Abstract
Protein interaction maps have provided insight into the relationships among the predicted proteins of model organisms for which a genome sequence is available. These maps have been useful in generating potential interaction networks, which have confirmed the existence of known complexes and pathways and have suggested the existence of new complexes and or crosstalk between previously unlinked pathways. However, the generation of such maps is costly and labor intensive. Here, we investigate the extent to which a protein interaction map generated in one species can be used to predict interactions in another species.
Protein interaction maps have provided insight into the relationships among the predicted proteins of model organisms for which a genome sequence is available (Froment-Racine et al. 1997, 2000; Flajolet et al. 2000; Ito et al. 2000, 2001; McCraith et al. 2000; Walhout et al. 2000; Rain et al. 2001). Those maps have been useful particularly in generating potential interaction networks. Such networks have confirmed the existence of known complexes and pathways and have suggested the existence of new complexes and or crosstalk between previously unlinked pathways (Froment-Racine et al. 1997, 2000; Uetz et al. 2000). While the knowledge gained from proteome-wide interaction maps is highly valuable, the generation of such maps is costly and labor intensive. Here, we investigate the extent to which a protein interaction map generated in one species can be used to predict interactions in another species. Systematic BLAST searches for pairs of potential orthologs of known interacting protein partners in Saccharomyces cerevisiae have been performed to identify potentially conserved interactions, or “interologs”, in Caenorhabditis elegans. Starting from a large number of published yeast two-hybrid interactions between yeast proteins, searches for candidate interologs identified networks of potential physical interactions among C. elegans proteins. At least 16% of protein interactions in these networks could be detected in a yeast two-hybrid system, suggesting that these interactions are indeed conserved. In addition, many true interologs were amenable to reverse two-hybrid selections (Vidal 1997). Thus, it should be possible to generate reagents such as interaction-defective alleles and/or interaction-dissociating compounds for the further experimental characterization of potential interologs. The above observations suggest that sequence searches for potential interologs and other “comparative proteomics” strategies performed using the protein interaction maps of model organisms will be useful for drug screening programs in parasites, pathogens, and humans.
The conceptual translation of complete genome sequences into predicted proteomes and the use of this information to provide a first approximation of functional interactions underlying molecular complexes and regulatory pathways are among the most important challenges of the postgenomic era. One general approach is to use in silico methods to compare the proteomes from two or more organisms and predict interactions based upon presumed or demonstrated protein properties. For example, the Rosetta Stone method predicts functional interactions based upon the observation that pairs of functionally related proteins in one organism are sometimes expressed as single-fusion polypeptides in other organisms (Marcotte et al. 1999). Similarly, phylogenetic profiling is based upon the presumption that groups of functionally interacting proteins are selected together during the course of evolution and may have a tendency to coexist in the same proteomes (Pellegrini et al. 1999).
The approach described here presumes that large numbers of physically interacting proteins in one organism have “coevolved” so that their respective orthologs in other organisms interact as well. This notion of conserved interactions, or “interologs” (Walhout et al. 2000a), is substantiated by the observation that many interactions in signal transduction pathways or molecular machines are conserved between different species. For example, ∼7% of interactions currently available in a C. elegans protein interaction database have interologs that already have been described in the literature (Walhout et al. 2000a). In addition, searches for potential interologs already have been used to identify the function of several genes. For example, a functional ortholog of the human retinoblastoma protein was found in Drosophila using an interolog-based approach (Du et al. 1996). Here, we investigate the extent to which large-scale systematic searches for interologs may be used to identify potential networks of interaction and discuss methods that could be used in studying such potential networks and/or generate therapeutic agents.
RESULTS AND DISCUSSION
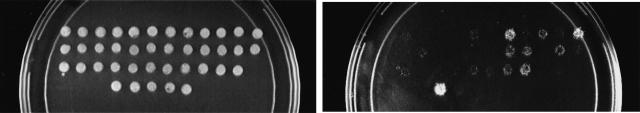
To determine the extent to which a protein interaction map generated in S. cerevisiae can be used to predict interactions in C. elegans, we utilized two large-scale, two-hybrid interaction maps of yeast proteins, which together contain 1195 interactions (Ito et al. 2000; Uetz et al. 2000). We identified potential C. elegans interologs by comparing the yeast proteins involved in these interactions to all predicted worm proteins as described in Methods. From 1195 yeast interactions, 257 potential worm interologs involving 282 proteins were identified (see below). Two hundred sixteen worm pairs (corresponding to a total of 276 worm open reading frames [ORFs]) were tested further. A sample of 71 of these potential interologs (corresponding to 72 yeast interactions) was used both to recapitulate the original yeast interactions and to experimentally test the corresponding worm protein interactions using the two-hybrid system (Table 1). The yeast and worm ORFs were cloned into both DNA binding (DB) and activation domain (AD) two-hybrid vectors and scored for interaction in both possible orientations (DB-X/AD-Y and DB-Y/AD-X) (see Methods). Each step of this analysis, beginning with the amplification of ORFs from the original clones, was performed in duplicate on two separate days. Protein pairs conferring at least one of the four yeast two-hybrid phenotypes (Vidal 1997) in both experiments were scored as positive interactions. Of the 72 potential yeast interactions tested, 26% (19) exhibited a detectable interaction in our version of the two-hybrid system (Table 1). It is likely that this number reflects differences in the conditions of yeast two-hybrid systems such as yeast strains, reporter genes, and the procedures for scoring interactions. Of these 19 interactions, six (31%) also scored positive with C. elegans proteins (Table 1). In addition, one more worm interaction was detected although the corresponding yeast interaction was undetected. Finally, we tested the remaining 145 worm potential interologs for their ability to mediate a two-hybrid interaction. After combining the two experiments, 35/216 potential interolog pairs (16%) exhibited a detectable interaction (Fig. 1A–D, Fig. 2, and Table 2)  .
.
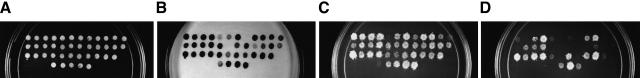
Figure 1.
Experimentally verified interactions between Saccharomyces cerevisiae and Caenorhabditis elegans. (A–D Yeast diploid cells expressing each of 35 C. elegans potential interologs. Pairs are arranged in the order described in Table 2. The five patches at the bottom are controls (negative control on the left side and controls of increasing interaction strength towards the right side). See Vidal (1997) for a detailed description of these controls. (B) β-Galactosidase assay to detect the expression of GAL1::lacZ. (C) Growth assay on SC-Leu-Trp-His, +20 mM 3AT plates to detect the expression of GAL1::HIS3. (D) Growth assay on SC-Leu-Trp-Ura plates to detect the expression of SPAL10::URA3. (E) Conservation of interactions. Each C. elegans protein pair tested was plotted according to two E-values. The first E-value corresponds to the conservation between the X (from yeast) and X‘ (from C. elegans) proteins while the second E-value corresponds to the conservation between the Y (from yeast) and Y‘ (from C. elegans) proteins. The smaller of the two E-values was plotted on the X-axis and the greater on the Y-axis. The C. elegans protein pairs that tested positive in the two-hybrid system are labeled in black.
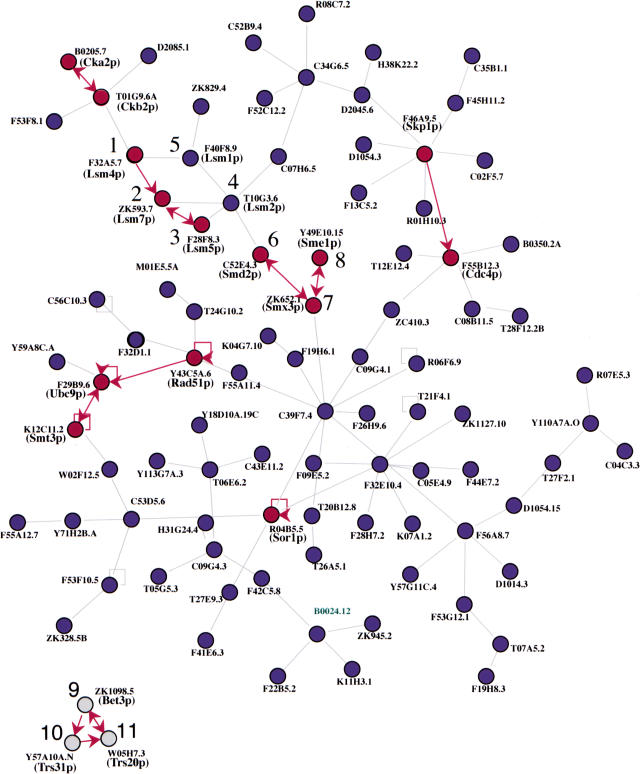
Figure 2.
A predicted network of Caenorhabditis elegans protein interactions involving a subset of the 216 pairs of protein interactions tested in our analysis. Corresponding yeast proteins names are shown in parentheses. Nodes represent proteins and edges represent an interaction between two yeast proteins. Interactions also detected in C. elegans are represented by red arrows. Interactions detected in both orientations are represented by double-headed red arrows. Interactions involving proteins 1–8 have been implicated in RNA processing in yeast and the interaction cluster (Nos. 9–11) involves three proteins whose potential yeast orthologs have been found in TRAPP complexes.
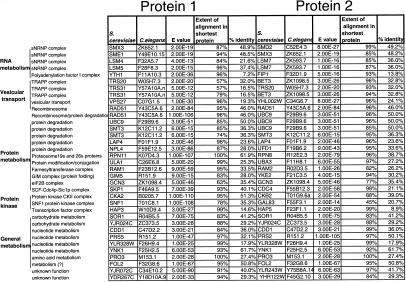
Table 2.
Summary of Yeast Two-Hybrid Results
The yeast protein pairs involved in the 35 conserved interactions could be divided into several general functional groups of core biological processes as described in the Yeast Protein Database YPD (http://www.proteome.com): (1) RNA metabolism, (2) vesicular transport, (3) protein metabolism (synthesis, modification, degradation), and (4) general metabolism. The C. elegans and S. cerevisiae protein names, their corresponding BLAST E-values, and the extent of the alignment in the shortest protein for each pair is shown.
This suggests that, using this approach, the minimal proportion of true interologs that can be detected between two species that are evolutionarily distant by about 900 million years is between 16% and 31%. By comparison, an average of five interactors per bait typically is obtained using a worm AD-cDNA library representing ∼19,000 genes (2.6×10−4) (Walhout et al. 2000a; Davy et al. 2001). Thus, the frequency of detection of interactions through searches for potential interologs is between 600- and 1100-fold higher than that obtained through conventional two-hybrid screens using random libraries. It should be noted that this approach allows direct testing of interactions that might otherwise be difficult to test in a random two-hybrid screen because of the biased representation of cDNA libraries toward highly expressed genes. In addition, the direct testing of an interaction between two proteins can be performed in 96 well setting and therefore is easily amenable to automation.
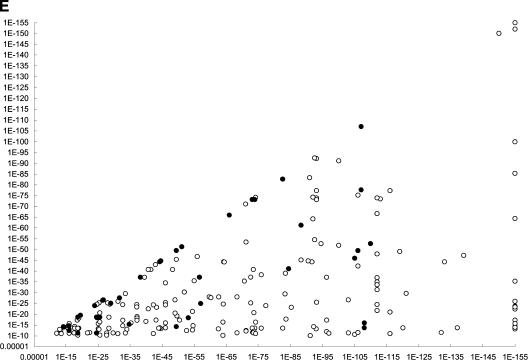
The E-value reported by BLAST when searching a database is a measure of the likelihood that the observed similarity could have occurred by chance. Thus, biologists can use this value to infer the homology likelihood. The BLAST E-value between the potential orthologous protein pairs tested in this analysis ranged between 10−10 and 10−151. One could expect that a potential interolog obtained with two low E-values is more likely to interact. Interestingly, no detectable correlation was found between the likelihood of homology (E-value) and the likelihood of an interaction being conserved between these two organisms (Fig. 1E). However, it is established that the three-dimensional structure of two proteins can be conserved despite considerable primary sequence divergence (Friedberg et al. 2000). In addition, it is possible that two interacting proteins may have coevolved such that only discrete interacting domains were conserved.
The data described above suggest that the approach of sequence-based searches for candidate interologs can be used globally to identify potential networks of interactions. However, such networks only can be considered as biological hypotheses. Hence, we investigated methods to generate reagents that can be used to study potential interaction networks identified by interolog searches. The reverse two-hybrid system provides a genetic selection that allows the rapid identification of cis-acting mutations or trans-acting molecules that dissociate potential interactions (Vidal 1997). The two-hybrid SPAL10::URA3 inducible reporter gene (Vidal et al. 1996) confers sensitivity to 5-Fluoroorotic acid (5-FOA). The dissociation of the yeast two-hybrid interaction confers a selective advantage allowing screens for dissociating compounds or for mutations that prevent the normal association of a protein pair using positive selection. Such reagents can be used back in vivo to characterize the role of the corresponding protein-protein interactions (Endoh et al. 2001).
To test the degree to which the reverse two-hybrid system can be applied to our network of identified interologs, we determined the percentage of the interactions described above that could be counter-selected on media containing 5-FOA (Vidal 1997). Starting from the 35 true worm interologs described above, 77% (27/35) of C. elegans interactions were detected as 5-FOA sensitive (Fig. 3). Because the reverse two-hybrid system can be automated (Endoh et al. 2001), it is possible that relatively large numbers of yeast two-hybrid interactions that emerge from interolog searches could indeed be tested back in the relevant biological settings.
Figure 3.
Assessment of the reverse two-hybrid system as a method to further study Caenorhabditis elegans protein interactions. Reverse two-hybrid system. Each yeast diploid expressing one of the 35 conserved interacting protein pairs from C. elegans was tested to identify those that are sensitive to 5-Fluoroorotic acid (5-FOA). Pairs are arranged in the order described in Table 2. Diploids were replica plated onto both SC-Leu-Trp (A) and SC -Leu, -Trp + 0.2% 5-FOA (B). Twenty-seven of thirty-five pairs (77%) were 5-FOA sensitive.
This work suggests that interaction maps from one species may be useful in predicting interactions in another species and may provide insight into the function of otherwise uncharacterized proteins. In addition, the identification of an interolog provides additional support for the validity of the initial interaction found in the “reference” species. This may be most meaningful if the only evidence for the original interaction comes, itself, from a high-throughput experiment. When the function of one of the proteins in the starting species has not been characterized, a “guilt by association” annotation also can be applied with more confidence. One potential therapeutic application of this approach would be to identify interactions conserved between a well-characterized model organism and a related pathogen. The majority of the interactions conserved between the distantly related species S. cerevisiae and C. elegans seem to be involved primarily in core metabolic processes. However, it is reasonable to predict that certain interactions conserved between more closely related organisms such as Drosophila and other insects such as mosquitoes, or C. elegans and other nematodes such as Ascaris lumbricoides, would be specific to the very closely related species. These interactions could be subjected to the reverse two-hybrid analysis (Vidal 1997) to identify reagents that are capable of dissociating them (Young et al. 1998). Such reagents might be valuable candidates for therapeutic agents.
METHODS
Potential Ortholog Identification
To identify potential orthologs of yeast proteins in C. elegans, we used BLASTP to search a C. elegans database generated from the ORF predictions in version WS7 of ACeDB. Only matches with E-values lower than 10−10 were considered. Although other studies (Snel et al. 1999) have considered as potential orthologs two proteins that are each other's best match in their respective genomes, we decided to systematically select as potential orthologs the best C. elegans matches for the yeast proteins. Indeed, orthology can be a one-to-many or many-to-many relationship (Tatusov et al. 1997). Thus, in the few cases where the same C. elegans protein is the best match for two distinct yeast proteins, we consider this C. elegans protein to be the potential orthologs for both yeast proteins. We recognize that this procedure allowed us to evaluate only one potential C. elegans ortholog per yeast protein.
PCR and Cloning
Complete C. elegans ORFs that correspond to 282 potential C. elegans orthologs were PCR amplified from a mixed-stage cDNA library (Walhout et al. 2000b). The control S. cerevisiae ORFs were amplified from S288C genomic DNA. PCR products were generated using ORF-specific primers designed by the program OSP (Hillier and Green 1991) and tailed with the AttB1 (5′) and AttB2 (3′) Gateway recombinational cloning sequences (Walhout et al. 2000a,b; Reboul et al. 2001). C. elegans and yeast ORFs were cloned into the Gateway Entry vector (INVITROGEN) as described with several modifications (Reboul et al. 2001).
Each ORF was PCR amplified from its corresponding Entry clone for subsequent cloning into both Gal4p DNA binding domain-fusion (DB) and Gal4p activation domain-fusion (AD) yeast two-hybrid vectors pGBT9 (Clontech) and pACT2 (Endoh 2000), respectively. ORFs to be cloned into the DB-fusion vector were amplified from Entry clones using the primers DB-B1: 5′TAGTAACAAAGGTCAAAGACAGTTGACTGTATCGTCGAGGTTGTACAAAAAAGCAGGCT-3′ and PGBT9.B2-TERM: 5′-AAATCAT AAATCATAAGAAATTCGCCCGGAATTAGCTTGGTTGTACAAGAAAGCTGGGT-3′. ORFs to be cloned into the AD vector were amplified from Entry clones using the primers AD-B1: 5′-CTATTCGATGATGAAGA TACCCCACCAAACCCAAAAAAAGAGTTGTACAAAAA AGCAGGCT-3′ and pACT2.B2-Term: 5′-TGAAGTGAACTTG CGGGGTTTTTCAGTATCTACGATTCATTTGTACAAG AAAGCTGGGT-3′.
Yeast Transformation/Gap-Repair and Mating
Yeast transformation/gap-repair reactions were performed in 96-well plates as described elsewhere (Walhout and Vidal 2001) with several modifications. MaV103 (MATa) cells were transformed with the 2 μm AD-fusion vector pACT2-GFP and the corresponding AD-PCR fragments. MaV203 (MATα) cells were transformed with the 2 μm DB-fusion vector pGBT9 and the corresponding DB-PCR fragments. Five microliters of each transformation mix were spotted in 96-spot format onto synthetic complete (SC) plates lacking leucine or tryptophane (SC-Leu or SC-Trp). Yeast carrying the AD-fusion constructs were mated to yeast cells carrying the DB-fusion constructs in “96-spot” format by replica plating both onto YEPD plates. To identify self-activator DB-fusion proteins, each transformant bearing a DB-fusion protein construct was mated to a transformant carrying an empty AD vector. Following 1 d of growth at 30°C, mating plates were replica plated to SC-Leu-Trp plates to select for diploids.
Yeast Two-Hybrid Analysis
Diploids were transferred to a 15-cm nylon filter on a YEPD plate for subsequent β-galactosidase assays and to an SC-Leu-Trp-Ura plate and an SC-Leu-Trp-His plate supplemented with 20 mM 3-aminotriazole (3AT) to assay for the SPAL10::URA3 and GAL1::HIS3 reporter gene activity, respectively. The identity of both hybrid proteins from diploids exhibiting a positive yeast two-hybrid phenotype was determined by sequencing the corresponding ORFs. The identity of all ORFs tested was verified by sequencing.
5-FOA Sensitivity
The two-hybrid inducible SPAL10::URA3 reporter gene drives expression of the URA3 gene, which is involved in uracil biosynthesis, and which also is able to catalyze the conversion of 5-FOA into a toxic compound. Thus, yeast diploids expressing interacting partners fused to AD and DB are able to grow on medium lacking uracil, but do not grow on medium containing 5-FOA. This allows for the screening of either dissociating compounds or interaction-defective alleles that will confer a positive growth selection. The 5-FOA negative selection is, however, dependent on the strength of the two-hybrid interaction. We identified interactions that are 5-FOA sensitive and thus suitable for reverse two-hybrid screening, by replica plating diploids onto both SC-Leu-Trp and SC-Leu-Trp + 0.2% 5-FOA.
Acknowledgments
We thank Marian Walhout and David Hill for helpful suggestions and critical reading of this manuscript. S.V. was supported by a fellowship from the Charles A. King Memorial Trust. This work was supported by grants 5R01HG01715–02 (NHGRI), P01CA80111–02 (NCI), 7 R33 CA81658–02 (NCI) and 232 (MGRI) awarded to M.V.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL marc_vidal@dfci.harvard.edu; FAX (617) 632-2425.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.205301.
REFERENCES
- Davy A, Bello P, Thierry-Mieg N, Vaglio P, Hitti J, Doucette-Stamm L, Thierry-Mieg D, Reboul J, Boulton S, Walhout AJM, et al. A protein-protein interaction map of the C. elegans 26S proteasome. EMBO Reports. 2001;2:821–828. doi: 10.1093/embo-reports/kve184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du W, Vidal M, Xie J-E, Dyson N. RBF, a novel RB-related gene that regulates E2F activity and interacts with cyclin E in Drosophila. Genes & Dev. 1996;10:1206–1218. doi: 10.1101/gad.10.10.1206. [DOI] [PubMed] [Google Scholar]
- Endoh H, Walhout AJM, Vidal M. GFP-reverse two-hybrid system: Application to the characterization of large numbers of potential protein-protein interactions. Methods Enzymol. 2000;328:74–88. doi: 10.1016/s0076-6879(00)28391-2. [DOI] [PubMed] [Google Scholar]
- Endoh, H., Vincent, S., Jacob, Y., Réal, E., Walhout, A.J.M., and Vidal, M. An integrated version of the reverse two-hybrid system for the “post-proteomic” era. Methods Enzymol. in press. [DOI] [PubMed]
- Flajolet M, Rotondo G, Daviet L, Bergametti F, Inchausp G, Tiollais P, Transy C, Legrain P. A genomic approach of the hepatitis C virus generates a protein interaction map. Gene. 2000;242:369–379. doi: 10.1016/s0378-1119(99)00511-9. [DOI] [PubMed] [Google Scholar]
- Friedberg I, Kaplan T, Margalit H. Evaluation of PSI-BLAST alignment accuracy in comparison to structural alignments. Protein Sci. 2000;9:2278–2284. doi: 10.1110/ps.9.11.2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromont-Racine M, Rain JC, Legrain P. Toward a functional analysis of the yeast genome through exhaustive two-hybrid screens. Nature Genetics. 1997;16:277–282. doi: 10.1038/ng0797-277. [DOI] [PubMed] [Google Scholar]
- Guthrie C. The spliceosome is a dynamic ribonucleoprotein machine. Harvey Lect. 1994;90:59–80. [PubMed] [Google Scholar]
- Hillier L, Green P. OSP: A computer program for choosing PCR and DNA sequencing primers. PCR Methods Appl. 1991;1:124–128. doi: 10.1101/gr.1.2.124. [DOI] [PubMed] [Google Scholar]
- Ito T, Tashiro K, Muta S, Ozawa R, Chiba T, Nishizawa M, Yamamoto K, Kuhara S, Sakaki Y. Toward a protein-protein interaction map of the budding yeast: A comprehensive system to examine two-hybrid interactions in all possible combinations between yeast proteins. Proc Natl Acad Sci. 2000;97:1143–1147. doi: 10.1073/pnas.97.3.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M, Sakaki Y. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc Natl Acad Sci. 2001;98:4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcotte EM, Pellegrini M, Ng HL, Rice DW, Yeates TO, Eisenberg D. Detecting protein function and protein-protein interactions from genome sequences. Science. 1999;285:751–753. doi: 10.1126/science.285.5428.751. [DOI] [PubMed] [Google Scholar]
- McCraith S, Holtzman T, Moss B, Fields S. Genome-wide analysis of vaccinia virus protein-protein interactions. Proc Natl Acad Sci. 2000;97:4879–4884. doi: 10.1073/pnas.080078197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrini M, Marcotte EM, Thompson MJ, Eisenberg D, Yeates TO. Assigning protein functions by comparative genome analysis: Protein phylogenetic profiles. Proc Natl Acad Sci. 1999;96:4285–4288. doi: 10.1073/pnas.96.8.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rain JC, Selig L, De Reuse H, Battaglia V, Reverdy C, Simon S, Lenzen G, Petel F, Wojcik J, Schachter V. The protein-protein interaction map of Helicobacter pylori. Nature. 2001;409:211–215. doi: 10.1038/35051615. [DOI] [PubMed] [Google Scholar]
- Reboul J, Vaglio P, Tzellas N, Thierry-Mieg N, Moore T, Jackson C, Shin-i T, Kohara Y, Thierry-Mieg D, Thierry-Mieg J, et al. Open-reading-frame sequence tags (OSTs) support the existence of at least 17,300 genes in C. elegans. Nat Genet. 2001;27:332–336. doi: 10.1038/85913. [DOI] [PubMed] [Google Scholar]
- Snel B, Bork P, Huynen MA. Genome phylogeny based on gene content. Nat Genet. 1999;21:108–110. doi: 10.1038/5052. [DOI] [PubMed] [Google Scholar]
- Tatusov RL, Koonin EV, Lipman DJA. A genomic perspective on protein families. Science. 1997;278:631–637. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- The C. elegans Genome Sequencing Consortium. Genome sequence of the nematode C. elegans: A platform for investigating biology. Science. 1998;282:2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight JR, Lockshon D, Narayan V, Srinivasan M, Pochart PA, et al. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- Vidal, M. 1997. The reverse two-hybrid system. In The Yeast Two-Hybrid System (eds. Bartel, P. and Fields, S.), pp. 109–147. Oxford University Press, New York.
- Vidal M, Endoh H. Prospects for drug screening using the reverse two-hybrid system. Trends in Biotechnology. 1999;17:374–381. doi: 10.1016/s0167-7799(99)01338-4. [DOI] [PubMed] [Google Scholar]
- Walhout AJM, Sordella R, Lu X, Hartley JL, Temple GF, Brasch MA, Thierry-Mieg N, Vidal M. Protein interaction mapping in C. elegans using proteins involved in vulval development. Science. 2000a;287:116–122. doi: 10.1126/science.287.5450.116. [DOI] [PubMed] [Google Scholar]
- Walhout AJM, Temple GF, Brasch MA, Hartley JL, Lorson MA, van den Heuvel S, Vidal M. Gateway recombinational cloning: Application to the cloning of large numbers of open reading frames or ORFeomes. Methods Enzymol. 2000b;328:575–592. doi: 10.1016/s0076-6879(00)28419-x. [DOI] [PubMed] [Google Scholar]
- Walhout AJM, Vidal M. High-throughput yeast two-hybrid assays for large-scale protein interaction mapping. Methods. 2001;24:297–306. doi: 10.1006/meth.2001.1190. [DOI] [PubMed] [Google Scholar]
- Young K, Lin S, Sun L, Lee E, Modi M, Hellings S, Husbands M, Ozenberger B, Franco R. Identification of a calcium channel modulator using a high-throughput yeast two-hybrid system. Nature Biotechnology. 1998;16:946–950. doi: 10.1038/nbt1098-946. [DOI] [PubMed] [Google Scholar]