Abstract
RNA molecules are important cellular components involved in many fundamental biological processes. Understanding the mechanisms behind their functions requires RNA tertiary structure knowledge. While modeling approaches for the study of RNA structures and dynamics lag behind efforts in protein folding, much progress has been achieved in the past two years. Here, we review recent advances in RNA folding algorithms, RNA tertiary motif discovery, applications of graph theory approaches to RNA structure and function, and in silico generation of RNA sequence pools for aptamer design. Advances within each area can be combined to impact many problems in RNA structure and function.
Keywords: RNA folding, RNA tertiary motifs, RNA graphs, in vitro selection
Introduction
Two of the most astonishing biological discoveries of the past decade have been the relatively small number of human genes and the fact that most of the human genome is transcribed and associated with regulatory RNAs. The latter has led to a paradigm shift in our understanding of biological regulation. Deciphering the functions of these regulatory RNAs presents a challenge for the new decade, with many biomedical and technological applications.
Complementing such functional interpretations are efforts to characterize the structures of RNAs over many functional classes spanning sizes from those associated with micro RNAs to large ribosomal systems. The greater structural diversity of RNAs compared to proteins – roughly 11 backbone torsional degrees of freedom for RNA building blocks compared to 2 for proteins –combined with the complex possible packing arrangements of RNA's many secondary-structural elements –double-stranded helices and single-stranded loops, bulges, and hairpins –poses a challenge to computation. Moreover, the sensitivity of RNA structures to ions, solvent, metabolites, and other biomolecules has made RNA structure determination at atomic resolution more difficult than for proteins.
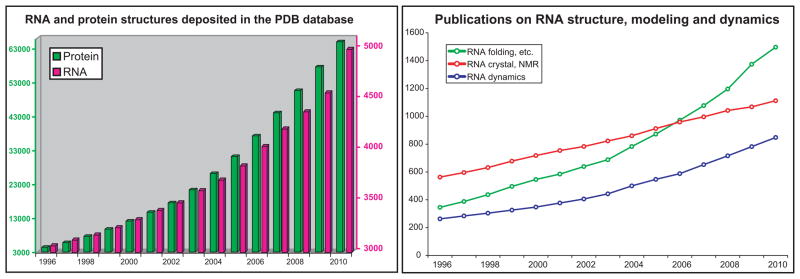
Yet, in recent years, the increasing recognition of RNA's prominence in gene control has led to impressive advances, on both the experimental and computational fronts, concerning RNA secondary and tertiary structure determination and motif analysis; synthetic and engineered RNA design; and applications of RNA to bioengineering, medicine, and nanotechnology [1–3]. See Figure 1 for a perspective of the increasing structure information for RNA as well as published papers in RNA modeling. In this review, we summarize progress in the past two years in the areas of computational approaches to RNA tertiary structure prediction, analysis of RNA tertiary motifs, graph theory approaches for RNA, and RNA design efforts that attempt to improve upon experimental in vitro selection for aptamer design. We also mention studies relevant to RNA structural comparison, since the notion of RMSD (root-mean-square-deviation) for RNA is insufficient at this stage where predictions are not accurate. For recent reviews on these topics, see [4–7], and for a general perspective on improvements in biomolecular modeling and simulation, see [8]. Note that there are many other areas of advances on RNA bioinformatics, for example secondary structure predictions [9], not covered here.
Figure 1.
(left) Number of RNA structures deposited in the NDB nucleic acid database (http://ndbserver.rutgers.edu/) as of December 2010. Note different scales used for protein (left) and RNA (right); (right) The number of scientific publications by year whose title contains the words “RNA folding”, “RNA structure prediction”, “RNA modeling”, or “modeling RNA structure” are colored in green; the words “RNA crystal” or “RNA NMR” are colored in red; and the words “RNA dynamics”, “RNA simulation”, “ribosome dynamics”, or “ribosome simulations” are colored in blue. The word search was done using the ISI web of knowledge (www.isiknowledge.com/).
RNA tertiary structure prediction
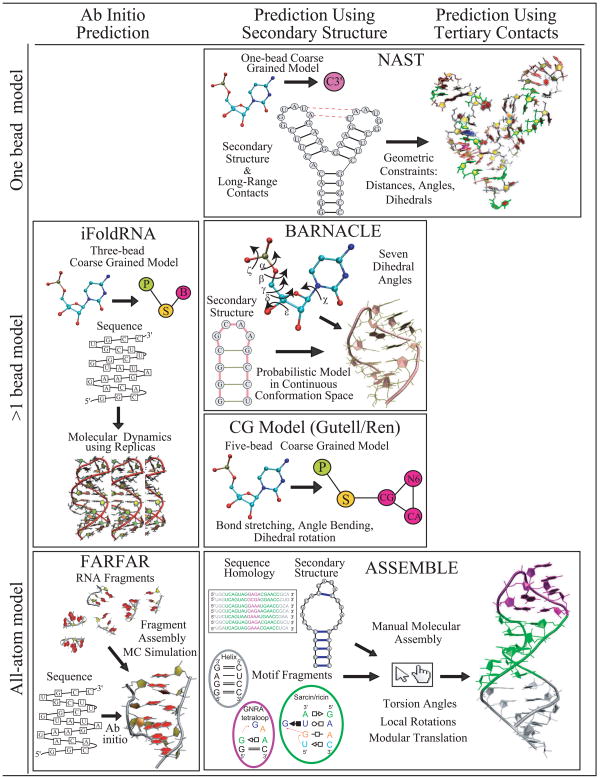
Compared to protein folding, the RNA folding problem is at an early stage: current 3D RNA folding algorithms require manual manipulation or are generally limited to simple structures in terms of size and topology. However, many groups have now been tackling this problem by a variety of techniques as represented in programs like NAST [10], BARNACLE [11], FARFAR [12], and others (see Fig. 2). These methods differ in the input data, prediction accuracy, and nucleotide representation – from one pseudo-atom per nucleotide model to all-atom detail. Our recent review [5] examined the performance of 3D structure prediction algorithms, namely iFoldRNA [13], FARNA [14], NAST [10], and MC-SYM [15], for an RNA dataset of 43 structures of various lengths and motifs. We found that most predictions have large root-mean square deviations from the crystal structure (e.g., RMSD>6Å). While the prediction accuracy improves with added knowledge from the 2D structure and 3D contacts, the lack of appropriate functions that favor compact RNA-like structure and the failure to detect long-range contacts remain clear challenges. Below we elaborate upon the most recent approaches.
Figure 2.
Examples of recent RNA 3D folding computer programs. The different algorithms are organized by their input data (ab initio or sequence, secondary structure, 3D contacts), as well as the level of model detail (from one-bead coarse grained models to all atom approaches).
The nucleic acid simulation tool or NAST developed by Jonikas et al. [10] is a molecular dynamics simulation tool consisting of a knowledge-based statistical potential function applied to coarse grained model with resolution of one bead per nucleotide residue. NAST requires secondary structure information and, if available, accepts tertiary contacts to direct the folding. NAST’s greatest strength is that it allows modeling of large RNA molecules (e.g., 160 nt), a limitation imposed by most programs. Overall, when only secondary structure information is considered, accurate prediction is limited to RNA structures with simple topologies such as hairpins with less than 34nt (8Å average RMSD) [5]. However, when input information from tertiary contacts is additionally provided, NAST can dramatically improve prediction accuracy.
The web-based program iFoldRNA by the Dokholyan group [13] predicts RNA structures using a coarse grained model of three beads per nucleotide through molecular dynamics (MD) sampling (by the replica exchange, or REMD method). iFoldRNA does not require secondary structure information and can rapidly predict structures for small RNAs (<50nt). However, as the RNA size increases, difficulties arise regarding long-range tertiary contacts (23Å average RMSD) [5]. The Dokholyan and Weeks groups recently improved their method by incorporating information regarding secondary and tertiary contacts based on experimental SHAPE chemistry to alleviate these limitations [16]. Prediction accuracy of 4Å RMSD for a 75 nt tRNA-Asp was reported as the best performance. This improvement is noteworthy, because a typical ab-initio prediction with iFoldRNA for a tRNA gives an accuracy of about 21Å RMSD [5].
Other coarse grained models such as recently presented by Fresllsen et al. [11] as an extension of their protein approach are one way to address conformational sampling limitations encountered in fragment assembly algorithms such as FARNA [14] and MC-SYM [15], which generate 3D structure models from small-residue fragments. Fresllsen et al. argue that the discrete nature of the fragment assembly method leads to sampling bottlenecks. Their alternative, a probabilistic model called BARNACLE, to RNA conformational space represents RNA conformational flexibility using circular analogues to Gaussian distributions (von Mises distributions) and multi-dimensional sampling. The continuity of the local conformational space allows for less biased sampling. Using secondary structure information, BARNACLE generates reasonable RNA-like structures (<10Å RMSD) for small RNA molecules (<50nt). However, most RNA structures with elaborate topologies such as junctions and long-range contacts are longer than 50nt and cannot be predicted due to an increase in complexity of the probabilistic model.
Another coarse grained approach by the Gutell/Ren groups [17] takes a more standard mesoscale (5 pseudo-atoms per nt) modeling approach in which simplified models are sampled by MD/simulated annealing with the guidance of atomistic, physics-based potential functions, here with optimized non-bonded parameters. Results for small RNAs (<30 nt) indicate great promise (3.36Å average RMSD). However, as in the case of BARNACLE, this method is limited to RNA molecules with relatively simple topologies.
Das et al. introduced a 2010 update to Rosetta's Fragment Assembly of RNAs (FARNA [14]) termed FARFAR that adds a refinement phase for atomic-level interactions [12]. Relying on the framework found successful for proteins using atomistic models and empirical potential functions assembled from conformational preferences represented in structural databases, FARFAR is only applicable to small RNA (6–20nt), and has variable accuracy, from excellent, less than 1Å RMSD, to more than 10Å (for 4-way junctions) when measured for cluster centers; results improve by up to 5Å when best clusters of refined conformations are used instead for final assessment. Standard sampling difficulties and convergence failures reflect both algorithmic and force-field limitations.
An alternative approach to automated programs is ASSEMBLE, a manual-input program by Jossinet et al. [18] that, like RNA2D3D (Martinez et al. [19]), uses secondary or tertiary structure information from homologous RNAs to build a first-order approximation RNA 3D model. Using an intuitive graphical interface, ASSEMBLE allows the manual insertion of base pairs and 3D motifs, as well as torsion angle modifications, rotations, and translations of modular elements. ASSEMBLE permits the input of electron density maps to improve the RNA model. While these user-input tools are practical, they rely on manual application of expert knowledge. Unfortunately, there are only a few of these experts.
In general, for RNA sequences of medium to large sizes (50-130 nt), even the best prediction methods lead to large RMSD values (20Å on average), considered poor for protein predictions. Alternatives to RMSD measurements for RNAs have thus been suggested for RNA structural comparisons. Parisien et al. recently introduced an interaction network fidelity measure that combines RMSD with counts of predicted base-pairing and base-stacking rates [20]. Similarly, Hajdin et al. [21] proposed assessing the global fold of an RNA at the nucleotide resolution by measuring a more “relative” RMSD value. For example, for a de novo prediction of 100nt RNA, the RMSD should be within 25Å of the accepted structure to reach a p-value of P ≤ 0.001 (the p-value is a measure of statistical significance). Such alternative comparison approaches should help distinguish successful models with RNA-like features from less successful predictions.
Overall, the accuracy of each program varies from structure to structure. For RNA sequences of small size (<20 nt), FARFAR can produce the best prediction model. If structural data obtained from experimental methods such as SHAPE chemistry is available, NAST and iFoldRNA can dramatically improve their prediction accuracy. For medium sized RNAs, MC-SYM can produce reasonable models, but accuracy is limited by the difficulty of predicting long-range contacts. Because the programs described above are recent, many improvements can be anticipated in the near future.
Tertiary motif discovery
Structural and comparative studies have suggested that RNA structure is largely modular composed of repetitive building blocks or motifs. These patterns appear at the primary, secondary, and tertiary structure levels. For instance, the secondary structure motifs such as hairpins, junctions, internal loops, and stems define hydrogen-bonding patterns. At the tertiary level, RNA tertiary (3D) motifs are recurrent structural elements subject to multiple RNA-RNA interactions constraints as described by Nasalean at el. [22]. RNA 3D motifs play an important role in RNA folding and biochemical functions, and determining 3D motifs provides a better understanding of the principles of organization of complex RNA structures, as well as serves as foundation for applications to synthetic biology and nanodesign [1,23,24].
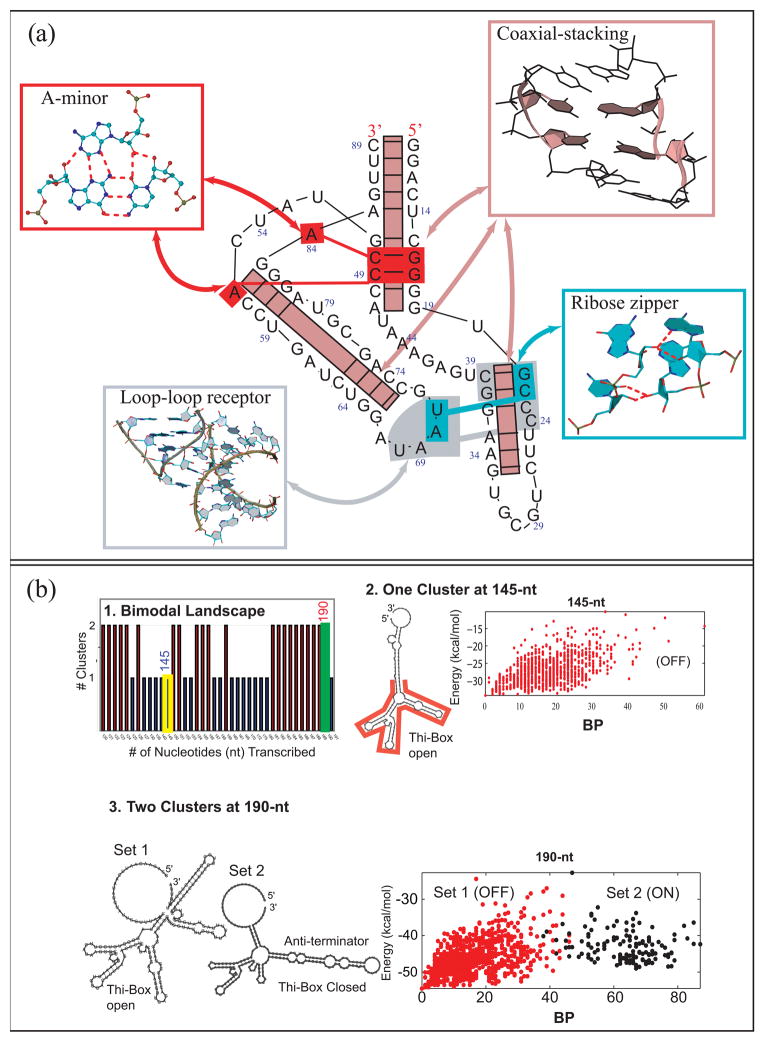
The RNA-RNA interactions that define 3D motifs include base-pairing, base-stacking, and base-backbone interactions [25]. In terms of base pairing patterns, RNAs possess remarkable versatility: base pair interactions can be classified into twelve geometric families in terms of pairs of interacting edges, which can be Watson and Crick, Hoogsteen, Sugar, and glycoside bond orientation cis and trans, as classified by Leontis and Westhof [26]. RNA base pairs have been recently analyzed in detail by Stombaugh et al. [27] in terms of their isosteric properties, that is, similar base pair interactions that can be substituted by compensatory mutations (e.g., a GC base pair can be substituted by an AU base pair), and a revised base pair catalog is now available [27]. Furthermore, base-backbone interactions are also common, and a recent classification model has been proposed by Zirbel et al. [28] based on phylogenetically conserved base-phosphate interactions. This study also determined 10 family types of base-phosphate interactions based on their hydrogen bond interaction patterns. Recent works by the Leontis [28], Schlick [29], and the James [30] groups have identified new RNA backbone interaction motifs involving both the sugar ribose and the phosphate group with the bases. These interactions have different functional roles including RNA-protein recognition sites and stabilization of the global RNA structure via helix-packing interactions. Such combinations of RNA-RNA interactions make up recognized 3D motifs, such as the A-minor, ribose zipper, and loop-loop receptor interactions (Fig. 3a).
Figure 3.
(a) Annotated diagram of the TPP riboswitch (PDB: 2GDI) shows several correlated motifs working in a cooperative way to stabilize RNA’s 3D conformation. These key motifs can be observed often in many other RNA structures. (b) TPP Riboswitch folding as a function of sequence elongation reveals: (1) either one or two conformational clusters of all suboptimal states for each sequence length. (2) At 145 nt, one cluster is apparent where the folding funnel (base pair difference against free energy plotted for the ensemble for all predicted suboptimal structures) shows a classic simple folding funnel landscape. (3) Near the full sequence length (190 nt), two clusters correspond to the two conformations. See [48] for details.
A new way of thinking about RNA 3D motifs – as elements of higher-order motifs (or supermotifs) –has also emerged from analysis of solved RNA structures by Xin et al. [31] and others [32] (Fig. 3a). Often, 3D motifs appear together, working cooperatively. For instance, the newly adenosine wedge motif by Gagnon and Steinberg [33] combines the along-groove packing, A-minor, and hook-turn motifs. Similarly, cooperation between A-minor and coaxial stacking motifs occur in most large RNAs, particularly in junctions as described by the Westhof [34] and Schlick groups [35].
The guiding and stabilizing roles that RNA motifs serve [32] has also emerged from studies of RNA junction topologies. Analyses by the Westhof [34] and Schlick [35] groups have described 3 and 9 major families for 3 and 4-way junctions, respectively. Higher-order junctions were also described as composed of these basic architecture motifs [29]. Helices within junctions tend to arrange in highly-ordered patterns (parallel and perpendicular), and conformations are stabilized using common 3D motifs like coaxial stacking, loop-helix interaction, and helix-packing interaction. Bailor et al. [36] further showed that secondary structure features such as loop size encode topological constraints on the 3D helical orientations of internal loops. This suggests that long-range contacts serve to stabilize specific helical conformations associated with the native structure within the topologically allowed ensemble.
As more structural information becomes available, a more complete repertoire of RNA motifs will help us better appreciate intricate interaction properties of RNA, which in turn will translate to a better understanding of RNA function.
Applications of graph theory to RNA
Graph theory is a field of mathematics widely used for analyzing various types of relationships (or networks), including chemical structures, genetic and biochemical objects. Graph theory has also proven valuable for analyzing RNA secondary structures, as pioneered by Waterman in 1978 and extended by many others, as well as for analyzing tertiary structures of RNAs. Graph representations of RNA can help address many fundamental biological questions concerning the organization, classification, and design of RNA motifs.
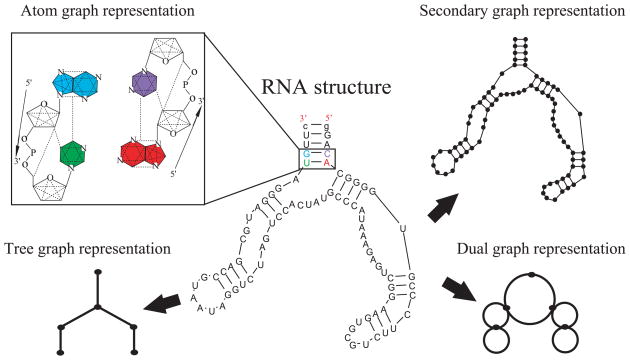
To represent RNA secondary structures, graph objects used include tree graphs, dual graphs, and secondary structure graphs (see Fig. 4); both dual and secondary structure graphs can represent RNA motifs with pseudoknots. The rules that define dual and tree graphs involve a translation of each secondary structure element (stems, bulges, junctions, and loops) into a vertex or an edge (Fig. 4). The edge/vertex specification is reversed for dual graphs with respect to tree graphs. Secondary structure graphs define instead each nucleotide as a vertex and the backbone and base pairs as edges. In addition, a finer atom graph representation for RNA tertiary structures is available, where vertices represent atoms and edges represent covalent, strong non-covalent bonds and angle constraints (Fig 4). The common advantage exploited by these approaches is that graph theory reduces the size of RNA space enormously, from the sequence space size of 4N where N is the number of nucleotides, to motif space, which is vastly smaller and grows much slower with size, as described, for example, by Gan et al. [37].
Figure 4.
Graphic rules for converting RNA 2D structures using tree (lower left), dual (lower right), and secondary structure (upper right) graphs. In addition the atom graph representation (upper left) can characterize RNA 3D structures.
One such RNA topology resource developed by the Schlick group, RAG (RNA-As-Graphs) (http://www.biomath.nyu.edu/rna/), is being used to classify/analyze topological characteristics of existing RNAs [38,39] and to design and predict novel RNA motifs [39-42]. Specifically, RAG and the associated graph theory framework for RNA [37,43] have been used to classify and catalog RNA motifs [38,39], estimate the size of RNA's structural/functional repertoire [39], detect structural and functional similarity among existing RNAs [44], identify RNA motifs of antibiotic-binding aptamers (found synthetically) in genomes [45,46], analyze the structural diversity of random pools used for in vitro selection of RNAs [47], simulate aspects of the process of in vitro selection in silico [40–42], and analyze RNA thermodynamics landscapes to better understand riboswitch mechanisms to ultimately enhance their design [48].
Cataloging based on graph theory enumeration suggests that the RNA structure universe is dominated (more than 90%) by pseudoknots, in agreement with available data by Kim et al. [39]. Cataloging has also led to RNA design [39], by a build-up procedure combined with clustering approaches. Specifically, statistical clustering techniques are employed to separate graphs that are “RNA-like” from those that do not resemble natural RNAs on the basis of quantitative graph descriptors. Such clustering, though highly approximate, can suggest new RNA-like motifs as design candidates. Candidate sequences that fold onto such RNA-like motifs can then be predicted using a build-up procedure that combines sequences for motif subsegments known from RNAs in nature with an algorithm for secondary prediction based on base-pairing thermodynamics [49]. Significantly, among 10 specific designed new RNA motifs, five have since been discovered (Kim et al., unpublished, see also Fig 7.13 of [50]). The large increase in solved RNAs in recent years has also made it possible to compare theoretical predictions of RNA-like and non-RNA-like motifs from 2004 to current RNA databases. Overall, the larger percentage of new RNAs from the theoretical RNA-like class (70%) compared to percentage of new RNAs from the theoretically-predicted non-RNA-like class (30%) shows promise in using graph-theory based cataloging and design of new RNA motifs based on a modular, build-up strategy.
Graph theory tools are also natural for comparing RNA structures to find existing RNA motifs within large RNAs based on graph isomorphisms [44]. This idea was applied to identify topological similarities among existing RNA classes and to define motifs of RNA within larger RNA topologies for major RNA classes (e.g., tRNA, tmRNA, hepatitis delta virus RNA, 5S, 16S, 23S rRNAs).
Inspired by RAG, Koessler et al. [51] implemented a predictive model to filter RNA-like structures from non-RNA-like structures that result from multiple solutions during RNA secondary structure prediction. The method uses tree graphs and computes graph theoretic values as input for a neural network to determine the best or most likely secondary structure candidates among all possible outcomes. Another web-based tool termed GraPPLE [52] applies secondary graph representations to identify and classify non-coding RNAs from sequences by using graph properties that capture structural features of functional RNAs.
Graph theory has been also useful for RNA structure prediction. For instance, Gillespie et al. [53] considered RNA backbones as polygonal curves on the 3D triangular lattices to represent RNA structures and simulated the folding of RNA structures, including pseudoknots, by considering only the 3D conformations that can realize pseudoknots structures in the 3D space given the base pair restrictions.
In a different application, Fulle and Gohlke [54,55] use the more detailed atom graph representation to analyze the flexibility of RNA structures by constraint counting. This is possible because sufficiently strong forces, which are included in the graph representation by edges, reflect rigidity and flexibility. Fulle and Gohlke applied constraint counting on this type of graph to reveal the static properties of the ribosomal exit tunnel and its functional role in cotranslational peptide folding [56]. Their method identifies large parts of the tunnel neighboring regions as rigid, with clusters of flexible tunnel components in the peptidyl transferase center, tunnel entrance and exit region [56]. Analyses of the rigidity of RNA structures, at both the local and global levels, can therefore help interpret biological functions of RNAs.
Graph theory is expected to continue to be a useful tool for representing, analyzing, and designing RNAs; such applications also provide exciting research opportunities in biology for mathematical scientists.
In Silico Generation of RNA Sequence Pools for Aptamer Design
The versatility of RNA structures and functions has also stimulated systematic efforts in the design of RNAs with tailored functions for a variety of medical and technological applications. In vitro selection technology has been widely used for discovering new synthetic RNAs of desired functions, such as high affinity and selectivity to a range of targets, including antibiotics, proteins, and even whole cells [57]. Essentially, this experimental procedure termed SELEX (Systematic Evolution of Ligands of Exponential Enrichment) [57,58] involves generation and screening of large (around 1015) sequence pools for binding and catalysis followed by amplification by PCR. However, practice quickly showed that RNA random pools are not structurally diverse as might be expected; this was also demonstrated computationally by generating random pools, “folding” them in 2D, and analyzing/grouping their folds by graph motifs [47]. Indeed, it was shown that simple topologies are favored, complex motifs are rare, and motif distribution depends on the sequence length (e.g., a 60-nt pool has a different distribution than a 100-nt pool). Novel approaches have thus been developed to expand the sampling of sequence space and thereby enhance motif diversity and complexity.
Random pool generation and analysis lends itself naturally to computation. For example, analytical frameworks for estimating motif probabilities were described by the Cedergren [59,60], and Schlick [45] groups. Motif scanning programs like RNAMotif [61] have been crucial to such efforts. The large size of the random pool, however, has been a challenge until recently, where several in silico approaches have begun to approach the experimental pool size (e.g., order 1014 sequences of size 60–100nt).
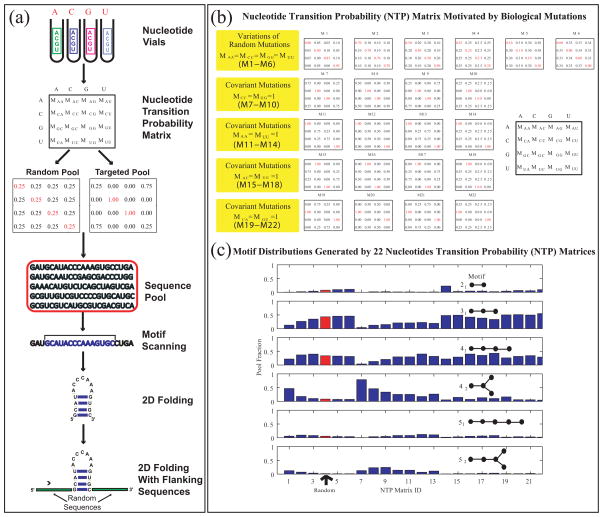
A simple mathematical approach using 4 × 4 “nucleotide transition probability” matrices (see Fig. 5a) to generate large pools of desired composition has been developed by the Schlick group [40], along with a web server tool, RAGPOOLS, (http://rubin2.biomath.nyu.edu) [42] that helps design structured pools for optimal yield of specific motifs. Such matrices specify the mixing ratios of nucleotides in the nucleotide vials (Fig. 5a) as applied to an initial sequence; different linear combinations of various matrices can be used to generate different motif distributions. That is, instead of uniform ratios in the nucleotide vials, different ratios can be introduced via design strategies (based on covariance mutations like AU to CG or conversion of AU to CG base pairs) produce different sequence pools (Fig. 5b) [42], for example aimed at specific motifs. Motif yield can be enhanced as desired by using more types of pools (basis matrices).
Figure 5.
(a) In Silico Approach to Pool Design. RNA sequence pool generation can be simulated using the nucleotide transition probability matrix, which specifies nucleotide mixtures in the nucleotide vials (or mutation rates for all nucleotide bases). The matrix composition can be defined to be a random matrix, corresponding to experiments, or to mimic specific biological situations, as shown in (b). The resulting sequences can be “folded”' into 2D structures using existing algorithms and analyzed further to screen and filter the candidates against a target motif. The non-random matrices shown in (b) form a basis of 22 probability matrices that generate a wide range of RNA motifs in silico [40], as shown in (c), where motifs yields are organized by RAG graph labels, and the yields of random matrices are shown in red.
Using this new tool and supercomputing (IBM Blue Gene) resources, very large pools of nucleotides (up to 1014) can be generated, screened, and filtered according to various 2D-structure similarity and flanking sequence analyses as shown by Kim et al. [41]. Such computational and theoretical yields agree for simple RNA motifs. For real aptamer targets, the in silico procedure overestimates the yields found experimentally, as expected, because experimental yields represent lower bounds and the screening does not yet involve 3D structural aspects. This targeted design approach has also shown promise for enhancing the selection of ligase enzymes [41].
Luo et al. [62] recently reported a complementary, directed-evolution-like approach for junction applications: “random filtering” to enrich 5-way junctions by repeatedly mutating junction motifs, and “genetic filtering” to produce pools with given motif distribution by a similar evolutionary approach. Similar in spirit is the patterned libraries approach by Ruff et al. [63] where a specific pattern like alternating purines and pyrimidines is generated by inserting random bases between single-stranded regions. It was shown to improve upon random pools in terms of yield and binding affinities. However, this approach requires many iterations of motif enrichment. Chushak & Stone [64] combine many of the above ideas in a multi-step large-pool generation approach for selection of RNA aptamers that uses 2D pattern searching, 3D structure generation, and screening for target binding using docking programs. They can thus reduce an initial set of order 1013–1014 candidates by nine orders of magnitude to propose RNA sequences which can be used for RNA microarray applications for aptamer design (e.g., [65]), an emerging alternative to in vitro selection technology.
Other recent interesting mathematical works that analyze aspects of in vitro selection include analyses of the dependence of aptamer affinity on magnesium ions and aptamer sequence patterns. The former study by Carothers et al. [66] reveals that tighter-binding aptamers are more robust, that is, less dependent on magnesium. The latter study by the Knight group [67] demonstrated that natural and artificial RNAs share similar sequence features, including a purine preference and GC bias.
Current challenges in the in silico approach to aptamer discovery and design remain efficient implementations for very large pool sizes, 3D structural characterization of the products to complement the 2D motif analysis, and estimating binding/catalysis properties. The pool size issue is straightforward to address with increasing computing speed and the ready availability of coupled networks for parallel computations. Advances in the prediction of tertiary RNA structure require more conceptual breakthroughs, to address the global positioning of RNA's secondary-structure elements. For evaluation of binding affinities and aptamer selectivity, standard molecular protocols for computing binding free energies and further assessment by molecular dynamics (MD) simulations have been used, but their reliance on approximate tertiary structures, free energy uncertainties, and limited sampling in MD [68] are sources of inaccuracies. With improvements, structural, flexibility and binding affinities could be systematically measured, as demonstrated by Anderson & Mecozzi [69] in an interesting application that sought to define the minimal RNA length required for selective binding to target aptamers by repeated rounds of sequence adjustments, MD simulations, and free energy calculations.
The ideas of directed evolution, sequence mutation, and tailoring presented above lend naturally to more global concepts of design involving the notion of energy landscapes. For proteins, statistical-mechanics frameworks based on density of states, pioneered by Frauenfelder, Wolynes, Dill, Onuchic, Thirumalai and others (e.g., [70-72]), have been invaluable in interpreting various protein kinetics and thermodynamics observations such as conformational substates, folding mechanisms, and function.
Recently, Pitt et al [73] introduced the notion of empirical RNA fitness landscapes by a combination of experiment and computation to analyze the optimization of typical SELEX products in terms of sequence/function relationships for small RNAs (up to 13nt). Such genotype/phenotype mapping are of general interest and practical importance.
Similar in spirit, though approached quite differently, are other computational approaches for riboswitch design. Riboswitches are RNAs which modulate gene expression by ligand-induced conformational changes [74,75]. However, the way in which their intrinsic sequences dictate such alternative folding pathways remains unclear. Shu et al [76] present a web tool that attempts to engineer temperature-sensitive allosteric RNAs through analysis of melting curves for designed sequences; the different minima on the melting curve correspond to two riboswitch conformations. Examining sequence length instead of temperature, Quarta et al [48] approach riboswitch design and analysis from a different point of view: identifying intrinsic sequence windows that favor one conformation over another as the enzyme elongates to full length, thereby mimicking the natural transcriptional process. They computed energy landscapes corresponding to secondary structures of RNA for the TPP riboswitch1 as a function of sequence length from 120 to 190nt (Fig. 3b); for each riboswitch length, the energy landscape is defined by the spread of accessible conformations at a range of energies: energy of that state vs. a distance measure between that conformation and all other conformations. Intriguingly, it was found that, depending on the sequence length, or time of transcription, the energy landscape may be populated by one or two configurational clusters, representing the opposing biological functions (Fig. 3b). This bimodal landscape suggests two low-energy states separated by an energy barrier; the metabolite acts to guide the RNA into one conformation by affecting the height of the energy barrier [48]. This thermodynamic switch, now found common to other riboswitches (Quarta & Schlick, unpublished), suggests a new avenue for riboswitch
Clearly, the many approaches described above can be used in concert with experimental technology to guide the procedures in a more targeted fashion and focused manner, to generate specific and/or complex RNA motifs. Laserson et al. have also shown that many of these designed synthetic RNAs have natural analogues [45], and this opens new avenues for discovery via genome analysis.
Conclusions
In recent years many computer algorithms have been reported in RNA modeling. Methodologies range from coarse grained spatial models to finer all-atom simulations, and the protocols can handle input data from none to secondary and partial tertiary contacts. It is encouraging that many of these advances have been achieved with relatively simple (coarse grained) models, which combine energy or statistical potentials, fragment assembly, and continuous conformational sampling techniques.
While all methods have strengths and weakness as recently reviewed [5], tertiary structures of small RNAs (<20nt) are better achieved with all-atom knowledge-based approaches (e.g., FARFAR) while those of large RNAs (>50nt) are better approached using coarse graining approaches (e.g., MC-SYM, NAST). Still, further developments and refinements of the existing models are needed. Specifically, for larger RNAs (>50 nt), programs are limited in predicting long-range contacts and helical arrangements. New strategies continue to emerge as RNA becomes more attractive to researchers. Indeed, the new computer game EteRNA (http://eterna.cmu.edu) uses ideas similar to Foldit to attract participants worldwide to develop new ways to design and fold RNA molecules.
Structural knowledge from observed RNA native structures in the form of 3D motifs is further needed for both automated and manual modeling approaches. In fact, the increasing availability of high resolution large RNA structures has made possible the identification and classification of RNA secondary and tertiary structure motifs, at different levels of detail. In particular, recent evidence supports unique combinations of 3D motifs that form larger architectural units of RNA, such as the adenosine wedge motif [33]. Such knowledge enhances RNA structure prediction, modeling, and design. Though we are still far from having a complete motif library, and also far from predicting the effect of 3D motifs in the long-range contacts that stabilize RNA 3D structures, graph theory approaches, as described here, can aid in the classification and prediction of RNA motifs.
Indeed, RNA’s modularity has been natural for the application of graph theory tools to estimate the size of RNA's structural/functional repertoire [39], detect structural and functional similarity among existing RNAs [44], classify and catalog RNA motifs [38,39], predict sequences that map into new RNA motifs [39], identify RNA motifs of antibiotic-binding aptamers (found synthetically) in genomes [45,46], analyze the structural diversity of random pools used for RNA in vitro selection [47], enhance in vitro selection for aptamer design [41], analyze RNA’s structural rigidity [54–56], and predict RNA 3D structures including pseudoknots [53].
In addition, new in silico approaches to aptamer discovery and design, as described here, also offer novel ways for efficient design. Because ultimate design depends on fine details of the tertiary structures, such designs will improve with better methods for tertiary structure algorithms. The combination of graph theory, 3D motif analysis, tertiary structure prediction, and general modeling and simulation improvements [8], will continue to advance RNA bioinformatics and RNA biology and chemistry.
Acknowledgments
This work was supported by NSF (EMT award CCF-0727001) and NIH (grants GM081410 and ES01269201).
Footnotes
This riboswitch is regulated by the metabolite TPP (thiamine pyrophosphate), so that the existence of this metabolite produces a termination hairpin (a secondary-structure element) that blocks transcription; without this metabolite, RNA polymerase can bind and transcription proceeds (Fig. 3b). design by combining, like the frameworks above, sequence mutation/evolution with energy landscape analysis.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the two year period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Afonin KA, Bindewald E, Yaghoubian AJ, Voss N, Jacovetty E, Shapiro BA, Jaeger L. In vitro assembly of cubic RNA-based scaffolds designed in silico. Nat Nanotechnol. 2010;5:676–682. doi: 10.1038/nnano.2010.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kasprzak W, Bindewald E, Kim TJ, Jaeger L, Shapiro BA. Use of RNA structure flexibility data in nanostructure modeling. Methods. 2010 doi: 10.1016/j.ymeth.2010.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sioud M. Ribozymes and siRnas: from structure to preclinical applications. Handb Exp Pharmacol. 2006:223–242. doi: 10.1007/3-540-27262-3_11. [DOI] [PubMed] [Google Scholar]
- 4.Hess H, Jaeger L. Nanobiotechnology. Curr Opin Biotechnol. 2010;21:373–375. doi: 10.1016/j.copbio.2010.08.005. [DOI] [PubMed] [Google Scholar]
- 5••.Laing C, Schlick T. Computational approaches to 3D modeling of RNA. Journal of Physics: Condensed Matter. 2010;22:283101. doi: 10.1088/0953-8984/22/28/283101. This review compares available 3D structure prediction algorithms from an RNA dataset of 43 structures of various lengths and motifs. The study finds that algorithms vary widely in terms of prediction quality; most predictions have large root-mean square deviations from the crystal structure (e.g., RMSD>6Å) due to the limitations in predicting long-range contacts. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marti-Renom MA, Capriotti E. Computational RNA Structure Prediction. Current Bioinformatics. 2008;3:32–45. [Google Scholar]
- 7.Schroeder R, Barta A, Semrad K. Strategies for RNA folding and assembly. Nat Rev Mol Cell Biol. 2004;5:908–919. doi: 10.1038/nrm1497. [DOI] [PubMed] [Google Scholar]
- 8.Schlick T, Collepardo-Guevara R, Halvorsen LA, Jung S, Xiao X. Biomolecular Modeling and Simulation: A Field Coming of Age. Quart Rev Biophys. 2010;43:1–38. doi: 10.1017/S0033583510000284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shapiro BA, Yingling YG, Kasprzak W, Bindewald E. Bridging the gap in RNA structure prediction. Curr Opin Struct Biol. 2007;17:157–165. doi: 10.1016/j.sbi.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 10•.Jonikas MA, Radmer RJ, Laederach A, Das R, Pearlman S, Herschlag D, Altman RB. Coarse-grained modeling of large RNA molecules with knowledge-based potentials and structural filters. RNA. 2009;15:189–199. doi: 10.1261/rna.1270809. This program for RNA structure prediction performs molecular dynamics simulations guided by a knowledge-based statistical potential functions. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11•.Frellsen J, Moltke I, Thiim M, Mardia KV, Ferkinghoff-Borg J, Hamelryck T. A probabilistic model of RNA conformational space. PLoS Comput Biol. 2009;5:e1000406. doi: 10.1371/journal.pcbi.1000406. Coarse-grained probabilistic model that allows efficient sampling of RNA conformations in continuous space. Modeling seven dihedral angles, BARNACLE can predict RNA-like features such as rotameric angles, and accurate helix lengths. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12••.Das R, Karanicolas J, Baker D. Atomic accuracy in predicting and designing noncanonical RNA structure. Nat Methods. 2010;7:291–294. doi: 10.1038/nmeth.1433. This all-atom prediction program FARFAR uses fragment library assemblies, Monte Carlo simulations, and standard potential functions to predict structures of small RNAs reasonably accurately. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sharma S, Ding F, Dokholyan NV. iFoldRNA: three-dimensional RNA structure prediction and folding. Bioinformatics. 2008;24:1951–1952. doi: 10.1093/bioinformatics/btn328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Das R, Baker D. Automated de novo prediction of native-like RNA tertiary structures. Proc Natl Acad Sci U S A. 2007;104:14664–14669. doi: 10.1073/pnas.0703836104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parisien M, Major F. The MC-Fold and MC-Sym pipeline infers RNA structure from sequence data. Nature. 2008;452:51–55. doi: 10.1038/nature06684. [DOI] [PubMed] [Google Scholar]
- 16.Gherghe CM, Leonard CW, Ding F, Dokholyan NV, Weeks KM. Native-like RNA tertiary structures using a sequence-encoded cleavage agent and refinement by discrete molecular dynamics. J Am Chem Soc. 2009;131:2541–2546. doi: 10.1021/ja805460e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17•.Xia Z, Gardner DP, Gutell RR, Ren P. Coarse-grained model for simulation of RNA three-dimensional structures. J Phys Chem B. 2010;114:13497–13506. doi: 10.1021/jp104926t. A five-bead coarse-grained model is developed to predict RNA structures using RNA secondary structure by MD sampling guided by statistical potential functions that score for bond stretching, angle bending, and dihedral rotations. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18••.Jossinet F, Ludwig TE, Westhof E. Assemble: an interactive graphical tool to analyze and build RNA architectures at the 2D and 3D levels. Bioinformatics. 2010;26:2057–2059. doi: 10.1093/bioinformatics/btq321. This computer tool constructs a 3D structure using the insertion of tertiary motifs, permitting manipulation of torsion angles, local rotations, and modular translations. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martinez HM, Maizel JV, Jr, Shapiro BA. RNA2D3D: a program for generating, viewing, and comparing 3-dimensional models of RNA. J Biomol Struct Dyn. 2008;25:669–683. doi: 10.1080/07391102.2008.10531240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parisien M, Cruz JA, Westhof E, Major F. New metrics for comparing and assessing discrepancies between RNA 3D structures and models. RNA. 2009;15:1875–1885. doi: 10.1261/rna.1700409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hajdin CE, Ding F, Dokholyan NV, Weeks KM. On the significance of an RNA tertiary structure prediction. RNA. 2010;16:1340–1349. doi: 10.1261/rna.1837410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nasalean L, Stombaugh J, Zirbel CL, Leontis NB. RNA 3D Structural Motifs: Definition, Identification, Annotation, and Database Searching. In: Walter NG, Woodson SA, Batey RT, editors. Non-Protein Coding RNAs. Springer; Berlin Heidelberg: 2009. pp. 1–26. [Google Scholar]; Springer Series in Biophysics. Vol. 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saito H, Inoue T. Synthetic biology with RNA motifs. Int J Biochem Cell Biol. 2009;41:398–404. doi: 10.1016/j.biocel.2008.08.017. [DOI] [PubMed] [Google Scholar]
- 24.Severcan I, Geary C, Chworos A, Voss N, Jacovetty E, Jaeger L. A polyhedron made of tRNAs. Nat Chem. 2010;2:772–779. doi: 10.1038/nchem.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leontis NB, Lescoute A, Westhof E. The building blocks and motifs of RNA architecture. Curr Opin Struct Biol. 2006;16:279–287. doi: 10.1016/j.sbi.2006.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leontis NB, Stombaugh J, Westhof E. The non-Watson-Crick base pairs and their associated isostericity matrices. Nucleic Acids Res. 2002;30:3497–3531. doi: 10.1093/nar/gkf481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27••.Stombaugh J, Zirbel CL, Westhof E, Leontis NB. Frequency and isostericity of RNA base pairs. Nucleic Acids Res. 2009;37:2294–2312. doi: 10.1093/nar/gkp011. A new measure is introduced for base pair isostericity which can be used to determine base pair substitutions that occur among homologous RNA due to compensatory mutations. Potential applications include prediction of 3D motifs by using comparative sequence alignments. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28•.Zirbel CL, Sponer JE, Sponer J, Stombaugh J, Leontis NB. Classification and energetics of the base-phosphate interactions in RNA. Nucleic Acids Res. 2009;37:4898–4918. doi: 10.1093/nar/gkp468. This analysis of base-phosphate interactions in solved RNAs reveals that such interactions are abundant and therefore important to RNA function and stability. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29•.Laing C, Jung S, Iqbal A, Schlick T. Tertiary motifs revealed in analyses of higher order RNA junctions. J Mol Biol. 2009;393:67–82. doi: 10.1016/j.jmb.2009.07.089. Exhaustive analysis of solved 3D RNA junctions reveals that higher-order junctions can be decomposed into sub-junctions resembling configurations found in 3 and 4-way junctions. This observation suggests a general modular design strategy for RNA junctions. New 3D motifs involved in helix-packing interactions are also reported. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ulyanov NB, James TL. RNA structural motifs that entail hydrogen bonds involving sugar-phosphate backbone atoms of RNA. New J Chem. 2010;34:910–917. doi: 10.1039/b9nj00754g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xin Y, Laing C, Leontis NB, Schlick T. Annotation of tertiary interactions in RNA structures reveals variations and correlations. RNA. 2008;14:2465–2477. doi: 10.1261/rna.1249208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Holbrook SR. Structural principles from large RNAs. Annu Rev Biophys. 2008;37 :445–464. doi: 10.1146/annurev.biophys.36.040306.132755. [DOI] [PubMed] [Google Scholar]
- 33.Gagnon MG, Steinberg SV. The adenosine wedge: a new structural motif in ribosomal RNA. RNA. 2010;16:375–381. doi: 10.1261/rna.1550310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lescoute A, Westhof E. Topology of three-way junctions in folded RNAs. RNA. 2006;12:83–93. doi: 10.1261/rna.2208106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35•.Laing C, Schlick T. Analysis of four-way junctions in RNA structures. J Mol Biol. 2009;390:547–559. doi: 10.1016/j.jmb.2009.04.084. Analysis of solved four-way junctions identified 9 major families by coaxial stacking patterns and helical configurations. Composite motifs involving A-minor and coaxial stacking are also reported. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36••.Bailor MH, Sun X, Al-Hashimi HM. Topology links RNA secondary structure with global conformation, dynamics, and adaptation. Science. 2010;327:202–206. doi: 10.1126/science.1181085. Evidence is provided that topological constraints observed at the secondary structure level are important in the 3D orientation of helices in internal loops. [DOI] [PubMed] [Google Scholar]
- 37.Gan HH, Pasquali S, Schlick T. Exploring the repertoire of RNA secondary motifs using graph theory; implications for RNA design. Nucleic Acids Res. 2003;31 :2926–2943. doi: 10.1093/nar/gkg365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gan HH, Fera D, Zorn J, Shiffeldrim N, Tang M, Laserson U, Kim N, Schlick T. RAG: RNA-As-Graphs database--concepts, analysis, and features. Bioinformatics. 2004;20:1285–1291. doi: 10.1093/bioinformatics/bth084. [DOI] [PubMed] [Google Scholar]
- 39.Kim N, Shiffeldrim N, Gan HH, Schlick T. Candidates for novel RNA topologies. J Mol Biol. 2004;341:1129–1144. doi: 10.1016/j.jmb.2004.06.054. [DOI] [PubMed] [Google Scholar]
- 40.Kim N, Gan HH, Schlick T. A computational proposal for designing structured RNA pools for in vitro selection of RNAs. RNA. 2007;13:478–492. doi: 10.1261/rna.374907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41••.Kim N, Izzo JA, Elmetwaly S, Gan HH, Schlick T. Computational generation and screening of RNA motifs in large nucleotide sequence pools. Nucleic Acids Res. 2010;38:e139. doi: 10.1093/nar/gkq282. A computational approach to generate and screen target motifs in large random pools with 10 14 sequences is developed and applied to ligases. The protocol combines target pool generation, motif scanning, and motif screening on RNA secondary structures to improve RNA design by simulating in silico the process of in vitro selection. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim N, Shin JS, Elmetwaly S, Gan HH, Schlick T. RagPools: RNA-As-Graph-Pools--a web server for assisting the design of structured RNA pools for in vitro selection. Bioinformatics. 2007;23:2959–2960. doi: 10.1093/bioinformatics/btm439. [DOI] [PubMed] [Google Scholar]
- 43.Fera D, Kim N, Shiffeldrim N, Zorn J, Laserson U, Gan HH, Schlick T. RAG: RNA-As-Graphs web resource. BMC Bioinformatics. 2004;5:88. doi: 10.1186/1471-2105-5-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pasquali S, Gan HH, Schlick T. Modular RNA architecture revealed by computational analysis of existing pseudoknots and ribosomal RNAs. Nucleic Acids Res. 2005;33:1384–1398. doi: 10.1093/nar/gki267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Laserson U, Gan HH, Schlick T. Exploring the Connection Between Synthetic and Natural RNAs in Genomes: A Novel Computational Approach. In: Barth TJ, Griebel M, Keyes DE, Nieminen RM, Roose D, Schlick T, editors. New Algorithms for Macromolecular Simulation. Springer; Berlin Heidelberg: 2005. pp. 35–56. [Google Scholar]; Lecture Notes in Computational Science and Engineering. Vol. 49 [Google Scholar]
- 46.Laserson U, Gan HH, Schlick T. Predicting candidate genomic sequences that correspond to synthetic functional RNA motifs. Nucleic Acids Res. 2005;33 :6057–6069. doi: 10.1093/nar/gki911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gevertz J, Gan HH, Schlick T. In vitro RNA random pools are not structurally diverse: a computational analysis. RNA. 2005;11:853–863. doi: 10.1261/rna.7271405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48•.Quarta G, Kim N, Izzo JA, Schlick T. Analysis of riboswitch structure and function by an energy landscape framework. J Mol Biol. 2009;393:993–1003. doi: 10.1016/j.jmb.2009.08.062. A computational framework that uses clustering analysis of folding energy landscapes at different nucleotide lengths is developed and applied to analyze the mechanics of transcription elongation in the TPP riboswitch. The study suggests that the riboswitch’s kinetics length-dependent, where two clusters can be observed during transcription elongation and where TPP's binding shifts the preference to one form. The “on” state serves to terminate transcription, and the “off” state correspond to an alternative riboswitch structure that activates transcription. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schlick T. Molecular Modeling and Simulation: An Interdisciplinary Guide. 2. Springer; 2010. [Google Scholar]
- 51.Koessler DR, Knisley DJ, Knisley J, Haynes T. A predictive model for secondary RNA structure using graph theory and a neural network. BMC Bioinformatics. 2010;11 (Suppl 6):S21. doi: 10.1186/1471-2105-11-S6-S21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Childs L, Nikoloski Z, May P, Walther D. Identification and classification of ncRNA molecules using graph properties. Nucleic Acids Res. 2009;37:e66. doi: 10.1093/nar/gkp206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gillespie J, Mayne M, Jiang M. RNA folding on the 3D triangular lattice. BMC Bioinformatics. 2009;10:369. doi: 10.1186/1471-2105-10-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fulle S, Gohlke H. Analyzing the flexibility of RNA structures by constraint counting. Biophys J. 2008;94:4202–4219. doi: 10.1529/biophysj.107.113415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fulle S, Gohlke H. Constraint counting on RNA structures: linking flexibility and function. Methods. 2009;49:181–188. doi: 10.1016/j.ymeth.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 56•.Fulle S, Gohlke H. Statics of the ribosomal exit tunnel: implications for cotranslational peptide folding, elongation regulation, and antibiotics binding. J Mol Biol. 2009;387:502–517. doi: 10.1016/j.jmb.2009.01.037. A graph theory approach to derive a measure of molecular flexibility is presented. This method can detect the interplay between the static properties of the ribosomal exit tunnel and the local zones of flexible nucleotides during the cotranslational process in the ribosome. [DOI] [PubMed] [Google Scholar]
- 57.Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 58.Joyce GF. Amplification, mutation and selection of catalytic RNA. Gene. 1989;82 :83–87. doi: 10.1016/0378-1119(89)90033-4. [DOI] [PubMed] [Google Scholar]
- 59.Bourdeau V, Ferbeyre G, Pageau M, Paquin B, Cedergren R. The distribution of RNA motifs in natural sequences. Nucleic Acids Res. 1999;27:4457–4467. doi: 10.1093/nar/27.22.4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Knight R, De Sterck H, Markel R, Smit S, Oshmyansky A, Yarus M. Abundance of correctly folded RNA motifs in sequence space, calculated on computational grids. Nucleic Acids Res. 2005;33:5924–5935. doi: 10.1093/nar/gki886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Macke TJ, Ecker DJ, Gutell RR, Gautheret D, Case DA, Sampath R. RNAMotif, an RNA secondary structure definition and search algorithm. Nucleic Acids Res. 2001;29:4724–4735. doi: 10.1093/nar/29.22.4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Luo X, McKeague M, Pitre S, Dumontier M, Green J, Golshani A, Derosa MC, Dehne F. Computational approaches toward the design of pools for the in vitro selection of complex aptamers. RNA. 2010;16:2252–2262. doi: 10.1261/rna.2102210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ruff KM, Snyder TM, Liu DR. Enhanced functional potential of nucleic acid aptamer libraries patterned to increase secondary structure. J Am Chem Soc. 2010;132:9453–9464. doi: 10.1021/ja103023m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64•.Chushak Y, Stone MO. In silico selection of RNA aptamers. Nucleic Acids Res. 2009;37:e87. doi: 10.1093/nar/gkp408. A multi-step large-pool generation approach is presented for selection of RNA aptamers that uses 2D pattern searching, 3D structure generation, and screening for target binding using docking programs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Aminova O, Disney MD. A microarray-based method to perform nucleic acid selections. Methods Mol Biol. 2010;669:209–224. doi: 10.1007/978-1-60761-845-4_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Carothers JM, Goler JA, Kapoor Y, Lara L, Keasling JD. Selecting RNA aptamers for synthetic biology: investigating magnesium dependence and predicting binding affinity. Nucleic Acids Res. 2010;38:2736–2747. doi: 10.1093/nar/gkq082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kennedy R, Lladser ME, Wu Z, Zhang C, Yarus M, De Sterck H, Knight R. Natural and artificial RNAs occupy the same restricted region of sequence space. RNA. 2010;16:280–289. doi: 10.1261/rna.1923210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schlick T. Molecular dynamics-based approaches for enhanced sampling of long-time, large-scale conformational changes in biomolecules. F1000 Biol Rep. 2009;1 :1–51. doi: 10.3410/B1-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Anderson PC, Mecozzi S. Minimum sequence requirements for selective RNA-ligand binding: a molecular mechanics algorithm using molecular dynamics and free-energy techniques. J Comput Chem. 2006;27:1631–1640. doi: 10.1002/jcc.20459. [DOI] [PubMed] [Google Scholar]
- 70.Dill KA, Ozkan SB, Shell MS, Weikl TR. The protein folding problem. Annu Rev Biophys. 2008;37:289–316. doi: 10.1146/annurev.biophys.37.092707.153558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Frauenfelder H, Sligar SG, Wolynes PG. The energy landscapes and motions of proteins. Science. 1991;254:1598–1603. doi: 10.1126/science.1749933. [DOI] [PubMed] [Google Scholar]
- 72.Wolynes PG. Recent successes of the energy landscape theory of protein folding and function. Q Rev Biophys. 2005;38:405–410. doi: 10.1017/S0033583505004075. [DOI] [PubMed] [Google Scholar]
- 73••.Pitt JN, Ferre-D'Amare AR. Rapid construction of empirical RNA fitness landscapes. Science. 2010;330:376–379. doi: 10.1126/science.1192001. A combination of computational analysis, next-generation sequencing, and in vitro selection methods is developed to generate an empirical fitness landscape for a ribozyme. The results show that optimal fitness is correlated with genotype abundance, and this information was used to generate a fitness landscape map of 107 unique RNA sequences. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mironov A, Epshtein V, Nudler E. Transcriptional approaches to riboswitch studies. Methods Mol Biol. 2009;540:39–51. doi: 10.1007/978-1-59745-558-9_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Montange RK, Batey RT. Riboswitches: emerging themes in RNA structure and function. Annu Rev Biophys. 2008;37:117–133. doi: 10.1146/annurev.biophys.37.032807.130000. [DOI] [PubMed] [Google Scholar]
- 76.Shu W, Liu M, Chen H, Bo X, Wang S. ARDesigner: a web-based system for allosteric RNA design. J Biotechnol. 2010;150:466–473. doi: 10.1016/j.jbiotec.2010.10.067. [DOI] [PubMed] [Google Scholar]