue was determined by examining the L4 vulva and restoration of fertility.
Western Blots.
Total membrane fractions from 2 ml of liquid cell culture were prepared by glass bead disruption followed by centrifugation at 100,000 g for 45 min. Membrane proteins were subjected to SDS/PAGE and electrotransfered to poly(vinylidene difluoride) membranes. After blocking with 3% gelatin, 1% milk, and 0.05% Tween 20, membranes were incubated with monoclonal anti-HA (1:1,000; Babco, Richmond, CA). Detection was performed by using horseradish peroxidase-conjugated mouse IgG (Promega) followed by chemiluminescence using Lumiglo (Kirkegaard & Perry Laboratories).
Subcellular Fractionation.
S. cerevisiae transformed with pPBO9 or pPBO9-sqv-7-HA were grown in SCM-URA liquid medium (31) to an OD600 of 4. The culture was chilled, centrifuged, and converted to spheroplasts as described (34) by using a total of 20 mg of Zymolyase 100 T (Seikagaku America, Rockville, MD) per 50 g of cells. This spheroplast suspension then was centrifuged at 450 g for 10 min. Cells were broken by suspending the pellet in 40 ml of 10 mM triethanolamine (pH 7.2), 0.8 M sorbitol, 1 mM EDTA and by drawing the cells rapidly several times into a narrow-bore serological pipette. Cell breakage was incomplete, but vesicle integrity was well maintained. The suspension was centrifuged successively at 450 g, 10,000 g, and 100,000 g to obtain pellet fractions P1, P2, and P3, respectively (35). The P3 fraction was enriched in Golgi apparatus-derived vesicles.
Nucleotide Sugar Translocation Assays.
The theoretical basis for the translocation assay of nucleotide derivatives into vesicles has been described (36). Golgi-enriched vesicles (P3 fraction, 0.5 mg of protein) were incubated at 30°C and 0°C for 3 min in 1 ml of 0.3 M sucrose, 30 mM triethanolamine (pH 7.2), 5 mM MgCl2, and 5 mM MnCl2 with the radioactive nucleotide sugar to be tested and, in parallel, with the standard vesicle nonpenetrator [3H]acetate (36). The vesicles were separated from the incubation medium by centrifugation at 100,000 × g for 40 min, and the total acid-soluble radioactive nucleotide sugars (St) associated with the washed vesicle pellets was determined. The amount of radioactive nucleotide sugars within vesicles (Si) was calculated by subtracting the estimated amount of radioactive nucleotide sugar outside the vesicles in the pellet (So) from St. So was calculated by multiplying the concentration of solutes in the incubation medium by the volume outside the vesicles in the pellet (Vo) of the nonpenetrator [3H]acetate (1.9 μl/mg of protein). Transport activity is defined as Si after incubation at 30°C minus Si after incubation at 0°C. For inhibition experiments, curves were computed by assuming that inhibition of transport follows simple saturation kinetics (37).
Generation of MDCK Stable Transfectants and Determination of Ricin Resistance.
MDCK RCAr cells (38) were transfected in OPTIMEM medium with 1 μg of plasmid DNA using Lipofectin (Life Technologies, Grand Island, NY) for 5 h and then grown for 48 h in complete medium (MEM containing 10% FCS and antibiotics). Cells were trypsinized and plated at a low density in complete medium containing geneticin (G418) (0.4 mg/ml). G418-resistant colonies were cloned, and ricin resistance was determined by growing 1,000–2,000 cells in 24-well plates containing variable amounts of RCA II (EY Laboratories, San Mateo, CA). After 7 days at 30°C, cell survival was determined by staining with methylene blue in 50% methanol.
Results
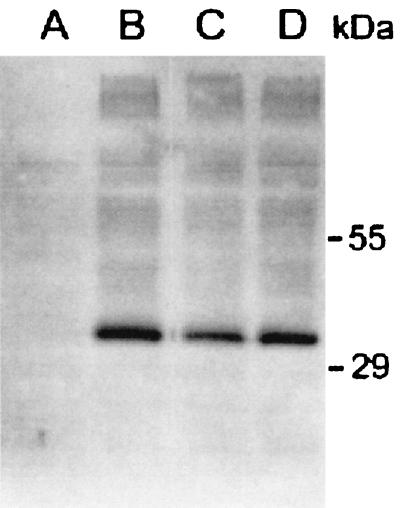
We obtained a sqv-7 cDNA by using RNA isolated from a mixed-stage population of C. elegans (see Materials and Methods). We generated a plasmid containing a sqv-7 HA translational construct and expressed the fusion protein in S. cerevisiae. A protein with an apparent mobility of about 34 kDa was detected by anti-HA antibodies in a total membrane fraction of sqv-7-HA transformed yeast (Fig. 1, lane B), consistent with a 37-kDa protein predicted from the sqv-7 cDNA. This protein was not detected in membranes from cells transformed with the vector alone (Fig. 1, lane A). We generated a C. elegans genomic DNA construct that encodes a SQV-7-HA fusion protein (see Materials and Methods) and determined that this construct can rescue the sqv-7 mutant phenotype (data not shown).
Figure 1.
Expression of SQV-7-HA in S. cerevisiae. Golgi-enriched fractions (P3, see Materials and Methods) were subjected to SDS/PAGE and analyzed by immunoblotting with a monoclonal anti-HA antibody. Lane A, pPBO9; lane B, pPBO9-sqv-7-HA; lane C, pPBO9-sqv-7(n2839)-HA; lane D, pPBO9-sqv-7(n2844)-HA.
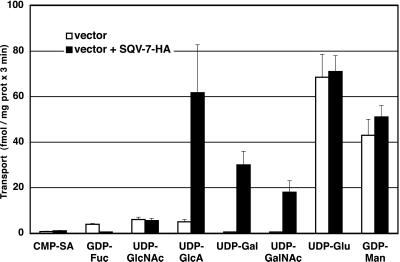
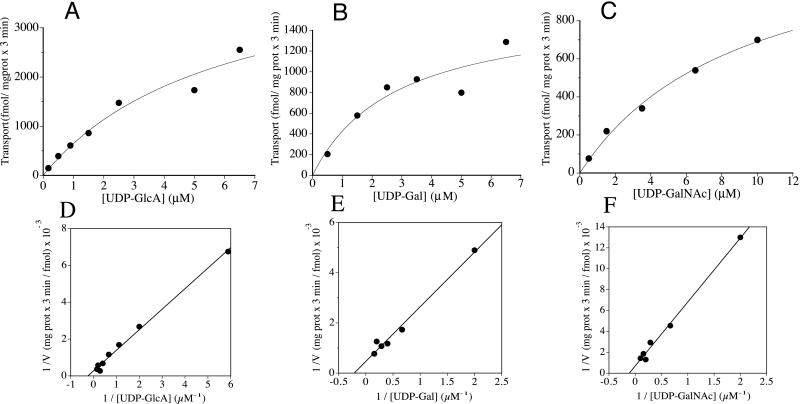
Golgi-enriched vesicles from S. cerevisiae expressing the SQV-7-HA fusion protein were isolated and assayed for the ability to transport radiolabeled nucleotide sugars in vitro. UDP-GlcA, UDP-Gal, and UDP-GalNAc were transported in a temperature-dependent manner into vesicles expressing the SQV-7-HA protein but not into vesicles isolated from cells transformed with vector alone (Fig. 2). GDP-Man and UDP-Glu, which are the only nucleotide sugars normally transported into Golgi and endoplasmic reticulum in yeast, were transported into vesicles from both the sqv-7-HA-transformed yeast and the vector-only-transformed yeast control, showing that the vesicles were of similar purity and integrity. Neither vesicle preparation transported CMP-SA, GDP-Fuc, or UDP-GlcNAc (Fig. 2). The uptake of the nucleotide sugars by SQV-7-HA fusion protein was saturable (Fig. 3). The Km for UDP-GlcA was 4.0 μM, for UDP-Gal 4.6 μM, and for UDP-GalNAc 8.7 μM. These low micromolar Km values of the transport of these nucleotide derivatives by SQV-7-HA are comparable to those of other nucleotide sugar transporters (11).
Figure 2.
Nucleotide sugar transport into Golgi vesicles by SQV-7. S. cerevisiae was transformed with pPBO9 (open bars) or pPBO9-sqv-7-HA (filled bars). Golgi-enriched (P3) fractions were isolated and assayed for the transport of different nucleotide sugars at 0.1 μM for 3 min at 30°C and 0°C. Transport activity is defined as Si (solutes inside vesicles) after incubation at 30°C minus Si after incubation at 0°C. Results shown indicate the averages of three independent assays and the standard error.
Figure 3.
Rates of nucleotide sugar transport by SQV-7 versus substrate concentration. The Golgi-enriched (P3) fraction prepared from S. cerevisiae expressing SQV-7-HA was incubated for 3 min at 30°C with different concentrations of UDP-GlcA (A and D), UDP-Gal (B and E), and UDP-GalNAc (C and F). Transport activities were analyzed for best fit to the Michaelis–Menten equation (A– C) and by linear regression of the double reciprocal plots (D–F).
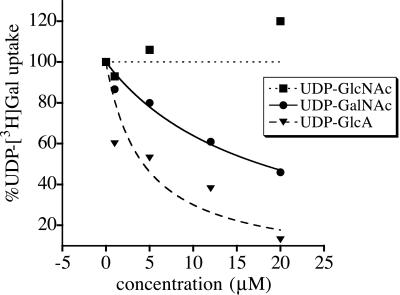
If all three nucleotide sugars bind to the same site on SQV-7, then the transport of one of these nucleotide sugar substrates should be inhibited in a competitive fashion by the other substrates. The transport of radiolabeled UDP-Gal was indeed inhibited in a concentration-dependent manner by the substrates UDP-GalNAc and UDP-GlcA but not by the nonsubstrate UDP-GlcNAc (Fig. 4). UDP-xylose did not inhibit UDP-Gal transport (data not shown), suggesting that it did not bind to the SQV-7 site mediating the transport activities we observed. Therefore, it seems unlikely that SQV-7 transports UDP-xylose. UDP-Gal transport was inhibited half-maximally by 4.3 μM UDP-GlcA and 17.7 μM UDP-GalNAc. These values are in close agreement with Kis calculated assuming UDP-GlcA and UDP-GalNAc are competitive, alternate substrates of UDP-Gal transport (4.1 μM and 8.9 μM, respectively). Competition analysis of UDP-GlcA inhibition of radiolabeled UDP-Gal transport was performed by determining transport of 2.0, 0.5, and 0.1 μM UDP-Gal in the presence of 0–20 μM UDP-GlcA. UDP-GlcA concentrations giving half-maximal inhibition of UDP-Gal transport at 2.0, 0.5, and 0.1 μM UDP-Gal were 16.9, 8.4, and 6.2 μM, respectively (data not shown). These values are consistent with calculated Kis of UDP-GlcA inhibition of UDP-Gal transport and support a model of alternate, competing substrates for SQV-7.
Figure 4.
UDP-GlcA and UDP-GalNAc are competitive substrates of SQV-7-HA-mediated UDP-Gal transport. Golgi-enriched (P3) fractions from S. cerevisiae expressing SQV-7-HA were assayed for transport of 0.1 μM UDP-[3H]Gal in the presence of variable concentrations of nonradiolabeled UDP-GlcA, UDP-GalNAc, or UDP-GlcNAc. Transport activities are plotted as the percentage of the initial rate of transport of UDP-Gal in the absence of competing substrates (15.0 ± 1.8 fmol/3 min per mg protein) and represent the average of two independent determinations. Curves were computed by assuming that inhibition of transport follows simple saturation kinetics and is characterized by the expression v = v(I=O) − {v(I=O) [I]/appKi + [I]}, where v is predicted UDP-Gal transport, v(I=O) is UDP-Gal transport in absence of competitor, and [I] is concentration of UDP-GlcA or UDP-GalNAc (37). Kis of the putative competing substrates on UDP-Gal transport were calculated as Kis = Km(S1) {1 + [UDP-Gal]/Km(S2)}, where Km(S1) is the Km for the UDP-GlcA or UDP-GalNAc transport and Km(S2) is the Km for the UDP-Gal transport (37).
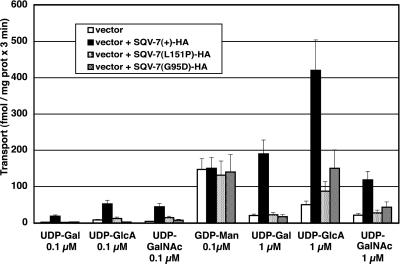
The two mutant sqv-7 alleles, n2839 (G95D) and n2844 (L151P), are missense mutations that cause nonconservative single amino acid substitutions. We generated sqv-7(n2839)-HA and sqv-7(n2844)-HA constructs and expressed the fusion proteins in S. cerevisiae. Both mutant constructs produced proteins of the same mobility in SDS/PAGE as that of wild-type SQV-7-HA (Fig. 1, lanes C and D). Transport of the three nucleotide sugars by both mutant SQV-7-HA proteins was impaired relative to that by SQV-7-HA (Fig. 5). Transport of UDP-GlcA into vesicles expressing SQV-7 (G95D)-HA was less impaired than into vesicles expressing SQV-7 (L151P)-HA. These results are consistent with previous observations that sqv-7(n2839) results in a less severe mutant phenotype than does sqv-7(n2844) (16).
Figure 5.
Nucleotide sugar transport into Golgi vesicles by SQV-7 and two SQV-7 mutant proteins. S. cerevisiae was transformed with pPBO9, pPBO9-sqv-7-HA, pPBO9-sqv-7(G95D)-HA, and pPBO9-sqv-7(L151P)-HA. Golgi-enriched vesicles were isolated (P3 fractions) and assayed for transport at 0.1 and 1 μM nucleotide sugar. Results shown indicate the averages of three independent determinations and the standard error.
We tested the ability of SQV-7 to transport UDP-Gal in vivo in the MDCK mutant cell line RCAr. This mutant cell line was isolated based on its resistance to ricin, the cytotoxic RCA, tolerating a 10 times higher concentration of the lectin than do wild-type cells (38). Ricin toxicity requires binding to terminal galactosyl residues, and the ricin-resistant phenotype of MDCK-RCAr cells correlates with a pleiotropic deficiency in galactosylation of glycoproteins and glycosphingolipids (12). The primary defect observed in MDCK-RCAr cells is impaired transport of UDP-Gal into the Golgi apparatus: Golgi vesicles from MDCK-RCAr cells translocate UDP-Gal at only 2% of the rate observed for vesicles from wild-type cells, resulting in reduced availability of UDP-Gal in the Golgi lumen (12). sqv-7-HA was stably transfected into MDCK-RCAr cells. SQV-7-HA was detectable when transfectants were grown at 30°C but not at 37°C (data not shown). MDCK-RCAr cells expressing SQV-7-HA were as sensitive to ricin as the wild-type cells (Table 1), indicating that expression of SQV-7 restored terminal Gal addition to glycoconjugates by compensating the UDP-Gal transport defect in vivo. MDCK-RCAr cells stably transfected with a CMP-SA transporter (39) remained resistant to ricin, demonstrating that the phenotypic correction mediated by expression of SQV-7 was specific.
Table 1.
Ricin sensitivity of MDCK ricin-resistant cells expressing SQV-7-HA
| MDCK cells | Ricin (ng/ml) LD10 |
|---|---|
| Wild type | 0.05 |
| RCAr | 0.50 |
| RCAr; SQV7-HA | 0.05 |
| RCAr; CMP-SA transporter | 0.50 |
Cells were grown at 30°C in the presence of different concentrations of RCA II as described in Materials and Methods. LD10 is the concentration of RCA II at which 10% of the cells survive.
Discussion
We have presented evidence that SQV-7, a C. elegans protein involved in vulval invagination and early embryonic development, is a multisubstrate nucleotide sugar transporter for UDP-GlcA, UDP-GalNAc, and UDP-Gal. SQV-7, a nucleotide sugar transporter protein, has been shown to transport the former two substrates. Transport of these nucleotide sugars by SQV-7 was temperature-dependent and saturable, with Km values in the low micromolar range, comparable to those of other nucleotide sugar transporters (11). UDP-Glu, CMP-SA, GDP-Fuc, and UDP-GlcNAc were not substrates for transport. SQV-7 was able to transport UDP-Gal in vivo, as shown by its ability to restore ricin sensitivity to MDCK RCAr, a cell line deficient in UDP-Gal transport into the Golgi (12).
The ability of UDP-GlcA and UDP-N-GalNAc to inhibit SQV-7-mediated UDP-Gal transport in a competitive manner is consistent with these three substrates being transported by SQV-7 using the same active site. UDP-GlcNAc, which was not transported into SQV-7 expressing vesicles, failed to inhibit UDP-Gal transport, indicating that inhibition by UDP-GlcA and UDP-GalNAc depends on the entire nucleotide sugar and not solely on the nucleotide moiety. This result is consistent with previous biochemical and genetic studies of nucleotide sugar transport showing that the nucleotide moiety is necessary for binding to the Golgi membrane but is not sufficient for transport: transport depends on the specific sugar bound to the nucleotide (40).
The two mutant alleles of sqv-7 encode proteins that are impaired in their abilities to transport all three nucleotide sugars, indicating a correlation between the in vitro transport activities and in vivo function. Furthermore, transport of UDP-GlcA appeared less affected in vesicles expressing the n2839 mutant allele of sqv-7 than those expressing the n2844 allele. C. elegans sqv-7(n2839) mutants have a less severe vulval mutant phenotype and a much smaller reduction in brood size than sqv-7(n2844) mutants (18). The in vitro transport activity presented here is consistent with the hypothesis that n2839 may cause only a partial loss of sqv-7 gene function (18).
Most known nucleotide sugar transporters translocate only a single nucleotide sugar. For example, the purified rat liver Golgi UDP-GalNAc transporter translocates only UDP-GalNAc among five UDP-sugars tested (27). Furthermore, mutant cell lines defective in one transport activity are normal for all other transport activities that have been tested (reviewed in ref. 11). For instance, Lec8 and clone 13 Chinese hamster ovary mutants and the RCAr MDCK mutant, which are all defective in UDP-Gal transport, translocate other nucleotide sugars at normal rates, suggesting they all have lesions in a dedicated UDP-Gal transporter. An exception is L. donovani LPG2, which has been reported to transport GDP-arabinose and GDP-Fuc in addition to GDP-Man (23). The multiple substrate specificities of SQV-7 and LPG2 are a novel feature that might have biological and evolutionary significance. These proteins might be specialized multisubstrate transporters dedicated to supplying several nucleotide sugars for the biosynthesis of a specific type of glycoconjugate. In addition, they might represent “ancestral” nucleotide sugar transporters with broad specificities, because all mammalian nucleotide sugar transporters characterized to date are mono-specific.
Transport of nucleotide sugars into the Golgi lumen is necessary for subsequent addition of the corresponding sugars to proteins, lipids, and GAGs in vivo and plays an important role in posttranslational modifications by regulating the availability of nucleotide sugars in the Golgi lumen (reviewed in ref. 11). The reduced availability of a nucleotide sugar can have a differential effect on the biosynthesis of different glycoconjugates. Thus, an MDCK mutant 98% deficient in UDP-Gal transport into the Golgi lumen shows significantly reduced levels of galactoproteins, galactosphingolipids, and keratan sulfate, a glycosaminoglycan that contains Gal in its polymeric repeating disaccharide units, but normal amounts of heparan sulfate and chondroitin sulfate, which have Gal only in the proteoglycan linkage region (12, 13). This differential effect might reflect a lower Km for the galactosyltransferases involved in the biosynthesis of the linkage region as compared with those involved in the biosynthesis of keratan sulfate, galactoproteins, and galactosphingolipids.
The biochemical defects we observed for the two SQV-7 missense mutant proteins should affect the availability of UDP-Gal, UDP-GlcA, and UDP-GalNAc in the lumen of the Golgi apparatus. These nucleotide sugars are required for posttranslational modifications of glycoproteins, glycolipids, and glycosaminoglycans. Indeed, it recently has been reported that the sqv-7(n2839) mutant has shorter polymer chains in chondroitin-modified proteoglycans and significant reduction in chondroitin- and heparan sulfate-derived disaccharides (41). The biochemical activities of the wild-type and mutant SQV-7 proteins we observed are consistent with these reductions in GAG biosynthesis. Given the multiple substrate specificity of SQV-7, defects in the biosynthesis of proteins and lipids containing GlcA, Gal, and GalNAc also seem likely. Whether such defects exist awaits further biochemical characterization of such molecules in C. elegans.
The sequence of the C. elegans genome predicts several putative nucleotide sugar transporter proteins, based on amino acid similarity to transporters from other organisms. It is possible that some of these putative transporters have specificities overlapping those of SQV-7. If so, then their inabilities to compensate the sqv-7 mutant phenotype may originate in their differential temporal or spatial patterns of expression.
In addition to causing a defect in vulval invagination, loss of sqv gene function results in a severe impairment in hermaphrodite fertility, as a consequence of defects in mutant oocyte development, function, and/or fertilization. Some sqv mutants produce eggs that arrest during embryogenesis, suggesting that the corresponding genes are required for embryonic development (17) as well as for vulval development. Our results, together with the biochemical characterization of other genes in the sqv pathway (41), suggest that a failure in GAG assembly of one or more proteoglycans may cause the sqv mutant abnormalities in vulval invagination and early development. Other sqv genes might be involved in glycosaminoglycan synthesis or might encode the protein acceptor of a glycosaminoglycan. Heparan sulfate glycosaminoglycans are critical components of Wingless and fibroblast growth factor signaling pathways in Drosophila, and defects in heparan sulfate GAG assembly are associated with profound effects on Drosophila embryonic development (6–9). That a chondroitin-modified proteoglycan is a major affected glycoconjugate in sqv-7 mutants (41) raises the possibility of an important role for these glycoconjugates in tissue development.
Acknowledgments
We thank Anthony Carruthers for assistance with kinetic calculations; Enrique Rodriguez-Boulan for wild-type and RCAr MDCK; Phil Robbins, Claudia Abeijon, David Carey, Brad Hersh, and Ewa Davison for helpful discussions; and Jacqueline Guinyard for secretarial support. This work was supported by National Institutes of Health Grants GM 30365 and GM 34396 (to C.B.H) and GM 24663 (to H.R.H.). H.R.H. is an Investigator at the Howard Hughes Medical Institute.
Abbreviations
- Man
mannose
- Fuc
fucose
- Glu
glucose
- SA
sialic acid
- MDCK
Madin-Darby canine kidney cells
- RCA
Ricinus communis agglutinin
- RCAr
RCA resistant
- Gal
galactose
- GlcA
glucuronic acid
- GalNAc
N-acetylgalactosamine
- GlcNAc
N-acetylglucosamine
- GAG
glycosaminoglycan
- HA
hemaglutinin
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Varki A, Marth J. Semin Dev Biol. 1995;6:127–138. [Google Scholar]
- 2.Crocker P R, Feizi T. Curr Opin Struct Biol. 1996;6:679–691. doi: 10.1016/s0959-440x(96)80036-4. [DOI] [PubMed] [Google Scholar]
- 3.McDowell G, Gahl W A. Proc Soc Exp Biol Med. 1997;215:145–147. doi: 10.3181/00379727-215-44121. [DOI] [PubMed] [Google Scholar]
- 4.Ioffe E, Stanley P. Proc Natl Acad Sci USA. 1994;91:728–732. doi: 10.1073/pnas.91.2.728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Metzler M, Gertz A, Sarkar M, Schachter H, Schrader J W, Marth J D. EMBO J. 1994;13:2056–2065. doi: 10.1002/j.1460-2075.1994.tb06480.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hacker U, Lin X, Perrimon N. Development (Cambridge, UK) 1997;124:3565–3573. doi: 10.1242/dev.124.18.3565. [DOI] [PubMed] [Google Scholar]
- 7.Binari R C, Staveley B, Johnson W A, Godavarti R, Sasisekharan R, Manoukian A S. Development (Cambridge, UK) 1997;124:2623–2632. doi: 10.1242/dev.124.13.2623. [DOI] [PubMed] [Google Scholar]
- 8.Tsuda M, Kamimura K, Nakato H, Archer M, Staatz W, Fox B, Humphrey M, Olson S, Futch T, Kaluza V, et al. Nature (London) 1999;400:276–280. doi: 10.1038/22336. [DOI] [PubMed] [Google Scholar]
- 9.Lin X, Perrimon N. Nature (London) 1999;400:281–284. doi: 10.1038/22343. [DOI] [PubMed] [Google Scholar]
- 10.Lind T, Tufaro F, McCormick C, Lindahl U, Lidholt K. J Biol Chem. 1998;273:26265–26268. doi: 10.1074/jbc.273.41.26265. [DOI] [PubMed] [Google Scholar]
- 11.Hirschberg C B, Robbins P W, Abeijon C. Annu Rev Biochem. 1998;67:49–65. doi: 10.1146/annurev.biochem.67.1.49. [DOI] [PubMed] [Google Scholar]
- 12.Brandli A W, Hansson G C, Rodriguez-Boulan E, Simons K. J Biol Chem. 1988;263:16283–16290. [PubMed] [Google Scholar]
- 13.Toma L, Pinhal M A, Dietrich C P, Nader H B, Hirschberg C B. J Biol Chem. 1996;271:3897–3901. doi: 10.1074/jbc.271.7.3897. [DOI] [PubMed] [Google Scholar]
- 14.Abeijon C, Mandon E C, Robbins P W, Hirschberg C B. J Biol Chem. 1996;271:8851–8854. doi: 10.1074/jbc.271.15.8851. [DOI] [PubMed] [Google Scholar]
- 15.Berninsone P, Miret J J, Hirschberg C B. J Biol Chem. 1994;269:207–211. [PubMed] [Google Scholar]
- 16.Lubke T, Marquardt T, von Figura K, Korner C. J Biol Chem. 1999;274:25986–25989. doi: 10.1074/jbc.274.37.25986. [DOI] [PubMed] [Google Scholar]
- 17.Herman T, Hartwieg E, Horvitz H R. Proc Natl Acad Sci USA. 1999;96:968–973. doi: 10.1073/pnas.96.3.968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herman T, Horvitz H R. Proc Natl Acad Sci USA. 1999;96:974–979. doi: 10.1073/pnas.96.3.974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Okajima T, Yoshida K, Kondo T, Furukawa K. J Biol Chem. 1999;274:22915–22918. doi: 10.1074/jbc.274.33.22915. [DOI] [PubMed] [Google Scholar]
- 20.Almeida R, Levery S B, Mandel U, Kresse H, Schwientek T, Bennett E P, Clausen H. J Biol Chem. 1999;274:26165–26171. doi: 10.1074/jbc.274.37.26165. [DOI] [PubMed] [Google Scholar]
- 21.Guillen E, Abeijon C, Hirschberg C B. Proc Natl Acad Sci USA. 1998;95:7888–7892. doi: 10.1073/pnas.95.14.7888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berninsone P, Eckhardt M, Gerardy-Schahn R, Hirschberg C B. J Biol Chem. 1997;272:12616–12619. doi: 10.1074/jbc.272.19.12616. [DOI] [PubMed] [Google Scholar]
- 23.Hong K, Ma D, Beverley S, Turco S. Biochemistry. 2000;39:2013–2022. doi: 10.1021/bi992363l. [DOI] [PubMed] [Google Scholar]
- 24.Mandon E C, Milla M E, Kempner E, Hirschberg C B. Proc Natl Acad Sci USA. 1994;91:10707–10711. doi: 10.1073/pnas.91.22.10707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Puglielli L, Mandon E C, Hirschberg C B. J Biol Chem. 1999;274:12665–12669. doi: 10.1074/jbc.274.18.12665. [DOI] [PubMed] [Google Scholar]
- 26.Puglielli L, Hirschberg C B. J Biol Chem. 1999;274:35596–35600. doi: 10.1074/jbc.274.50.35596. [DOI] [PubMed] [Google Scholar]
- 27.Pugielli L, Mandon E C, Rancour D M, Menon A K, Hirschberg C B. J Biol Chem. 1999;274:4474–4479. doi: 10.1074/jbc.274.7.4474. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 29.Land M, Islas-Trejo A, Rubin C S. J Biol Chem. 1994;269:14820–14827. [PubMed] [Google Scholar]
- 30.Hill J E, Myers A M, Koerner T J, Tzagoloff A. Yeast. 1986;2:163–167. doi: 10.1002/yea.320020304. [DOI] [PubMed] [Google Scholar]
- 31.Sherman F. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- 32.Brenner S. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mello C C, Kramer J M, Stinchcomb D, Ambros V. EMBO J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abeijon C, Yanagisawa K, Mandon E C, Hausler A, Moremen K, Hirschberg C B, Robbins P W. J Cell Biol. 1993;122:307–323. doi: 10.1083/jcb.122.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goud B, Salminen A, Walworth N C, Novick P J. Cell. 1988;53:753–768. doi: 10.1016/0092-8674(88)90093-1. [DOI] [PubMed] [Google Scholar]
- 36.Perez M, Hirschberg C B. Methods Enzymol. 1987;138:709–715. doi: 10.1016/0076-6879(87)38061-9. [DOI] [PubMed] [Google Scholar]
- 37.Segel I H. Enzyme Kinetics: Behavior and Analysis of Rapid Equilibrium and Steady-State Enzyme Systems. New York: Wiley; 1975. [Google Scholar]
- 38.Meiss H K, Green R F, Rodriguez-Boulan E J. Mol Cell Biol. 1982;2:1287–1294. doi: 10.1128/mcb.2.10.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eckhardt M, Muhlenhoff M, Bethe A, Gerardy-Schahn R. Proc Natl Acad Sci USA. 1996;93:7572–7576. doi: 10.1073/pnas.93.15.7572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Capasso J M, Hirschberg C B. Biochim Biophys Acta. 1984;777:133–139. doi: 10.1016/0005-2736(84)90505-4. [DOI] [PubMed] [Google Scholar]
- 41.Bulik D A, Wei G, Toyoda H, Kinoshita-Toyoda A, Waldrip W R, Esko J D, Robbins P W, Selleck S B. Proc Natl Acad Sci USA. 2000;97:10838–10843. doi: 10.1073/pnas.97.20.10838. [DOI] [PMC free article] [PubMed] [Google Scholar]