Abstract
LINE-1 (L1) elements play an important creative role in genomic evolution by distributing both L1 and non-L1 DNA in a process called retrotransposition. A large percentage of the human genome consists of DNA that has been dispersed by the L1 transposition machinery. L1 elements are not randomly distributed in genomic DNA but are concentrated in regions with lower GC content. In an effort to understand the consequences of L1 insertions, we have begun an investigation of their genomic characteristics and the changes that occur to them over time. We compare human L1 insertions that were created either during recent human evolution or during the primate radiation. We report that L1 insertions are an important source for the creation of new microsatellites. We provide evidence that L1 first strand cDNA synthesis can occur from an internal priming event. We note that in contrast to older L1 insertions, recent L1s are distributed randomly in genomic DNA, and the shift in the L1 genomic distribution occurs relatively rapidly. Taken together, our data indicate that strong forces act on newly inserted L1 retrotransposons to alter their structure and distribution.
The non-long terminal repeat (LTR) retrotransposons are an ancient family of mobile elements that have played a major role shaping eukaryotic genomes for over 600 million years (Malik et al. 1999). The recent publication of the nearly complete human genome sequence highlighted the need to understand the dynamics of these elements and their effects on human biology (Lander et al. 2001). In the human genome the LINE-1 element (L1) is by far the most abundant and the only active member of this family. Early estimates based on hybridization experiments suggested that 4000 full-length 6-kb L1 elements and 100,000 L1 fragments exist in human DNA (Adams et al. 1980; Grimaldi et al. 1984; Hwu et al. 1986). More recent analyses performed by computational methods that permit the identification of more highly diverged, and therefore older, L1 elements indicate that 500,000 fragments reside in the human genome (Smit 1996; Lander et al. 2001). Altogether, LINE-1 elements constitute an estimated 17% of human DNA (Smit 1996; Lander et al. 2001).
Although most L1s in the human genome are ancient and transpositionally inert, an estimated 40 elements continue to produce new L1 copies (Sassaman et al. 1997). All or most of the transposition-competent human L1s belong to a subset of elements called Ta. Subset Ta elements (L1Hs- Ta) can be identified by the presence of the sequence “ACA” at position 5930–5932 (numbers refer to the active element LRE-1) in the 3′ untranslated region (UTR) where older elements most commonly have the sequence “GAG” (Skowronski et al. 1988). A recent detailed investigation concluded that subset Ta elements first appeared ∼4 million years ago and that most of these elements are 3 million years old or younger (Boissinot et al. 2000). When new L1 sequences insert into the genome they produce new genetic markers and sometimes disease (Kazazian et al. 1988). Genomic loci that can be found in the human population in two forms, with and without an L1 insertion, are called LINE-1 insertion dimorphisms, or LIDs (Dombroski et al. 1993; Holmes et al. 1994; Sassaman et al. 1997; Sheen et al. 2000). L1 transposition has been frequent enough during human evolution to make LIDs an important contributor to human genetic variation and a valuable resource for investigating the structure of modern human populations (Boissinot et al. 2000; Sheen et al. 2000). An efficient molecular method for identifying LIDs, called L1 display, has been described recently (Sheen et al. 2000).
Historically, several different observations indicated that human and mammalian L1s are not distributed randomly in the genome. These studies concurred that L1s are found more frequently in genomic regions characterized by relatively low average levels of G + C nucleotides and less commonly in regions of high GC (Soriano et al. 1983; Korenberg and Rykowski 1988; Moyzis et al. 1989; Boyle et al. 1990; Baker and Kass 1994). The recently completed draft human genome sequence confirms these results (Lander et al. 2001). The distribution of L1s in human DNA stands in marked contrast to the distribution of Alu elements in the human genome. Alus are most concentrated in genomic regions of high GC and less concentrated in DNA low in GC. These differences are most perplexing in light of the major similarities between these two types of transposons. Both L1s and Alus transpose via an RNA intermediate. Both elements insert into the genome followed by poly(A) tails and the production of short target site duplications, and both are believed to transpose by making use of the L1-encoded transposition machinery (Dombroski et al. 1991; Jurka 1997).
There are two general pathways by which the uneven distribution of human L1s in the genome could have been derived. The first pathway involves the favored transposition of L1s into GC-poor genomic regions. L1 transposition is believed to proceed by a mechanism called target primed reverse transcription (TPRT) (Luan et al. 1993). The working model for TPRT is based primarily on the results of experiments performed on the R2Bm element of Bombyx mori. According to this model, transposition begins with the nicking of the antisense DNA strand at the insertion site by an element-encoded endonuclease (Xiong and Eickbush 1988). The newly created free 3′-hydroxyl group is then used to prime first strand cDNA synthesis. This model predicts that the selection of insertion sites would primarily be a function of the endonuclease although other components of the transposition machinery may also play important roles. Indeed, one class of non-LTR retrotransposons (including R2Bm itself) encodes site-specific endonucleases that strictly determine their insertion sites (Yang et al. 1999). The human L1-encoded endonuclease does not appear to be a site-specific enzyme although it does greatly favor A-T rich cleavage sites with the consensus sequence 3′-AATTTT-5′ (Feng et al. 1996; Jurka 1997; Cost and Boeke 1998). It is possible that this sequence preference, along with other as yet undefined components of the transposition machinery, induces a site selection bias that gives rise to the uneven distribution of human L1s (Cost and Boeke 1998).
The second pathway by which the observed distribution of human L1s in the genome could be derived depends on events that take place after transposition has occurred. This pathway predicts that the selection of insertion sites is virtually random except for the sequence of several nucleotides immediately surrounding the cleavage site. Once transposition has occurred, however, L1s in regions of high GC content would be lost from the genome at a greater rate than L1s in regions of low GC content. This could result either from an increased loss of L1s from regions of high GC or by the specific retention of L1s in regions of low GC. Evidence that Alus, which are believed to transpose via the L1-encoded machinery, insert randomly into the genome (Arcot et al. 1998) supports this pathway.
Much remains to be learned about the molecular events associated with the insertion and long-term residence of L1s in the human genome. In this study we have characterized many genomic characteristics of L1s and have analyzed how they change over time of residence in the genome. We identified many recent L1 insertions by L1 display and compared characteristics of their insertion sites to those of a group of older L1s identified from a GenBank search. We report that the poly(A) tails of L1 elements shorten with age and are a rich source for the birth of new microsatellites. We observe that the poly(A) addition signal of L1s degrade rapidly after insertion thereby mitigating the potentially disruptive effects on transcription caused by L1 insertion into introns. We identify a case of internal priming of first strand L1 cDNA synthesis that supports the TPRT mechanism and has possible implications for the targeting of L1 insertions. Our findings also indicate that in contrast to older L1s, recent L1s are found dispersed randomly in human genomic DNA. This suggests that the current distribution of L1s in the human genome is the result of events that occur after transposition.
RESULTS
Identification of Recent and Ancient L1 Insertions
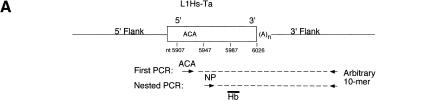
We used L1 display to randomly identify a group of recently inserted human L1 elements. L1 display is a PCR-based method specifically designed to identify LIDs. The method, which has been described in detail (Sheen et al. 2000), is summarized in Fig. 1A. Genomic DNA is amplified with a primer specific for L1Hs-Ta elements (ACA primer) and a second 10-bp arbitrary primer. The 3′ ends of L1Hs-Ta elements located near binding sites for the arbitrary primer are amplified along with the intervening 3′ flanking regions. The products of the first round of PCR are reamplified using the same arbitrary primer and a L1 nested primer (NP), and the products of this second PCR amplification are Southern blotted and hybridized with a probe (Hb) specific for L1 3′ UTRs (Fig. 1A). Bands that are revealed in the Southern blot (Fig. 1B) consist of the terminal 80 bp of L1 sequence (64 bp of amplified L1 sequence plus 16 bp from primer NP), an A-rich region that is derived from the elements' poly(A) tail, and a variable length of 3′ flanking DNA. Each L1 display reaction searches a fraction of genomic DNA for the presence of L1Hs-Ta elements. Performing L1 display with a large number of arbitrary primers queries a greater portion of the genome for the presence of insertions. Elements identified by L1 display will be selected nearly at random because the method relies on the use of very short (10 bp) primers with arbitrary sequences.
Figure 1.
L1 display, method, and results. (A) L1 display. A L1Hs-Ta insertion (rectangle) is depicted surrounded by flanking DNA (solid lines). The broken lines represent the products of two rounds of PCR amplifications. The arrows below indicate the relative positions and orientations of the primers. (B) L1 display results. A typical L1 display experiment performed with a single decamer on genomic DNA from 42 individuals is shown. One fixed (solid line) and two polymorphic (broken lines) L1Hs-Ta insertions can be seen.
We performed L1 display with 50 arbitrary primers on a panel of 91 genomic DNA samples representing several different geographic regions. A total of 152 bands were detected. Some of the bands were present in all of the samples but many were present in relatively few samples. Sixty-two bands were selected for further analysis. DNA sequencing of the cloned fragments revealed that 58 were unique. One sequence had been cloned twice and a second had been cloned four times. In several cases this resulted from the amplification of an L1 insertion by inappropriate priming of the NP primer in the flanking DNA during the nested PCR. Of the remaining sequences, one did not contain an L1 3′ UTR and four represented insertions containing the nucleotides GAG that are diagnostic of older non-Ta elements. In total, 53 of 62 cloned bands (85%) represented unique L1Hs-Ta insertions. These were subjected to further analysis.
The RepeatMasker program (Smit and Green 2000) revealed that for seven of the clones, the 3′ flanking sequence consisted only of repeat sequence DNA. Of the remaining 46 clones, we unambiguously identified the GenBank entry for 39 (85%) of them by searching the database for the clones' 3′ flanks with the BLASTN program (Altschul et al. 1990). We also successfully identified the GenBank locus of one of the clones that had a fully masked sequence; in this case the 3′ flanking DNA was sufficiently unique to allow for an unambiguous assignment. In 15 of the 40 (38%) GenBank loci an L1Hs-Ta element was located at the position indicated by the clone. For the remaining 25 GenBank loci, no L1 insertion was represented in the database entry, suggesting that the average gene frequency of the L1Hs-Ta clones was low and that the L1 insertions had occurred during relatively recent human evolution. This result is in concert with the relatively low representation of many of the L1 display bands in the genomic samples and confirms the utility of the method for identifying recent and polymorphic L1Hs-Ta insertions. From the information present in the GenBank entries and other public databases, we successfully identified chromosomal assignments for 32 of the clones. The 32 L1Hs-Ta insertions were located on 15 different chromosomes (Table 1). This distribution of insertion sites was not statistically different from a random pattern of insertion (see Methods).
Table 1.
Chromosomal Distribution of the L1Hs-Ta Elements
| Chromosome | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | X | Y |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. of elements | 3 | 1 | 0 | 3 | 4 | 4 | 0 | 2 | 0 | 1 | 1 | 2 | 1 | 0 | 1 | 0 | 3 | 0 | 0 | 1 | 0 | 0 | 4 | 1 |
Although subset Ta elements are defined by the sequence “ACA” in their 3′ UTRs, most L1 elements in the human genome bear the sequence “GAG” at the same position (5930–5932 according to the numbering of LRE-1, an element that has transposed recently) (Dombroski et al. 1991). In addition, all known L1Hs-Ta elements have a “G” at LRE-1 position 6015 whereas most elements not belonging to subset Ta have an “A” at this position (Skowronski et al. 1988; Boissinot et al. 2000). To date, no human L1 element with the sequence “GAG” has been identified that has undergone de novo transposition or is polymorphic in the human population. All of the GAG L1s (L1-GAG) in the human genome are likely therefore to be incapable of transposition and fixed in the human population. To select a control group of older human L1 insertions, we searched the GenBank nonredundant database with a 113-bp fragment of the L1 3′ UTR (bp 5914–6026 of LRE-1) that had the “GAG” and “A” bases that are characteristic of older L1 elements. From the list of matches with the highest similarities to the query, we selected 30 L1-GAG elements with >95% sequence identity to the query sequence. All of these elements had both the “GAG” at 5930–5932 and the “A” at 6015 and therefore did not belong to subset Ta. RepeatMasker analysis (Smit and Green 2000) confirmed that this group of L1 insertions belonged to older subfamilies of L1 elements including 19 in subfamily L1PA2, seven in L1PA3, three in L1PA4, and one in L1PA5 (Smit et al. 1995).
Features of the 3′ UTRs of L1 Insertions
To determine the degree of sequence divergence of the recent and older groups of elements, we aligned the terminal 58 bp of L1 DNA of each of the clones. The L1 poly(A) addition signals were excluded from this analysis because, as described below, they were found to be extremely variable. To measure the relative ages of the two groups of elements we counted the number of nucleotide positions at which at least one element differed in sequence from the group consensus. We chose only to count the number of divergent positions instead of the total number of mutational events because of the possibility that more than a single member of each of the L1 groups may have been derived from the repeat transposition of individual “master” elements. Counting the total number of mutational events under these circumstances might lead to an overestimation of the average divergence of the group of elements. In the L1-GAG group of elements, 17 nucleotide positions were polymorphic whereas only 12 nucleotide positions were polymorphic in the L1Hs-Ta group of elements (data not shown). This difference was highly significant (Fisher's Exact Test, 2-tailed, P = 0.004) and confirmed that the L1- (GAG elements have been residing in the human genome for a longer time than the L1Hs-Ta group.
L1 elements have been noted previously to have nonstandard poly(A) addition signals (PAS) (Skowronski et al. 1988). Unlike the consensus PAS in which the signal is separated from the poly(A) tail by 20–30 bp of intervening DNA, the PAS of human L1s (and many L1-like elements in other species as well) is followed immediately by a poly(A) tail. The L1 PAS also appears to be weak; RNA transcription of L1s often proceeds into 3′ flanking DNA until a second PAS is found. This situation sometimes gives rise to the transduction of 3′ flanking DNA during the transposition process (Holmes et al. 1994; Goodier et al. 2000; Pickeral et al. 2000). It has been proposed that the possession of a weak PAS by L1s is advantageous both for the L1 and for the “host” organism (Moran et al. 1999). A weak PAS might allow a gene into which an L1 has inserted in the sense orientation to continue to be transcribed to completion, albeit at a reduced level, despite the presence of a new internal L1 PAS. The degradation of the PAS of recently inserted L1 elements by random mutagenesis or DNA polymerase “slipping” would also serve this purpose by removing the PAS and its effect on diminishing proper transcription termination. We therefore compared the PAS of the recent and older groups of L1s. Canonical AATAAA PAS sequences were found in 44 of the 53 (83%) L1Hs-Ta insertions but in only 14 of the 30 (47%) L1-GAG insertions. This confirms that the PAS of L1 insertions degrade more rapidly than the rest of the L1 sequence after transposition.
We also examined the length of the poly(A) tails of the two groups of elements. Insertions in the L1Hs-Ta group had poly(A) tails that averaged 13.9±9 bp in length whereas the insertions in the L1-GAG group had poly(A) tails that were only 3.7±3 bp long. This difference was highly significant (Student's T-test, P = 0.0001). Batzer and colleagues (Arcot et al. 1995) have reported previously that the poly(A) tails of Alu elements become shorter with time after insertion into the genome. Our results confirm that the same process occurs for L1 elements. The relative instability of the L1 poly(A) tails was further highlighted by our observation of frequent differences in the poly(A) tails of our L1 display clones and their GenBank counterparts. Of the 15 L1 display clones whose GenBank loci also contained a L1 insertion, 10 had differences in the length or sequences of their poly(A) tails and A-rich regions (data not shown).
Features of the 3′ Flanking Sequences
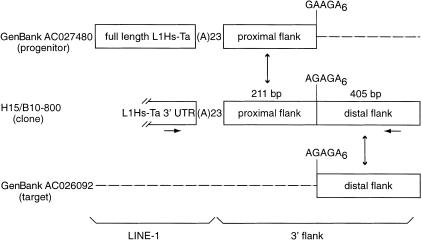
Recent reports have highlighted the ability of L1s to transpose along with a variable length of 3′ flanking sequence (Holmes et al. 1994). This event, called 3′ transduction, may occur in 15%–23% of all human L1 transposition events (Goodier et al. 2000; Pickeral et al. 2000). L1 3′ transduction may have shuffled as much as 0.6%–1% of the human genome and has been proposed to be a mechanism for shuffling exons (Moran et al. 1999). The characteristics of an L1 insertion associated with a 3′ transduction is the presence of target site duplications surrounding both the L1 insertion and a segment of non-L1 3′ flanking DNA. Between the L1 and the transduced DNA resides the remnants of the poly(A) tail from the parental L1 insertion, and a second poly(A) tail is located at the integration site where the transduction event occurred. We detected at least two likely 3′ transduction events among our 53 L1Hs-Ta insertions (see Methods). In the first, 138 bp of 3′ flanking DNA was transduced, and both the parent L1 (accession no. AC005798) and the progeny L1 insertions (accession no. AL353153) were represented in GenBank. In the second case (insertion H15/B10–800, Fig. 2), 211 bp of the proximal 3′ flanking sequence of the L1 display clone matched a GenBank entry (accession no. ACO27480) in which a full-length L1 was inserted (Fig. 2). This locus probably represents the parent L1. A second GenBank entry (accession no. AC026092) matched the L1 display insertion exactly from the end of the 211 bp of proximal flanking DNA till the end of the sequence of the L1 display insertion (another 405 bp of DNA, Fig. 2). PCR amplifications confirmed the identity of this second locus as the L1 display clone insertion site and indicated that the locus contained a 1.5-kb truncated L1 insertion (data not shown). Interestingly, no poly(A) tail was found in the L1 display sequence between the transduced region and the new 3′ flanking DNA (the integration site). Instead, an A-rich sequence was present between the proximal and distal 3′ flanking regions. This same sequence was present at the beginning of the homologous distal flanking region in sequence AC026092 and a similar sequence was present following the proximal flanking region in sequence AC027480 (Fig. 2). These results strongly support the following proposed sequence of events. Transcription of the parental L1 (AC027480) proceeded beyond the proximal flanking region. The target site at locus AC026092 was cleaved immediately upstream of the distal flanking region either by the L1 endonuclease or by another mechanism. First strand cDNA synthesis was then initiated from the nicked strand by the pairing of the A-rich regions in the RNA and AC026092 instead of from the L1 poly(A) tail. These data lend further support to the hypothesis that L1–Hs transposition occurs via TPRT (Luan et al. 1993) and suggests that cDNA priming during L1 transposition can occur internally.
Figure 2.
Organization of the L1 insertion H15/B10-800. The L1 display clone (H15/B10-800) contains the 3′ end of a L1Hs-Ta followed by a 23-bp poly(A) tail and 616-bp of 3′ flanking DNA. GenBank accession no. AC027480 contains a full-length L1Hs-Ta element followed by a 23-bp long poly(A) tail and the proximal 211 bp of 3′-flanking DNA from the H15/B10-800 L1 insertion. More distal 3′ flanking DNA is not homologous to the distal 3′ flanking DNA from H15/B10-800. GenBank accession no. AC026092 is the locus where H15/B10-800 inserted. It contains only the distal 405 bp of the H15/B10-800 3′ flanking DNA. The horizontal arrows indicate the positions of the PCR primers that were used to confirm the structure of the H15/B10-800 integration. The A-rich sequences present at each of the loci between the proximal and distal 3′-flanking DNA are indicated.
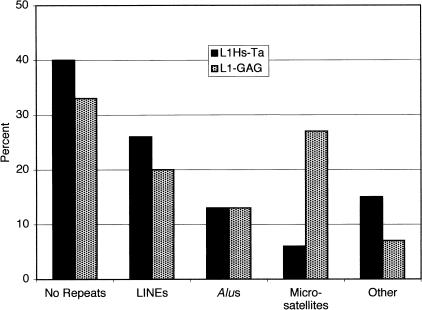
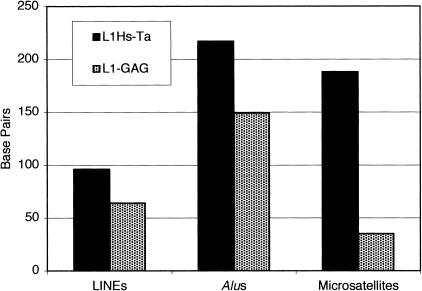
Next we compared the features of the 3′ flanking sequences of the younger L1Hs-Ta and the older L1-GAG insertions. Counting from the first base after the poly(A) tail, the 53 unique L1Hs-Ta clones identified by L1 display contained a total of 26,481 bp of 3′ flanking DNA or an average of 500 bp (S.D. = 227) per clone. For comparison we analyzed 500 bp of 3′ flanking DNA from each of the L1-GAG insertions. We ascertained the presence or absence of various types of repeat sequences in the 3′ flanking DNA of each of the clones by using the RepeatMasker program (Smit and Green 2000). A larger percentage of L1Hs-Ta elements had either no repeats or L1 repeats in their 3′ flanking DNA than did the L1-GAG clones (Fig. 3). These differences were not statistically significant. Alu elements were equally common in the 3′ flanks of the two groups of insertions. We also found that, when present, L1 and Alu sequences in the 3′ flanking regions tended to be located slightly farther from the PAS of the L1Hs-Ta insertions than the L1-GAG insertions (Fig. 4). These differences were also not statistically significant. In contrast, however, microsatellites were much more frequent in the 3′ flanking DNA of L1-GAG elements than L1Hs-Ta elements (Fisher's Exact Test, 2-tailed, P = 0.01) and, when present, they were located closer to the PAS (Figs. 3 and 4). These data indicate that new microsatellites may arise from the poly(A) tails of L1 insertions as they reside in the human genome. A similar situation has been observed for Alu elements (Arcot et al. 1995). These data suggest that L1 and Alu poly(A) tails represent a rich source for the creation of new human microsatellites. This mechanism of creating new microsatellites from the poly(A) tails of L1 insertions may also be responsible for many microsatellites that are not located near obvious L1 fragments because most L1 insertions are 5′-truncated and free poly(A) tails without adjacent L1 sequences may potentially result from truncated L1 transposition events.
Figure 3.
Presence of various types of DNA repeat sequences in the 3′-flanking DNA of L1 insertions. The presence of repeat sequences in the 3′-flanking DNA of L1 insertions was determined by the RepeatMasker program. L1Hs-Ta insertions are represented in the black bars; L1-GAG insertions are represented in the stippled bars.
Figure 4.
Distance from the poly(A) addition signal to the nearest repeat sequence in 3′-flanking DNA. The distance was calculated in bp from the end of the poly(A) addition signal. L1Hs-Ta insertions are represented in the black bars; L1-GAG insertions are represented in the stippled bars.
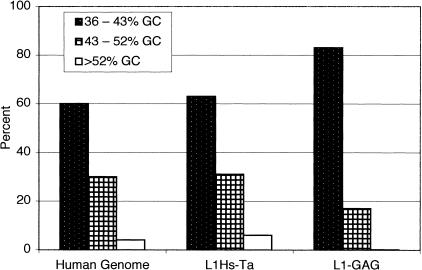
Finally, we compared the GC content of the 3′ flanking DNA of the two groups of L1 insertions. Bernardi and colleagues have suggested that the genomic DNA of eukaryotic organisms is organized into regions of relatively uniform GC content (for review, see Bernardi 1995). These genomic regions are, on average, >300 kb in length and are called “isochores.” Results from the recently published draft human genome sequence confirm that significant long-range variations in GC content exist although the concept of isochores may require redefinition (Lander et al. 2001). We were concerned about the possibility that our calculation of the GC content of the L1 3′-flanking sequences would be affected by the presence of fossils of L1 poly(A) tails. As noted above, the poly(A) tails of L1 elements undergo deterioration and shortening after they are inserted into the genome by transposition. The de novo L1 insertions should therefore be the group of elements with the longest poly(A) tails. Data on the poly(A) tails of de novo germ-line L1 transpositions are currently available for seven insertions (see GenBank accession no. AF149422 and Kazazian et al. 1988; Narita et al. 1993; Holmes et al. 1994; Meischl and Roos 1998; Yoshida et al. 1998). These insertions have poly(A) tails that are 15, 24, 41, 57, 67, 71, and 77 nucleotides long. Accordingly, we calculated the GC content of the 3′ flanking sequences for each of the L1 insertions starting 150 bp after the PAS to ensure that fossil poly(A) tail DNA was not included. Each of the 3′ flanks was then assigned to one of three bins based on its GC content (Fig. 5). The fraction of recent L1Hs-Ta elements in the different bins was proportional to the fraction of human genomic DNA in the same bins (Lander et al. 2001). In contrast, older L1-GAG elements were overrepresented in DNA of low GC content, and underrepresented in DNA of high GC content. These results suggest that older GAG and younger L1Hs-Ta L1s are not similarly distributed in human genomic DNA.
Figure 5.
Distribution of L1 insertions in genomic DNA of different GC content. The GC content of the 3′ flanks of L1 insertions was calculated starting 150 bp after the poly(A) addition signal. Each insertion was assigned to bins of either 36%–43% GC (stippled), 43%–52% GC (checkered), or >52% GC (open box).
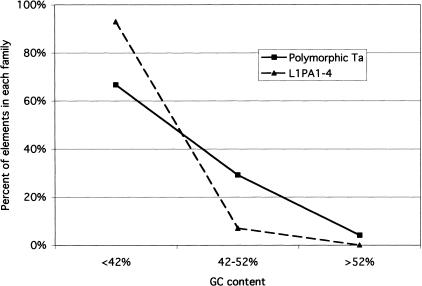
The International Human Genome Sequencing Consortium reported no difference in the distribution of younger and older L1s. One of the differences between the methods used in the two studies is the length of the DNA sequences analyzed. Lander et al. (2001) analyzed the distribution of L1s in 50-kb windows of genomic DNA, whereas we were limited, as a result of the identification of young elements by L1 display, to the analysis of only short regions of DNA. Because both long and short distance variations in GC content are known to exist in the human genome (Lander et al. 2001) we analyzed the distribution of L1s over greater genomic distances. The genomic distribution of a set of polymorphic L1Hs-Ta elements whose insertions were present in GenBank was compared to the distribution of an expanded set of GAG L1s (see Methods). As shown in Figure 6, a marked difference in the distribution of the older and younger L1s was evident when 10 kb of flanking DNA (5 kb on either side of the insertion site) was analyzed. Younger polymorphic L1s were distributed randomly with respect to total human DNA whereas older elements were preferentially located in GC-poor DNA. Similar results were obtained when 2-kb or 20-kb regions of flanking DNA were analyzed. We conclude that recent L1 insertions are distributed randomly among human genomic DNA whereas older L1 insertions are not.
Figure 6.
Comparison of the distribution of younger and older L1 insertions in genomic DNA of different GC content. The GC content of 10 kb surrounding the insertion sites (5 kb of both the 5′ and 3′ flanks) of polymorphic L1Hs–Ta and GAG elements was calculated. Each insertion was assigned to different bins of GC content. For this analysis we analyzed 24 polymorphic insertions, and 57 GAG insertions; the available flanking sequences of one GAG and five polymorphic elements were too short to be included.
DISCUSSION
In this study we have analyzed several characteristics of the flanking regions of recent human L1 insertions and compared them to a group of older insertions. The recent L1 insertions were selected by L1 display whereas the older insertions were identified by a database search. We chose these methods of L1 identification for several reasons. The conclusion that L1 elements are stable residents in the human genome and remain in place for millions of years is based mainly on the observation of older elements (Smit et al. 1995). This is so because most L1s in the human genome are very old and only limited data about LIDs are available. When Ta elements are identified by searching human sequence databases, relatively few elements with very low gene frequencies will be recovered, and the average age of these elements will be higher than for the Ta elements present in the human population as a whole. Rapid changes that may occur to recent L1 insertions would be missed if only older elements were to be analyzed. By selecting recent L1 insertions with L1 display we ensured that our collection would be enriched for younger elements with lower gene frequencies. We chose not to use L1 display to identify older (non-Ta) L1 insertions because the method is less successful at amplifying bands representing older L1s because of the great number of these loci. In addition, we wished to compare the characteristics of the most recent L1 insertions with the L1P elements, a group of elements that were amplified during the primate radiation (Smit et al. 1995). These elements are older than the Ta elements but not so old that they have lost many of their characteristic features. L1 display is successful at identifying Ta elements because they differ from older elements by the ACA trinucleotide; no similar molecular tag is available to distinguish L1P elements from still older L1s.
Although using different methods to identify recent and older L1s could potentially bias our results, we do not believe that this is a significant problem. L1 display identifies Ta elements on the basis of the hybridization of 10-bp primers to the flanking DNA (Sheen et al. 2000). These primers are short enough that little bias is introduced into the selection process. In addition, empiric observation indicates that successful amplifications by L1 display occur with the hybridization of 7–10 of 10 possible matches to the primers (G. Swergold, unpubl.). Indeed, most amplifications occur from binding sites with less than perfect matches. Furthermore, the nonmatching nucleotides may occur anywhere in the primer sequence except the two nucleotides at the 3′ end. These results further diminish any potential bias introduced by the primers. Finally, we have recently investigated a group of L1 insertions that are older than the Ta group but still human-specific. These elements, which were identified by database searches, have characteristics that are intermediate between the Ta elements and the L1P elements including the GC content of their 3′ flanks (I. Ovchinnikov and G. D. Swergold, in prep.). These results indicate that no significant ascertainment bias exists in the present work.
Our results indicate that several changes occur to L1s after integration. First, L1s integrate randomly with respect to the GC content of the target site DNA. To our knowledge this is the first demonstration of a remodeling of the L1 distribution in the human genome. We also report that the length of the L1 poly(A) tails shortens over time, probably as the result of both mutation and replication slippage. A similar observation has been reported for Alu elements (Arcot et al. 1995). In addition, we observed that the L1 PAS degrades more rapidly than the rest of the L1 sequence. As noted above, the 3′ ends of L1 insertions, and presumably of L1 preintegration RNA, is nonstandard in that the poly(A) tail appears to follow immediately after the PAS. An attractive hypothesis has been advanced for this, namely that the L1 PAS is weak and that this allows the genome to better tolerate L1 integrations that occur within introns in the sense orientation (Moran et al. 1999). This hypothesis also helps explain the frequent occurrence of 3′ flanking transductions during L1 integrations (Goodier et al. 2000; Pickeral et al. 2000) and the experimental finding that alternative PAS's are preferred when available in in vitro transposition assays (Moran et al. 1996). Our finding that the L1 PAS is relatively unstable after integration is consistent with this hypothesis because any detrimental effects that may occur from the L1 PAS after integration will be mitigated by its degradation.
We were only able to find evidence for 3′-transduction events in two of the 53 L1Hs-Ta integrations, significantly fewer than expected (Goodier et al. 2000; Pickeral et al. 2000). We believe that the primary reasons for this are the relatively short 3′ flanking sequences that are cloned by the L1 display technique (500 bp), and the absence of 5′ flanking DNA in the L1 display clones. In one of the likely transduction events, first strand cDNA synthesis was primed not from the L1 poly(A) tail but instead from an A-rich sequence that was present in the transduced region and that was homologous to the sequence at the integration site. This represents the first reported example of an L1Hs transposition that occurred as the result of an internal priming event. Furthermore, it suggests that the ability of the L1 transposition machinery to prime cDNA synthesis from a homologous internal binding site may permit the development of L1-based vectors that can be targeted to specific insertion sites.
The young L1Hs-Ta elements discovered during the course of this work were distributed randomly both with respect to chromosome and to GC content. Recent studies reported that the X and Y chromosome are abundant in young L1s (Bailey et al. 2000; Lander et al. 2001). Although our data appear to contradict these findings, the power of the present study to detect a nonrandom chromosomal distribution of L1Hs-Ta elements is low due to the relatively small number of insertions (32) that we analyzed distributed on 15 different chromosomes (Table 1). Although the distribution of the L1Hs-Ta elements did not differ statistically from a random distribution, we note that a relatively high number (five) were present on the sex chromosomes. Future analyses with a greater number of polymorphic Ta elements are needed to determine whether L1 insertions are targeted to the X and Y chromosome or whether insertions are more stable when they occur there.
In contrast, our data strongly support the random insertion of human L1s with respect to the GC content of the target sites. These data derive from two separate analyses. In the first, the distribution of 53 L1Hs-Ta insertions was compared to the distribution of 30 older GAG elements. This analysis was performed on relatively short stretches of 3′ flanking sequences due to the limitations of our L1Hs-Ta discovery method. In the second, the GC distribution of a set of polymorphic L1Hs-Ta elements present in GenBank was compared to the distribution of an expanded set of GAG L1s. This analysis, which was performed over much greater spans of genomic DNA, confirmed that recent L1s appear to be randomly distributed with respect to GC content whereas older L1s are not. The findings reported here are further supported by other studies that indicate that older but still human-specific L1 subfamilies have characteristics that are intermediate between the L1Hs-Ta and L1-GAG elements reported here (I. Ovchinnikov and G.D. Swergold, in prep.).
Lander et al. (2001) reported that both older and younger L1s are preferentially located in GC-poor genomic regions. They concluded that L1 target site selection is not random but favors GC-poor DNA, possibly as the result of the preference of the L1 endonuclease for AT-rich sequences. Why did they not observe a distribution shift for L1s? We propose that it likely resulted from their choice of which elements to include in the “young” category. This group included both human-specific and primate-specific L1s and therefore included elements that were >30 million years old (Smit et al. 1995). The signal from the young elements was probably overwhelmed by the older elements and therefore was not evident in the data. In contrast, our group of young elements is greatly enriched for polymorphic elements and is therefore likely to be <250,000 years old. We note that our L1-GAG group of elements is distributed nonrandomly and is similar in age to the group of young elements analyzed by Lander et al (2001). These data suggest that the shift in the L1 distribution occurs relatively rapidly, in contrast to the shift of the distribution of Alus that appears to occur more slowly (Lander et al. 2001). They also suggest that the preference of the L1 endonuclease for AT-rich cleavage sites does not impose a strong bias on target site selection. The ability of L1Hs-Ta elements to integrate randomly into the genome also suggests that L1-induced insertional mutagenesis may be a useful tool for creating mouse mutant “libraries.”
Our data do not address the question of what mechanism is responsible for the alteration in L1 distribution over time. We favor the hypothesis that the shift in the distribution of L1 elements occurs as a result of selection either for the retention of L1s in regions of low GC or for the loss of L1s from regions of high GC. Lander et al. (2001) suggested that the shift of the distribution of Alus towards DNA of high GC was a result of the positive selection for Alus located near genes (Lander et al. 2001). It is interesting to note that extremely old L1s in the human genome appear to be distributed more randomly with respect to GC (Lander et al. 2001). One possible unifying hypothesis is that active, full-length L1 elements are selected against when they are present in GC-rich (and therefore transcriptionally active) DNA. Extremely old L1s, which have accumulated lethal mutations and are therefore no longer transpositionally active, would no longer be subject to this selective process and may become randomly distributed again over long periods of evolutionary time. Indeed, Boissinot et al. (2001) have recently reported evidence that full-length L1s are selectively lost from the human genome. If this process selectively removes L1s that are potentially active, it may explain how L1s may be selectively lost from GC-rich DNA, because actively transcribed genes are more commonly located in these regions. The possibility that only transpositionally active L1 elements located in GC-rich DNA are selected against can be tested.
An alternative explanation for the origin of different distributions of L1Hs-Ta and GAG L1s in the human genome is that the selection of insertion sites during L1 transposition has changed over time. According to this model, older elements were inserted when L1 transposition favored GC-poor DNA regions, but modern L1 transposition occurs randomly with respect to GC content. We do not favor this model for several reasons. First, few diagnostic nucleotide changes define the different classes of L1s that have inserted into the human genome during the last 20–30 million years (Smit et al. 1995; Boissinot et al. 2000; I. Ovchinnikov and G.D. Swergold, in prep.). It is unlikely that these few mutations have induced a major change in the selection of target sites during transposition. Second, L1 subfamilies of intermediate age have intermediate patterns of distribution with respect to GC content, further supporting a process of “remodeling” (I. Ovchinnikov and G.D. Swergold, in prep.). Third, Alu elements are inserted randomly into the human genome and are redistributed towards GC-rich DNA over the course of 60–100 million years (Arcot et al. 1998; Lander et al. 2001). The L1 transposition machinery is believed to mobilize Alu elements (Dombroski et al. 1991; Jurka 1997) and it is difficult to reconcile the difference in the kinetics of the redistribution of Alu and L1 elements in the human genome on the basis of a change in target site selection. Still, although we have presented evidence that young and old L1's are distributed differently in human genomic DNA, the question of whether this difference is the result of a selective process must await future studies.
METHODS
DNA Samples
The DNA samples used for LID discovery by L1 display included the six samples described previously (Sheen et al. 2000). In addition, the samples included (1) 38 samples from the Coriell repository (10 Northern Europeans, 10 African Americans, four Amish, and four Druze); (2) 17 samples from the repository of the National Laboratory for the Genetics of Israeli Populations, Tel Aviv University (three samples each of Sephardic Jews, Palestinians, Iraqi Jews, Yemenite Jews, Ethiopian Jews, and two samples of Ashkenazi Jews); (3) 12 samples kindly donated by M. Stoneking, Max Planck Institute for Evolutionary Anthropology (six Indonesians and six Papua New Guineans); (4) two Chinese samples kindly donated by Li Jin (University of Texas); and (5) 16 samples (four Northern Europeans, six Hispanics, four African Americans, and two Asians) collected from patients under a protocol approved by the Columbia Presbyterian Medical Center Institutional Review Board. DNA was either purchased or extracted from blood samples using standard protocols.
Discovery of L1 Insertions
To collect a random sample of L1 elements that were recently inserted into the human genome, we performed L1 display on 91 samples of DNA from individuals representing many geographic regions. L1 display was performed as described previously with the following changes. PCR reactions were performed in 96-well plates along with negative controls that had no genomic DNA added. Amplifications were run in MJ Research model PTC 200 DNA Engines. PCR fragments that were selected for further analysis were isolated from agarose gels by the Wizard PCR Prep DNA Purification kit (Promega) and cloned into the pGEM-T vector (Promega). Automated DNA sequencing of the clones was performed on both strands with the ABI PRISM BigDye Termination Cycle Sequencing protocol by the DNA Sequencing Core Laboratory of the Herbert Irving Comprehensive Cancer Center of Columbia University. GenBank accession numbers of the sequences are AF417122–AF417164.
A group of older L1 insertions was collected as follows. A BLASTN search of the nonredundant GenBank database was performed using a query sequence that consisted of bp 5914–6026 of LRE-1 in which the ACA sequence (bp 5930–5932) was replaced with GAG and the 6015 G was replaced with A. This query sequence was chosen to favor the identification of L1 insertions that did not belong to subset Ta but nevertheless were relatively recent insertions into the human genome. Thirty clones with >95% sequence identity to the query and with the “GAG” and 6015 “A” nucleotides were chosen. The accession numbers of the clones are: AC006984, HS1100E15, AC002556, AP001432, HS581F12, ACOO3099, HS260B21, AC004065, AC008012, CNS0000I, HSDJ581P3, HS82J11, AC007198, AC002564, AC007320, AB019437, AC005820, AB020870, HUAE000659, AF017104, AC002385, HSDJ80E14, HSBG54N10, HS134N8, HSDJ828H9, CNS01DRZ, HS23K20, AL022166.1, AC004053, and HS466P17.
To analyze the GC content of L1-flanking DNA over long genomic distances, we required sets of older and younger elements that (1) had identifiable target site duplications, and (2) were present in GenBank. Several of the older GAG elements described above did not have identifiable target site duplications. To the group of 20 elements that did, we added an additional 38 GAG elements, identified as described above, with identifiable target site duplications. A set of younger elements was assembled that (1) belonged to subset Ta,(2) were known to be polymorphic in the human genome, (3) were represented in the GenBank database, and (4) had identifiable target site duplications. These elements were drawn from the literature (15 from Boissinot et al. [2000], and six from Sheen et al. [2000]) and from the present study (eight elements, data not shown).
Informatics
BLAST searches were performed either with the Web BLAST server or with the MacVector program (version 6.5.3, Oxford Molecular Group). Genomic loci representing the insertion sites for the L1Hs-Ta insertions identified by L1 display were identified by searching the nucleotide and high-throughput genomic sequence GenBank databases with the 3′ flanking sequences of the insertions. The presence of repeat sequences in the 3′ flanks of L1 insertions was determined by the RepeatMasker program (Smit and Green 2000). The GC content of the 3′ flanks was calculated by the MacVector program. The possibility that an insertion represented a transduction event was considered when the 3′ flanking DNA had nearly perfect matches to two different genomic loci, and one of the loci contained a correctly positioned, full-length L1Hs-Ta element with intact open reading frames (ORFs).
Statistical Analysis
The L1Hs-Ta and L1-GAG groups were compared using t-tests for continuous factors and Fisher's exact tests for dichotomous factors, using a significance level of 0.05. We also assessed the continuous factors using the nonparametric Wilcoxon rank-sum test but found no differences in results. The placement of L1s among the chromosomes was tested using a goodness-of-fit test for the multinomial distribution. The lengths of the chromosomes were taken into account.
Acknowledgments
The authors thank Drs. H. Kazazian, J. Goodier, and E. Ostertag for helpful discussions.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL gs314@columbia.edu; FAX (212) 342-5316.
Article published on-line before print: Genome Res., 10.1101/gr.194701.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.194701.
REFERENCES
- Adams JW, Kaufman RE, Kretschmer PJ, Harrison M, Nienhuis AW. A family of long reiterated DNA sequences, one copy of which is next to the human beta globin gene. Nucleic Acids Res. 1980;8:6113–6128. doi: 10.1093/nar/8.24.6113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Arcot SS, Adamson AW, Risch GW, LaFleur J, Robichaux MB, Lamerdin JE, Carrano AV, Batzer MA. High-resolution cartography of recently integrated human chromosome 19—specific Alu fossils. J Mol Biol. 1998;281:843–856. doi: 10.1006/jmbi.1998.1984. [DOI] [PubMed] [Google Scholar]
- Arcot SS, Wang Z, Weber JL, Deininger PL, Batzer MA. Alu repeats: A source for the genesis of primate microsatellites. Genomics. 1995;29:136–144. doi: 10.1006/geno.1995.1224. [DOI] [PubMed] [Google Scholar]
- Bailey JA, Carrel L, Chakravarti A, Eichler EE. Molecular evidence for a relationship between LINE-1 elements and X chromosome inactivation: The Lyon repeat hypothesis. Proc Natl Acad Sci. 2000;97:6634–6639. doi: 10.1073/pnas.97.12.6634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker RJ, Kass DH. Comparison of chromosomal distribution of a retroposon (LINE) and a retrovirus-like element mys in Peromyscus maniculatus and P. leucopus. Chromosome Res. 1994;2:185–189. doi: 10.1007/BF01553318. [DOI] [PubMed] [Google Scholar]
- Bernardi G. The human genome: Organization and evolutionary history. Annu Rev Genet. 1995;29:445–476. doi: 10.1146/annurev.ge.29.120195.002305. [DOI] [PubMed] [Google Scholar]
- Boissinot S, Chevret P, Furano AV. L1 (LINE-1) retrotransposon evolution and amplification in recent human history. Mol Biol Evol. 2000;17:915–928. doi: 10.1093/oxfordjournals.molbev.a026372. [DOI] [PubMed] [Google Scholar]
- Boissinot S, Entezam A, Furano AV. Selection against deleterious line-1-containing loci in the human lineage. Mol Biol Evol. 2001;18:926–935. doi: 10.1093/oxfordjournals.molbev.a003893. [DOI] [PubMed] [Google Scholar]
- Boyle AL, Ballard SG, Ward DC. Differential distribution of long and short interspersed element sequences in the mouse genome: Chromosome karyotyping by fluorescence in situ hybridization. Proc Natl Acad Sci. 1990;87:7757–7761. doi: 10.1073/pnas.87.19.7757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cost GJ, Boeke JD. Targeting of human retrotransposon integration is directed by the specificity of the L1 endonuclease for regions of unusual DNA structure. Biochemistry. 1998;37:18081–18093. doi: 10.1021/bi981858s. [DOI] [PubMed] [Google Scholar]
- Dombroski BA, Mathias SL, Nanthakumar E, Scott AF, Kazazian HH., Jr Isolation of an active human transposable element. Science. 1991;254:1805–1808. doi: 10.1126/science.1662412. [DOI] [PubMed] [Google Scholar]
- Dombroski BA, Scott AF, Kazazian HH., Jr Two additional potential retrotransposons isolated from a human L1 subfamily that contains an active retrotransposable element. Proc Natl Acad Sci. 1993;90:6513–6517. doi: 10.1073/pnas.90.14.6513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Q, Moran JV, Kazazian HH, Jr, Boeke JD. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell. 1996;87:905–916. doi: 10.1016/s0092-8674(00)81997-2. [DOI] [PubMed] [Google Scholar]
- Goodier JL, Ostertag EM, Kazazian HH., Jr Transduction of 3′-flanking sequences is common in L1 retrotransposition. Hum Mol Genet. 2000;9:653–657. doi: 10.1093/hmg/9.4.653. [DOI] [PubMed] [Google Scholar]
- Grimaldi G, Skowronski J, Singer MF. Defining the beginning and end of the KpnI family segments. EMBO J. 1984;3:1753–1759. doi: 10.1002/j.1460-2075.1984.tb02042.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes SE, Dombroski BA, Krebs CM, Boehm CD, Kazazian HH., Jr A new retrotransposable human L1 element from the LRE2 locus on chromosome 1q produces a chimaeric insertion. Nat Genet. 1994;7:143–148. doi: 10.1038/ng0694-143. [DOI] [PubMed] [Google Scholar]
- Hwu HR, Roberts JW, Davidson EH, Britten RJ. Insertion and/or deletion of many repeated DNA sequences in human and higher ape evolution. Proc Natl Acad Sci. 1986;83:3875–3879. doi: 10.1073/pnas.83.11.3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurka J. Sequence patterns indicate an enzymatic involvement in integration of mammalian retroposons. Proc Natl Acad Sci. 1997;94:1872–1877. doi: 10.1073/pnas.94.5.1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazazian HHJ, Wong C, Youssoufian H, Scott AF, Phillips DG, Antonarakis S. Haemophilia A resulting from de novo insertion of L1 sequences represents a novel mechanism for mutation in man. Nature (London) 1988;332:164–166. doi: 10.1038/332164a0. [DOI] [PubMed] [Google Scholar]
- Korenberg JR, Rykowski MC. Human genome organization: Alu, Lines, and the molecular structure of metaphase chromosome bands. Cell. 1988;53:391–400. doi: 10.1016/0092-8674(88)90159-6. [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, Fitzhugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Luan DD, Korman MH, Jakubczak JL, Eickbush TH. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: A mechanism for non-LTR retrotransposition. Cell. 1993;72:595–605. doi: 10.1016/0092-8674(93)90078-5. [DOI] [PubMed] [Google Scholar]
- Malik HS, Burke WD, Eickbush TH. The age and evolution of non-LTR retrotransposable elements. Mol Biol Evol. 1999;16:793–805. doi: 10.1093/oxfordjournals.molbev.a026164. [DOI] [PubMed] [Google Scholar]
- Meischl C, Roos D. The molecular basis of chronic granulomatous disease. Springer Semin Immunopathol. 1998;19:417–434. doi: 10.1007/BF00792600. [DOI] [PubMed] [Google Scholar]
- Moran JV, Holmes SE, Naas T, DeBerardinis P, Boeke RJ, Kazazian JD, Jr HH. High-frequency retrotransposition in cultured mammalian cells. Cell. 1996;87:917–927. doi: 10.1016/s0092-8674(00)81998-4. [DOI] [PubMed] [Google Scholar]
- Moran JV, DeBerardinis RJ, Kazazian HH., Jr Exon shuffling by L1 retrotransposition. Science. 1999;283:1530–1534. doi: 10.1126/science.283.5407.1530. [DOI] [PubMed] [Google Scholar]
- Moyzis RK, Torney DC, Meyne J, Buckingham JM, Wu J-R, Burks C, Sirotkin KM, Goad WB. The distribution of interspersed repetitive DNA sequences in the human genome. Genomics. 1989;4:273–289. doi: 10.1016/0888-7543(89)90331-5. [DOI] [PubMed] [Google Scholar]
- Narita N, Nishio H, Kitoh Y, Ishikawa Y, Ishikawa Y, Minami R, Nakamura H, Matsuo M. Insertion of a 5′ truncated L1 element into the 3′ end of exon 44 of the dystrophin gene resulted in skipping of the exon during splicing in a case of Duchenne muscular dystrophy. J Clin Invest. 1993;9:1862–1867. doi: 10.1172/JCI116402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickeral OK, Makaowski W, Boguski MS, Boeke JD. Frequent human genomic DNA transduction driven by LINE-1 retrotransposition [In Process Citation] Genome Res. 2000;10:411–415. doi: 10.1101/gr.10.4.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassaman DM, Dombroski BA, Moran JV, Kimberland ML, Naas TP, DeBerardinis RJ, Gabriel A, Swergold GD, Kazazian HH., Jr Many human L1 elements are capable of retrotransposition. Nat Genet. 1997;16:37–43. doi: 10.1038/ng0597-37. [DOI] [PubMed] [Google Scholar]
- Sheen F, Sherry ST, Risch GM, Robichaux M, Nasidze I, Stoneking M, Batzer MA, Swergold GD. Reading between the LINEs: Human genomic variation induced by LINE-1 retrotransposition. Genome Res. 2000;10:1496–1508. doi: 10.1101/gr.149400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skowronski J, Fanning TG, Singer MF. Unit-length line-1 transcripts in human teratocarcinoma cells. Mol Cell Biol. 1988;8:1385–1397. doi: 10.1128/mcb.8.4.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit AFA. The origin of interspersed repeats in the human genome. Curr Opin Genet Dev. 1996;6:743–748. doi: 10.1016/s0959-437x(96)80030-x. [DOI] [PubMed] [Google Scholar]
- Smit, A.F.A. and Green, P. 2000. RepeatMasker at http://ftp.genome.washington.edu/RM/RepeatMasker.html.
- Smit AFA, Toth G, Riggs AD, Jurka J. Ancestral mammalian-wide subfamilies of LINE-1 repetitive sequences. J Mol Biol. 1995;246:401–417. doi: 10.1006/jmbi.1994.0095. [DOI] [PubMed] [Google Scholar]
- Soriano P, Meunier-Rotival M, Bernardi G. The distribution of interspersed repeats in nonuniform and conserved in the mouse and human genomes. Proc Natl Acad Sci. 1983;80:1816–1820. doi: 10.1073/pnas.80.7.1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong YE, Eickbush TH. Functional expression of a sequence-specific endonuclease encoded by the retrotransposon R2Bm. Cell. 1988;55:235–246. doi: 10.1016/0092-8674(88)90046-3. [DOI] [PubMed] [Google Scholar]
- Yang J, Malik HS, Eickbush TH. Identification of the endonuclease domain encoded by R2 and other site-specific, non-long terminal repeat retrotransposable elements. Proc Natl Acad Sci. 1999;96:7847–7852. doi: 10.1073/pnas.96.14.7847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K, Nakamura A, Yazaki M, Ikeda S, Takeda S. Insertional mutation by transposable element, L1, in the DMD gene results in X-linked dilated cardiomyopathy. Hum Mol Genet. 1998;7:1129–1132. doi: 10.1093/hmg/7.7.1129. [DOI] [PubMed] [Google Scholar]