Abstract
Deducing the structure of the DNA double helix in 1953 implied the mode of its replication: Watson-Crick base pairing might instruct an enzyme, now known as the DNA polymerase, during the synthesis of a daughter stand complementary to a single strand of the parental double helix. What has become increasingly clear in the last sixty years, however, is that adducted and oxidatively generated DNA bases are ubiquitous in physiological DNA, and all organisms conserve multiple DNA polymerases specialized for DNA synthesis opposite these damaged templates. Here, we review recent crystal structures depicting replicative and bypass DNA polymerases encountering two typical lesions arising from the oxidation of DNA: abasic sites, which block the replication fork, and the miscoding premutagenic lesion 7,8-dihydro-8-oxoguanine.
Introduction
Aerobic organisms must actively protect their genomes, not only from a milieu of toxic exogenous environmental agents, but also from the reactive byproducts of metabolism. Reactive oxygen species produce an array of DNA lesions that can confound replication if left unrepaired. It is therefore not surprising that a diverse set of genomes, ranging in complexity from that of some viruses to Homo sapiens, conserve enzymes able to recognize and remove all types of oxidant damage in DNA [1]. Despite these precautions, however, a low level of DNA damage inevitably escapes repair and persists to later confront the replication machinery with a chemically modified substrate. This occurrence necessitates a process of translesion DNA synthesis (TLS), the fidelity of which depends substantially on the incorporation specificity and bypass parameters of the specific DNA polymerase recruited to catalyze this special-case reaction [2].
The human genome encodes 14 template-dependent DNA polymerases belonging to 4 different sequence families (A, B, X, and Y) and the conservation of these enzymes supports an important role for each in DNA replication and repair [3,4]. Although high-fidelity replicative polymerases catalyze the majority of DNA synthesis at the replication fork, it is believed that polymerase switching occurs when the DNA template is damaged [5,6]. Replicative DNA polymerases impose strict geometric constraints during selection of the correct nucleotide complementary to the templating base [7]. High-fidelity is also a consequence of closely monitoring the DNA minor groove during translocation of the nascent base pair, which can trigger the 3'–5' exonuclease proofreading activity associated with replicative polymerases if a mismatch or abnormal base pair is sensed. Therefore, when a site of damage violates these quality control measures, causing either persistent misincorporation or failed translocation, the replication fork stalls and it becomes necessary to recruit a specialized polymerase able to synthesize DNA opposite the modified template and bypass the lesion.
Specialized polymerases capable of bypassing DNA lesions generally lack both the tight fitting active site and 3'–5' exonuclease activity associated with replicative polymerases. While polymerases from all sequence families share an approximate topology likened to a right hand—with thumb, palm, and fingers domains—replicative polymerases especially rely on a large conformational change to achieve high-fidelity [8]. After selecting the correct nucleotide, the fingers domain of the replicative polymerase rotates toward the palm, thereby forming the fully assembled solvent-excluded active site, which aligns the α-phosphate of the incoming nucleotide for nucleophilic attack by the primer 3' terminus [9]. In contrast, compiling all available structures depicting family Y polymerases binding DNA and nucleotides does not produce a single example of a substantial conformational change for this class of bypass polymerases, despite the varied configurations these DNA complex structures represent [10]. For this reason, current research portrays the active sites of these specialized bypass polymerases as open and preformed, therefore able to accommodate certain bulky adducts or DNA lesions without extensive reliance on induced fit [11].
In the last decade, dozens of crystal structures were deposited into the Protein Data Bank (PDB) depicting complexes between DNA polymerases binding modified primer•templates. Moreover, contributions of the last several years have substantially expanded our understanding of TLS catalyzed by specialized bypass polymerases, especially from the family Y. With this increasing set of atomic resolution models describing a varied set of DNA polymerases caught deploying different strategies to process the same DNA lesion, we are now coming to fully appreciate the diversity of DNA polymerases conserved in eukaryotic genomes. Reviewed here are a collection of recent crystal structures that facilitate this closer look at the adaptations of bypass polymerases allowing these specialized enzymes to successfully process aberrant templates unsuitable for replicative polymerases. This discussion will focus primarily on two of the most common forms of damage arising due to the oxidation of DNA: abasic or apurinic/apyrimidinic (AP) sites and the product of the oxidation of guanine, 7,8-dihydro-8-oxoguanine (8-oxoG).
Abasic sites stall replicative polymerases
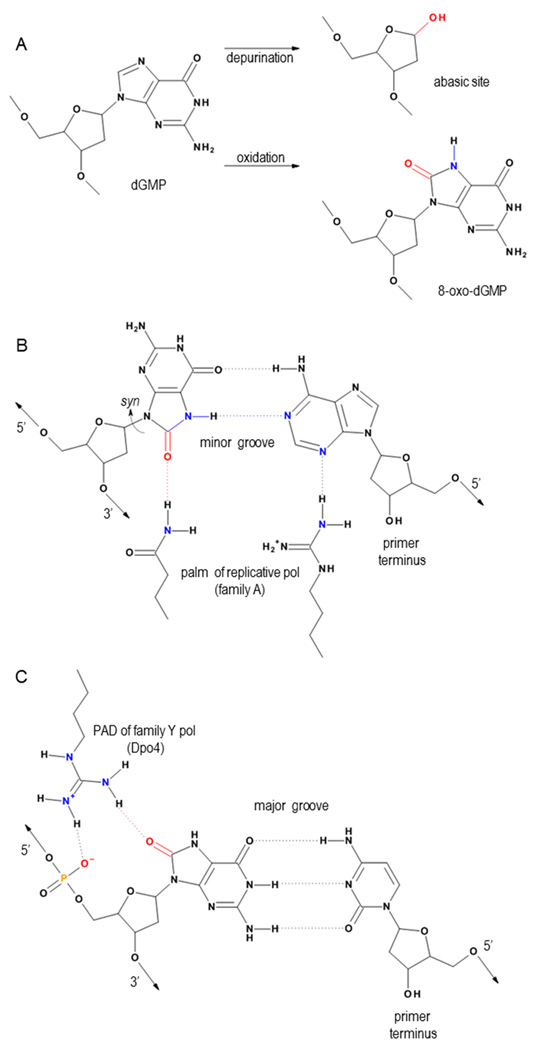
The deoxyribose sugar of the DNA backbone is susceptible to hydroxyl radical attack such that the base is liberated, leaving a fragmented sugar moiety in place of the intact nucleotide. This type of DNA damage is referred to as an abasic site and is analogous to the product resulting from cleavage of the N-glycosylic bond by DNA glycosylases when these enzymes excise a damaged base and initiate the base excision repair (BER) pathway [12]. A third mode of abasic site formation occurs primarily at guanines in the DNA, which can depurinate spontaneously (Figure 1A). DNA synthesis opposite an AP site is inherently error prone, since this lesion carries no genetic information. Interestingly, without instruction from a templating base, various polymerases process abasic sites differently, leaving behind certain mutational spectra. Polymerases from families A and B obey the A-rule, preferentially inserting dAMP without instruction from the template [13]. This trend is attributed to the preferential stacking and desolvation characteristics of dATP over other nucleoside triphosphates [14,15]. Replicative polymerases also share an inability to translocate a nucleotide paired with an abasic site, which leaves the primer terminus vulnerable to exonuclease proofreading and subsequent cycles of dAMP incorporation and excision. Crystal structures of the family B replicative polymerase from bacteriophage RB69 (gp43) illustrated a loss of minor groove contacts and a move toward proofreading when binding DNA containing a templating tetrahydrofuran, a stable abasic site analog [16,17]. Accordingly, abasic sites provide strong blocks to replication forks that might prohibit DNA synthesis and otherwise kill dividing cells lacking the specialized enzymes able to bypass these DNA lesions.
Figure 1.
(A) Premutagenic DNA lesions form at guanine nucleotides by several mechanisms. Spontaneous depurination constitutes one such mechanism giving rise to abasic sites. 8-oxoG, the product of oxidation of the guanine nucleobase, becomes protonated at the N7. (B) When paired with dAMP, replicative polymerases translocate the 8-oxoG mismatch without triggering 3’→5’ exonuclease activity. The extraneous O8 mimics the N3 of an undamaged purine and is therefore able to interact appropriately with conserved H-bond donors in the palm domain. (C) Y-family polymerases encode an additional domain, the PAD, which facilitates TLS. This example depicts Dpo4 stabilizing the anti form of 8-oxoG by juxtaposing an arginine residue between the 5’ phosphate and the O8 of 8-oxoG such that the lesion pairs with dCMP.
Side chains can govern nucleotide incorporation opposite abasic sites
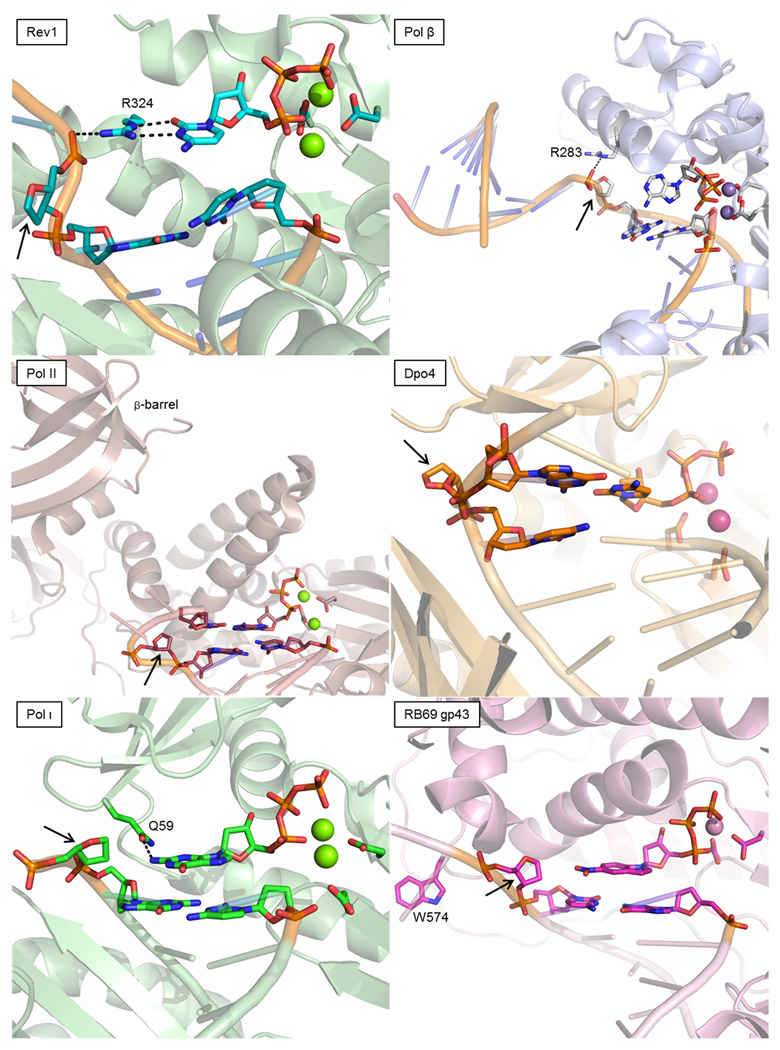
The notion that side chains could dictate the incorporation specificity of a DNA polymerase was realized with the first crystal structure of Rev1, in which an arginine residue provides for this family Y enzyme's propensity to incorporate dCMP opposite a wide array of templates [18]. A recent crystal structure verified the prediction that the active site arginine (R324) of Rev1 could hydrogen bond to the Watson-Crick (WC) face of dCTP while an extruded abasic site resided in the templating position (Figure 2, top left) [19]. Given the predominance of spontaneous depurination of dGMP, Rev1 is believed to catalyze the incorporation step of “best guess” TLS opposite abasic sites, relying on the associated polymerase ζ to catalyze the extension step [20]. Although Rev1 appears unique in its incorporation specificity, side chains might direct incorporation opposite abasic sites in other enzymes as well, by both providing direct contacts to the incoming nucleotide or perhaps by more indirect means that impinge on conformational changes. The family X polymerase β (pol β), which is a specialized gap filling enzyme participating in BER, follows the A-rule when the abasic site is paired opposite a single nucleotide gap [21]. In a recent crystal structure, pol β was observed stabilizing the template strand by stacking an arginine (R283) against the templating furan and hydrogen bonding to the 5’ phosphate (Figure 2, top right). This particular conformation had not been observed previously for R283, which usually interacts with the minor groove of the template during normal DNA synthesis. When pol β binds a mismatched nucleoside triphosphate, however, R283 occupies the template binding pocket, discouraging assembly of the fully closed complex [22]. This more recent crystal structure of pol β therefore correlated a novel R283 conformation with dATP binding opposite furan templates, providing the hypothesis that R283 played a role in pol β‘s A-rule without actually contacting the incoming dATP. Mutational analysis supported this hypothesis and demonstrated that the R283A variant forfeited the ability to efficiently incorporate dAMP opposite an abasic site. This difference was mostly due to a reduction in the catalytic rate, rather than by a loss of dATP binding, inferring that R283 stabilizes a conformation of pol β able to properly align dATP opposite an abasic site for nucleophilic attack.
Figure 2.
Replicative and specialized polymerases process abasic sites (indicated by black arrows) differently by partitioning the template strand into alternative conformations. Rev1 employs H-bonds (dashed lines) from R357 to direct incorporation of dCTP, while extruding the furan template (PDBID 3OSP [19]). Polymerase β requires R283 to stack under the abasic site for efficient selection of dATP and alignment of the primer terminus of this gapped duplex DNA substrate (PDBID 3ISD [21]). A single hydrogen bond is also observed form R283 to the phosphate backbone. Polymerase II flips out three nucleotides, including the abasic site, and utilizes an upstream (+3) nucleotide for incorporation of dATP in this particular example (PDBID 3K5L [28]). Dpo4, like pol II, flips out the abasic site and allows an upstream (+1) guanine to pair with dCTP in the polymerase active site (PDBID 1S0N [27]). Polymerase ι squeezes the active site such that the incoming nucleotide stacks with the template strand (−1 base) and engages in van der Waals contacts with the lesion itself (PDBID 3G6X [33]). Pol ι inserts dGTP slightly more readily than other nucleotides due the H-bond depicted between Q59 and the exocyclic N2. The replicative polymerase RB69 gp43 does not utilize any of these template rearrangements. 5-NITP, a non natural dATP analog, protrudes its nitro moiety into the void created by the presence of the abasic site (PDBID 2OZM [15]). This substituent stacks with the template (−1 base), which is reminiscent of nucleotide incorporation opposite an abasic site by pol ι.
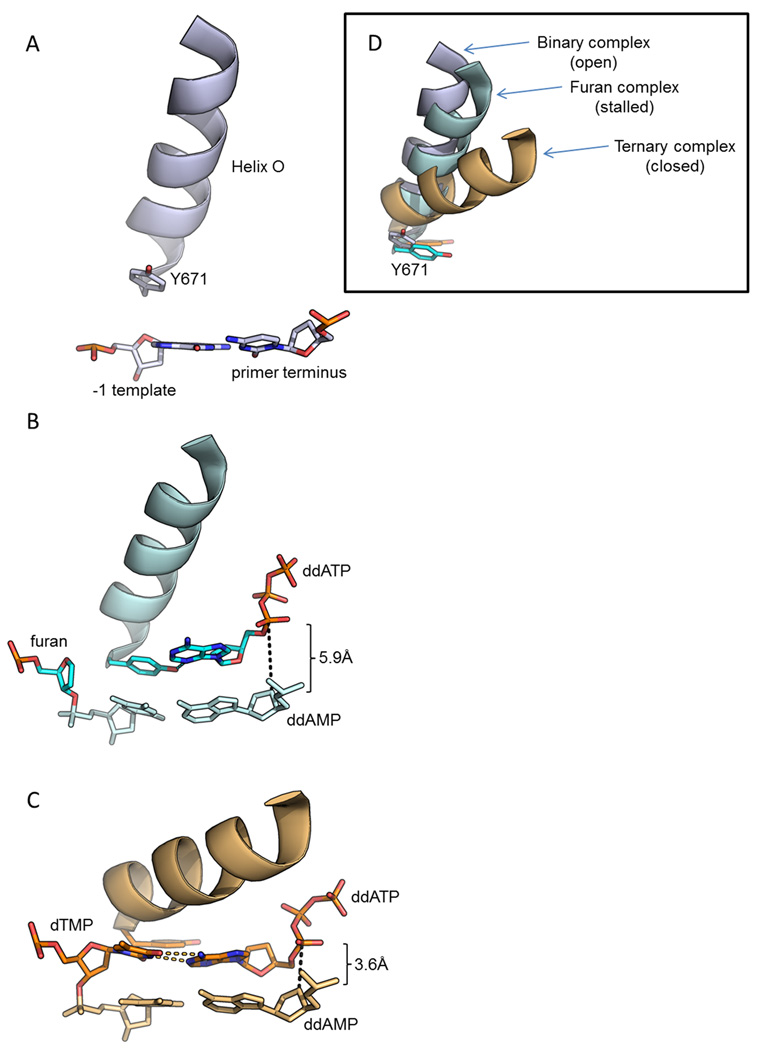
The catalytic cycle of high-fidelity family A and B polymerases differs slightly, due to the presence of a preinsertion site in family A polymerases. A tyrosine or phenylalanine residue conserved in the helix O stacks with the translocated −1 template base (Figure 3A) during nucleotide binding in family A polymerases while the templating base remains extrahelical in the pre-insertion site, possibly providing a safe-guard against template slippage and frameshift mutations [23]. Therefore, before the incoming nucleotide is interrogated by the template, a conformational change in the fingers domain must displace the aromatic residue from its position stacking with the templating strand and allow the templating base to flip into the polymerase active site (Figure 3C). Recently, Klentaq was crystallized in complex with furan containing DNA and dideoxy-dATP (ddATP), providing the first glance of DNA synthesis opposite an abasic in a family A DNA polymerase [24]. Klentaq, a thermostable DNA polymerase from Thermus aquaticus, shares extensive homology to the analogous Klenow fragment, which was originally derived by proteolytic cleavage of Escherichia coli DNA polymerase I to eliminate the 5’→3’ exonuclease domain. In the crystal structure, Klentaq oriented the DNA such that the furan resided in the pre-insertion site and the fingers occupied a conformation intermediate between open and closed (Figure 3D, insert). The triphosphate tail of the nucleotide contacted the conserved basic residues of the fingers domain, R659 and K663, as expected. The N3 of the adenine base, however, formed a H-bond with the hydroxyl of the conserved Y671 (Figure 3B). Due to the intermediate conformation in the fingers domain, the furan complex of Klentaq depicted the α-phosphate 2.3 Å farther from the primer terminus than its position in the previously solved fully assembled closed complex with templating dTMP [25]. Since the primer terminus conformation is identical in these two structures, it appears unlikely that nucleophilic attack could span the additional space, suggesting that the new Klentaq structure represents a stalled complex. Although the authors proposed that the H-bond to Y671 implied a structural basis for selection of adenine, it is unclear why the N3 of guanine would not allow dGTP to bind similarly across the abasic site. Because Klentaq binds dGTP with a higher affinity than dATP opposite furan [24], the chemical step assumes great importance in explaining the higher efficiency (Kd/kpol) for incorporation of dATP. Therefore, given that the furan complex structure does not likely represent the fully assembled catalytically competent complex, the hypothesis that preferable stacking and desolvation characteristics of dATP explains the A-rule for replicative polymerases likely applies to Klentaq as well [15].
Figure 3.
Klentaq stalls while inserting dATP opposite an abasic site. (A) The binary complex illustrates the conformation of helix O and Y671 while the fingers are fully open (PDBID 4KTQ [56]). (B) In the stalled complex where furan occupies the preinsertion site, a hydrogen bond is formed between Y671 and dATP (PDBID 3LWL [24]). At this point, approximately 5.9Å separate the α-phosphate and the C3’ at the primer terminus, making nucleophilic attack unlikely in this intermediate conformation. (C) In the closed ternary complex, the distance between the α-phosphate and the primer terminus is 3.6 Å, showing that full closure of the fingers domain reduces this distance by ~2.3 Å from the stalled complex (PDBID 1QSY [25]). (D, insert) The superposition of binary, furan, and ternary structures shows that the furan complex represents a stalled complex. The closure of the fingers domain provides a twisting motion at Y671 that moves this conserved residue away from the insertion site such that the template can interrogate the nucleotide in the closed complex. Binary and furan complexes were superimposed onto the ternary complex, achieving an RMSD on protein atoms in core regions of 0.97 and 0.68 Å, respectively.
Template rearrangements facilitate DNA synthesis opposite abasic sites
The adherence of pol β to the A-rule represents a special-case scenario for this enzyme with gapped DNA primer•templates. In situations where the template strand is not involved in WC base pairing downstream of the insertion site, pol β is able to direct incorporation opposite an abasic site by template slippage, matching the incoming nucleotide to the template base 5' to the abasic site [26]. A similar strategy was observed for the Y family polymerase Dpo4 from Sulfolobus solfataricus, which has been observed bypassing abasic sites with both −1 and +1 frameshifts (Figure 2, middle right) [27]. In 2009, an additional example demonstrating this replication strategy emerged when a series of crystal structures depicting E. coli polymerase II (pol II) showed how family B polymerases can also utilize template flipping and slippage in TLS [28]. With the exception of pol II and the eukaryotic polymerase ζ, family B polymerases are generally recognized for high-fidelity, sharing homology with the eukaryotic replicative polymerases δ and ε. Unlike high-fidelity family B polymerases, pol II harbors a unique β-barrel at its N-terminus. The presence of this β-barrel shifts the overall arrangement of protein domains slightly, in particular reducing the relative distance between the palm and N-terminal domains. Despite this shift, pol II and gp43 conserve in length a linker strand that runs parallel to the template DNA from the N-terminal domain to the palm. This segment becomes more relaxed in pol II, given that its linker spans a shorter distance with a similar number of protein residues. Coupled with the fact that the pol II linker harbors fewer bulky side chains than RB69 gp43, pol II is able to loop out DNA lesions into extrahelical cavities at several positions upstream of the active site during TLS (Figure 2, middle left).
The β-barrel insertion of pol II appears to also induce shifts between the N-terminal and exonuclease domains, causing the β-hairpin of the exonuclease domain to deviate 10 Å from its location in RB69 gp43. In family B polymerases, the β-hairpin loop facilitates primer switching from the polymerase active site to the exonuclease active site [29]. This role of the β-hairpin was explored with crystal structures of an RB69 gp43 variant bearing a truncation in the loop region of the hairpin [30]. The resulting polymerase exhibited reduced proofreading and crystallized in complex with templating furan such that the primer strand failed to switch to the exonuclease active site, as had been observed earlier when the β-hairpin remained intact [16]. In pol II, the β-hairpin is shifted away from the DNA such that it mimics the shortened loop of the β-hairpin truncation variant and, likewise, pol II exhibits relatively weak exonuclease proofreading, thus encouraging its function in error-prone TLS.
Yet another strategy for TLS opposite abasic sites is employed by polymerase ι (pol ι). This Y family polymerase exhibits very low-fidelity and does not follow the A-rule. Rather, pol ι inserts all nucleotides opposite an abasic site, albeit with dGMP incorporation being slightly more efficient than the rest. Pol ι is especially adept at forming Hoogsteen base pairs because the active site cleft squeezes the primer and template together, imposing a reduced distance between the deoxyribose sugars [31,32]. Due to the reduced space, purines in the active site of pol ι adopt the syn conformation. The recent crystal structures of pol ι captured in ternary complex with dGTP, dTTP or dATP opposite an abasic site demonstrated how its constricted active site serves to accommodate this specific type of DNA damage (Figure 2, bottom left) [33]. As the abasic site remained intrahelical, the base of the incoming nucleotide approached close enough to the furan moiety to engage in weak van der Waals interactions. This configuration also allows the incoming nucleotide to obtain additional stabilization by stacking under the −1 templating base adjacent to the abasic site, a situation analogous to that observed in RB69 gp43 when the nitro moiety of a non natural nucleotide (5-nitro-indolyl-triphosphate, 5-NITP) stacked similarly across from templating furan (Figure 2, bottom right) [15]. High-fidelity polymerases incorporate 5-NITP opposite furan anywhere from 1 (pol β [21]) to 1000 (T4 gp43 [14]) times more efficiently than dATP, which suggests an importance for nucleotide-template interactions during DNA synthesis opposite abasic sites. Pol ι appears adept at exploiting this type of stabilization in order to conduct DNA synthesis opposite abasic sites without reliance on template slippage, thus reducing the potential for frameshifts in favor of point mutations stemming from Hoogsteen base pairing.
8-oxoG, a common oxidation product of guanine, can pair with dCMP and dAMP
High-fidelity replicative DNA polymerases achieve mutation frequencies such that one mistake occurs in every 104 to 106 nucleotides synthesized opposite undamaged templates [34]. This fidelity becomes much reduced under conditions of oxidative stress, however, because of the inevitable encounter of the polymerase with unrepaired premutagenic DNA lesions. DNA synthesis opposite 8-oxoG becomes especially problematic because this lesion evades 3’→5’ exonuclease proofreading in the replicative polymerase when paired with dAMP. Oxidation at the C8 of guanine alters the protonation state of the N7, which assists in forming Hoogsteen base pairs opposite dAMP (Figure 1B) [35]. Hoogsteen base pairing with dAMP requires 8-oxoG to adopt the syn conformation, placing the WC edge of the damaged base into the major groove and leaving the O8 poised in the minor groove, where it accepts hydrogen bonds from amino acid side chains conserved in the palm domain of replicative polymerases. These hydrogen bonds provide feedback concerning the minor groove geometry about the purine N3 and pyrimidine exocyclic O2 and are required for successful translocation as the primer strand is elongated [9]. Because the O8 of syn 8-oxoG adeptly mimics the N3 of an unmodified anti purine, replicative polymerases translocate 8-oxoG•dAMP without triggering 3’–5’ exonuclease proofreading (Figure 1B). The potency of 8-oxoG as a premutagenic DNA lesion is, therefore, explained in terms of both the propensity of 8-oxoG to pair with dAMP and the ease with which replicative polymerases extend this mismatch.
Although the unmodified WC face of 8-oxoG allows this lesion to form a cognate base pair with dCMP, replicative polymerases extend dCMP•8-oxoG inefficiently. Crystal structures of the large fragment of DNA polymerase I from Bacillus fragment (BF), a high-fidelity family A repair polymerase, indicate perturbation of the DNA and a loss of minor groove contacts when dCMP is paired with 8-oxoG in the −1 post-insertion site [35]. The O8 clashes unfavorably with the deoxyribose and approaches van der Waals contact with the 5’ phosphate, causing distortion of the DNA in this translocated complex. However, at the insertion site the incorporation preference varies among high-fidelity polymerases, where enzymes such as Klenow fragment, T7 DNA polymerase, and RB69 gp43 insert dCMP more efficiently than dAMP opposite 8-oxoG. For this reason, mutating these enzymes has often been a prerequisite for capturing the closed catalytically active complex describing the Hoogsteen base pair between dATP and 8-oxoG (syn) at the insertion site. A lysine residue of the fingers domain of T7 DNA polymerase (K536) was mutated to alanine to increase the relative binding of dATP and allow crystallization of the closed complex [36]. Mutations in RB69 gp43 DNA polymerase adding volume to the active site (L561A or Y567A) provided similar observations of Hoogsteen base pairing with dATP in this family B polymerase [37,38]. On the other hand, family B polymerases α and δ from eukaryotic organisms appear more adept at incorporating dAMP opposite this particular lesion [39]. In the polymerase family X, alternatively, pol β readily accommodates dCMP opposite 8-oxoG, due to conformational freedom in the template backbone that allows the phosphate 5’ to the lesion to flip 180° and avoid clash with the O8 of anti-8oxoG [40]. A structure of pol β has more recently demonstrated how oxidized dGTP can also lead to mutation. In nucleoside triphosphate form, 8-oxo-dGTP exists primarily in the syn conformation, which explains why this damaged nucleotide is incorporated most efficiently opposite templating dAMP via Hoogsteen base pairing [41].
Dpo4 and polymerase η favor canonical base pairing between 8-oxoG and dCMP
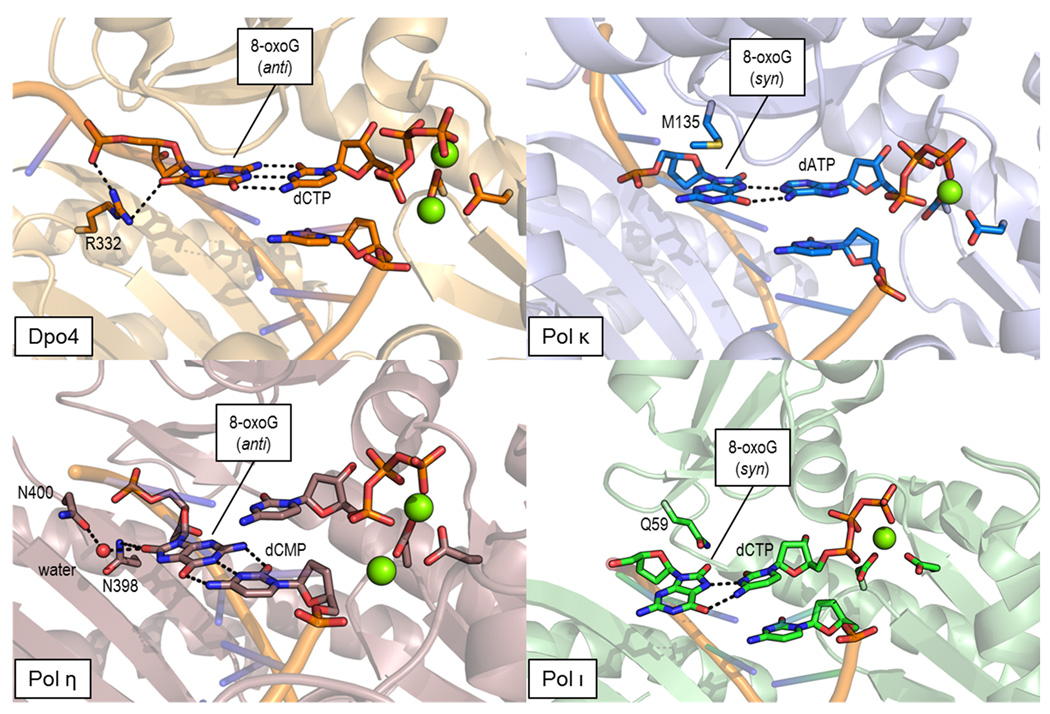
Dpo4 and polymerase η (pol η) promote error-free bypass of UV induced cis-syn thymine dimers [42]. Accordingly, humans with genetic deficiencies in pol η incur a variant of xeroderma pigmentosum (XPV) and therefore harbor a predisposition for skin cancers [43,44]. In addition to efficient processing of thymine dimers, these polymerases achieve error-free bypass of 8-oxoG, which was previously seen as a conundrum since 8-oxoG foils proofreading by so-called high-fidelity polymerases [45]. Recently, crystal structures of Dpo4 and pol η unveiled the molecular mechanisms underlying competent 8-oxoG bypass. Like other family Y polymerases, Dpo4 harbors an accessory palm associated domain (PAD or little finger domain) believed to facilitate TLS. The PAD contributes side chains in the vicinity of the polymerase active site and the DNA major groove, allowing family Y polymerases to interrogate the DNA from more angles than replicative polymerases. Crystal structures demonstrate that Dpo4 manipulates the configuration of 8-oxoG templates by presenting the guanidinium group of an arginine residue (R332) of the PAD between the O8 and next 5' phosphate group (Figure 4, top left) [46,47]. These contacts provide stabilization of the anti form and allow Dpo4 to preferentially bind and extend cognate base pairs to 8-oxoG even more efficiently than G•C, where the potential for a contact to the O8 is absent. Mutational analysis has verified that R332 is required for preserving the processivity of Dpo4 during the subsequent extension steps, which supports the observation in the crystal structure that Dpo4 maintains contacts from R322 to 8-oxoG following translocation of the lesion to the −1 position [48]. Given the additional six basic residues from the PAD contacting the DNA, Dpo4 appears not only especially adapted to ensure faithful nucleotide selection, but also well prepared to extend the desired base pair for completion of error-free TLS.
Figure 4.
Dpo4 contributes R332 to stabilize 8-oxoG in the anti conformation, such that the Watson-Crick face of the lesion is available for hydrogen bonding with dCMP (PDBID 2XCP [57]). Although the PAD of polymerase η does not contact the nucleotide in the insertion site, pol η makes two contacts from asparagine residues to the O8 of 8-oxoG in the translocated complex (PDBID 3OHB [50]). This stabilization is likely important for extension of the error-free incorporation product. Polymerase κ accommodates 8-oxoG in its active site, except the syn form is preferred (PDBID 3IN5 [53]). This establishes Hoogsteen base pairing with dATP, which can lead to G→T transversion mutations. Polymerase ι achieves error-free insertion opposite 8-oxoG (syn) by a unique mode of Hoogsteen base pairing with dCTP. Polymerase ι has a propensity to bind purines in the syn configuration in its very narrow active site cleft (PDBID 3Q8P [55]).
As an archaeal homolog, Dpo4 has served to model eukaryotic pol η in the absence of available DNA complex structures. Although the sequences of these enzymes are somewhat divergent, the comparison has been accepted on the grounds that both polymerases treat damaged templates similarly. Two laboratories recently achieved the feat of crystallizing eukaryotic pol η with DNA, after prerequisite amino acid substitutions removed undesirable lattice contacts characteristic of crystals grown from the wild type proteins [49,50]. One of these variants from Saccharomyces cerevisiae crystallized in complex with damaged DNA containing 8-oxoG, providing a closer look at how pol η achieves its specialized high-fidelity [51]. The mutations K140A and S144W succeeded in breaking a non-physiological dimer found in the asymmetric unit of the apo structure, in which interactions between the PAD and palm domain interfered with proper DNA binding [52]. Interestingly, pol η supplies very few contacts to the O8 of 8-oxoG at the insertion site, negating a hypothesis that there might be a conserved basic residue in the PAD homologous to R322 of Dpo4 [48]. The striking aspect of the pol η structure concerned the ease with which the enzyme accommodates 8-oxoG template in the anti conformation by avoiding a clash between the O8 and the phosphate backbone [50]. The torsion angles at the lesion deviate substantially from those of B-form DNA, providing just slightly less than 1 Å of additional clearance for the O8. This increased distance is also obtained when an undamaged guanine occupies the templating position, suggesting that pol η enforces this DNA conformation regardless of the nature of the templating base. Following translocation to the −1 post-insertion site, the clearance for the O8 is maintained, but the PAD also contributes a water-mediated interaction between N400 and the O8. An additional direct contact to the O8 is observed from N398, thus fully stabilizing 8-oxoG in the anti conformation during translocation (Figure 4, bottom left).
Polymerase κ bypasses 8-oxoG with errors
In addition to Dpo4 and pol η, polymerase κ (pol κ) readily bypasses 8-oxoG, albeit with much reduced fidelity. Like pol η, the spacious active site of pol κ accommodates 8-oxoG without significant rearrangements in protein residues over the active site conformation observed for a canonical G•C base pair [53]. However, pol κ prefers to bind 8-oxoG in the error-prone syn configuration, allowing the lesion to Hoogsteen base pair with A. The crystal structure of pol κ revealed that a methionine residue (M135) in the fingers domain stacks in van der Waals contact with the rotated templating lesion (Figure 4, top right). When the mutation M135A is introduced, the polymerase retains its incorporation specificity but the overall catalytic activity is reduced [53]. The full catalytic efficiency of pol κ also depends on the presence of an additional N-terminal domain referred to as the N-clasp (residue 25–74). The N-clasp, which is not conserved in pol η or pol ι, features a helix-turn-helix motif that allows pol κ to fully encircle the DNA. Like other family Y polymerases, however, pol κ contacts the DNA near the active site with its PAD. Consistent with the propensity of pol κ to bind 8-oxoG in the syn conformation, structural superpositions reveal that the basic residue R233 of Dpo4 implicated in error-free bypass aligns with L508 in pol κ. Interestingly, the L508K mutation in pol κ increases the error-free bypass of 8-oxoG, supporting the hypothesis that diversity in the PAD of family Y polymerases is essential for their varying incorporation specificities during TLS [54].
Polymerase ι utilizes Hoogsteen base pairing for error-free incorporation opposite syn 8-oxoG
Although the syn configuration leads to mismatch formation in all other known examples, the squeezed active site of pol ι excludes dATP and facilitates a unique Hoogsteen base pair allowing two hydrogen bonds between dCTP and 8-oxoG (Figure 4, bottom right) [55]. The glutamine (Q59) of the fingers domain, which accounts for the small preference in this polymerase for incorporation of dGMP opposite abasic sites, lies in close proximity to the extraneous O8. However, due to hydrogen bonding between the side chain amide of Q59 and a nearby backbone carbonyl, the side chain of Q59 appears unable to rotate and contact 8-oxoG specifically. Regardless, Q59 maintains a role in selecting dCTP, which was demonstrated by the acquired ability of the Q59A variant to incorporate dAMP opposite 8-oxoG in primer extension assays, suggesting that this side chain might be necessary to fully exclude dATP from the active site of pol ι [55].
Conclusions and perspectives
The recent availability of crystal structures of polymerases bound to lesion containing DNA creates a nearly complete picture, focused at the DNA template, describing the mechanics of TLS. Representative structures of all the eukaryotic family Y polymerases—Rev1, polymerases ι, η and κ—provide atomic resolution detail of their encounter with DNA lesions. This review focused on 8-oxoG in the context of family Y polymerases, where the active sites of these enzymes accommodate lesions without clash or perturbation of protein side chains. These structures describe the preformed nature of the family Y active site. Although the PAD seems to modulate the dynamics of the DNA conformation, especially as was observed in the complex of Dpo4 with 8-oxoG, discrete contacts to the lesion account for only a portion of the story. Pol η, for example, does not require contacts to 8-oxoG at the insertion site to achieve a DNA conformation orienting the lesion in the error-free anti configuration. Likewise, pol κ bypasses 8-oxoG without unique contacts recognizing the lesion, albeit in an error-prone fashion. Pol ι, on the other hand, utilizes a strategically placed glutamine residue to exclude dATP from its active site, in favor of dCTP, rather than to specifically recognize the lesion.
Abasic sites challenge DNA polymerases to select a nucleotide without instruction from the template. Single-stranded DNA containing an abasic site requires an error-prone strategy to avoid catastrophic failure of DNA synthesis due to a perpetually stalled replication fork. Rev1 potentially evolved to anticipate spontaneous depurination of dGMP by inserting dCMP opposite abasic sites. However, this specificity is unique among bypass polymerases where bypass efficiency seems paramount. In addition to side chain-directed incorporation, bypass polymerases employ strategies of template slippage and looping out of abasic sites. Although Rev1 relies on polymerase ζ for catalyzing the extension step, other bypass polymerases could catalyze both incorporation and extension by manipulating the parental DNA strand such that the abasic site never actually templates in the polymerase active site. Allowing the DNA template to form alternative conformations, however, introduces a route for frameshift mutations. On the other hand, pol ι does not achieve high fidelity. Rather, the direct utilization of abasic sites as templates by pol ι could constitute the preferable strategy for TLS within a coding region where a point mutation might be less deleterious than a frameshift.
We now understand that different DNA polymerases process the same DNA lesions via alternative strategies, at times requiring non-canonical base pairs. With this diversity more so exposed, our concept of the replication of physiological DNA is enriched, though still incomplete. We anticipate that the current expansion of crystal structures will continue into the years ahead. Although the gaps in this body of knowledge are decreasing rapidly, several DNA polymerases from the human genome, particularly those polymerases belonging to family A, have yet to be captured in crystal form. These developments now loom on the horizon, poised to fortify our understanding of how DNA polymerases orchestrate variations on a mechanistic theme to deliver competent replication and repair of physiological DNA.
Acknowledgements
Our work on DNA replication is supported by NIH grant CA52040, awarded by the National Cancer Institute.
Abbreviations
- 5-NITP
5-nitro-indolyl-triphosphate
- 8-oxoG
7,8-dihydro-8-oxoguanine
- TLS
Translesion synthesis
- AP
Apurinic/apyrimidinic
- BER
Base excision repair
- BF
Bacillus stearothermophilus DNA polymerase I fragment
- ddATP
dideoxy-dATP
- PAD
Palm associated domain
- PDB
Protein Data Bank
- WC
Watson-Crick
- XPV
Xeroderma pigmentosum variant
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
• of special interest
•• of outstanding interest
- 1.Bandaru V, Zhao X, Newton MR, Burrows CJ, Wallace SS. Human endonuclease VIII-like (NEIL) proteins in the giant DNA Mimivirus. DNA Repair (Amst) 2007;6:1629–1641. doi: 10.1016/j.dnarep.2007.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Waters LS, Minesinger BK, Wiltrout ME, D'Souza S, Woodruff RV, Walker GC. Eukaryotic translesion polymerases and their roles and regulation in DNA damage tolerance. Microbiol Mol Biol Rev. 2009;73:134–154. doi: 10.1128/MMBR.00034-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Loeb LA, Monnat RJ., Jr DNA polymerases and human disease. Nat Rev Genet. 2008;9:594–604. doi: 10.1038/nrg2345. • This well-executed and thorough review describes the polymerases of the human genome inclusively, presenting some compelling figures to accompany the discussion.
- 4.Bebenek K, Kunkel TA. Functions of DNA polymerases. Adv Protein Chem. 2004;69:137–165. doi: 10.1016/S0065-3233(04)69005-X. [DOI] [PubMed] [Google Scholar]
- 5.Lovett ST. Polymerase switching in DNA replication. Mol Cell. 2007;27:523–526. doi: 10.1016/j.molcel.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 6.Guo C, Kosarek-Stancel JN, Tang TS, Friedberg EC. Y-family DNA polymerases in mammalian cells. Cell Mol Life Sci. 2009;66:2363–2381. doi: 10.1007/s00018-009-0024-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kool ET. Active site tightness and substrate fit in DNA replication. Annu Rev Biochem. 2002;71:191–219. doi: 10.1146/annurev.biochem.71.110601.135453. [DOI] [PubMed] [Google Scholar]
- 8.Steitz TA. DNA polymerases: structural diversity and common mechanisms. J Biol Chem. 1999;274:17395–17398. doi: 10.1074/jbc.274.25.17395. [DOI] [PubMed] [Google Scholar]
- 9.Doublié S, Ellenberger T. The mechanism of action of T7 DNA polymerase. Curr Opin Struct Biol. 1998;8:704–712. doi: 10.1016/s0959-440x(98)80089-4. [DOI] [PubMed] [Google Scholar]
- 10.Vaisman A, Ling H, Woodgate R, Yang W. Fidelity of Dpo4: effect of metal ions, nucleotide selection and pyrophosphorolysis. EMBO J. 2005;24:2957–2967. doi: 10.1038/sj.emboj.7600786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang W, Woodgate R. What a difference a decade makes: insights into translesion DNA synthesis. Proc Natl Acad Sci U S A. 2007;104:15591–15598. doi: 10.1073/pnas.0704219104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.David SS, O'Shea VL, Kundu S. Base-excision repair of oxidative DNA damage. Nature. 2007;447:941–950. doi: 10.1038/nature05978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shibutani S, Takeshita M, Grollman AP. Translesional synthesis on DNA templates containing a single abasic site. A mechanistic study of the "A rule". J Biol Chem. 1997;272:13916–13922. doi: 10.1074/jbc.272.21.13916. [DOI] [PubMed] [Google Scholar]
- 14.Reineks EZ, Berdis AJ. Evaluating the contribution of base stacking during translesion DNA replication. Biochemistry. 2004;43:393–404. doi: 10.1021/bi034948s. [DOI] [PubMed] [Google Scholar]
- 15.Zahn KE, Belrhali H, Wallace SS, Doublié S. Caught bending the A-rule: crystal structures of translesion DNA synthesis with a non-natural nucleotide. Biochemistry. 2007;46:10551–10561. doi: 10.1021/bi7008807. [DOI] [PubMed] [Google Scholar]
- 16.Hogg M, Wallace SS, Doublié S. Crystallographic snapshots of a replicative DNA polymerase encountering an abasic site. EMBO J. 2004;23:1483–1493. doi: 10.1038/sj.emboj.7600150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Freisinger E, Grollman AP, Miller H, Kisker C. Lesion (in)tolerance reveals insights into DNA replication fidelity. EMBO J. 2004;23:1494–1505. doi: 10.1038/sj.emboj.7600158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nair DT, Johnson RE, Prakash L, Prakash S, Aggarwal AK. Rev1 employs a novel mechanism of DNA synthesis using a protein template. Science. 2005;309:2219–2222. doi: 10.1126/science.1116336. [DOI] [PubMed] [Google Scholar]
- 19. Nair DT, Johnson RE, Prakash L, Prakash S, Aggarwal AK. DNA synthesis across an abasic lesion by yeast Rev1 DNA polymerase. J Mol Biol. 2010 doi: 10.1016/j.jmb.2010.12.016. •• This paper confirms an important prediction made previously by the authors that Rev1 directs incorporation of dCMP with an arginine residue of the N-digit. The prior hypothesis was based on a structure where an estranged dGMP occupied templating position. In this new paper, the furan complex is presented.
- 20.Nelson JR, Lawrence CW, Hinkle DC. Deoxycytidyl transferase activity of yeast REV1 protein. Nature. 1996;382:729–731. doi: 10.1038/382729a0. [DOI] [PubMed] [Google Scholar]
- 21. Beard WA, Shock DD, Batra VK, Pedersen LC, Wilson SH. DNA polymerase beta substrate specificity: side chain modulation of the "A-rule". J Biol Chem. 2009;284:31680–31689. doi: 10.1074/jbc.M109.029843. • Pol β, as a gap filling polymerase, must kink the duplex DNA to expose the primer terminus. This paper demonstrates that pol β depends on an arginine residue to maintain compliance with the A-rule.
- 22.Batra VK, Beard WA, Shock DD, Pedersen LC, Wilson SH. Structures of DNA polymerase beta with active-site mismatches suggest a transient abasic site intermediate during misincorporation. Mol Cell. 2008;30:315–324. doi: 10.1016/j.molcel.2008.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson SJ, Taylor JS, Beese LS. Processive DNA synthesis observed in a polymerase crystal suggests a mechanism for the prevention of frameshift mutations. Proc Natl Acad Sci U S A. 2003;100:3895–3900. doi: 10.1073/pnas.0630532100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Obeid S, Blatter N, Kranaster R, Schnur A, Diederichs K, Welte W, Marx A. Replication through an abasic DNA lesion: structural basis for adenine selectivity. EMBO J. 2010;29:1738–1747. doi: 10.1038/emboj.2010.64. • Klentaq is a high-fidelity repair polymerase of prokaryotic origin. Here, the first structure of a family A polymerase conducting DNA synthesis opposite an abasic site is described.
- 25.Li Y, Mitaxov V, Waksman G. Structure-based design of Taq DNA polymerases with improved properties of dideoxynucleotide incorporation. Proc Natl Acad Sci U S A. 1999;96:9491–9496. doi: 10.1073/pnas.96.17.9491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Efrati E, Tocco G, Eritja R, Wilson SH, Goodman MF. Abasic translesion synthesis by DNA polymerase beta violates the "A-rule". Novel types of nucleotide incorporation by human DNA polymerase beta at an abasic lesion in different sequence contexts. J Biol Chem. 1997;272:2559–2569. doi: 10.1074/jbc.272.4.2559. [DOI] [PubMed] [Google Scholar]
- 27.Ling H, Boudsocq F, Woodgate R, Yang W. Snapshots of replication through an abasic lesion; structural basis for base substitutions and frameshifts. Mol Cell. 2004;13:751–762. doi: 10.1016/s1097-2765(04)00101-7. [DOI] [PubMed] [Google Scholar]
- 28. Wang F, Yang W. Structural insight into translesion synthesis by DNA Pol II. Cell. 2009;139:1279–1289. doi: 10.1016/j.cell.2009.11.043. •• Pol II’s specialized function in TLS seems to rely on a unique N-terminal insertion of a β-barrel. Although much work focuses on discrete interactions between the polymerase and the bound primer•template pair, the authors here suggest that evolution can operate at a distance from the polymerase active site, impinging on the relative organization of protein domains to bear a bypass polymerase sharing structural homology with replicative family B polymerases.
- 29.Subuddhi U, Hogg M, Reha-Krantz LJ. Use of 2-aminopurine fluorescence to study the role of the beta hairpin in the proofreading pathway catalyzed by the phage T4 and RB69 DNA polymerases. Biochemistry. 2008;47:6130–6137. doi: 10.1021/bi800211f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hogg M, Aller P, Konigsberg W, Wallace SS, Doublié S. Structural and biochemical investigation of the role in proofreading of a beta hairpin loop found in the exonuclease domain of a replicative DNA polymerase of the B family. J Biol Chem. 2007;282:1432–1444. doi: 10.1074/jbc.M605675200. [DOI] [PubMed] [Google Scholar]
- 31.Nair DT, Johnson RE, Prakash L, Prakash S, Aggarwal AK. An incoming nucleotide imposes an anti to syn conformational change on the templating purine in the human DNA polymerase-iota active site. Structure. 2006;14:749–755. doi: 10.1016/j.str.2006.01.010. [DOI] [PubMed] [Google Scholar]
- 32.Nair DT, Johnson RE, Prakash S, Prakash L, Aggarwal AK. Replication by human DNA polymerase-iota occurs by Hoogsteen base-pairing. Nature. 2004;430:377–380. doi: 10.1038/nature02692. [DOI] [PubMed] [Google Scholar]
- 33. Nair DT, Johnson RE, Prakash L, Prakash S, Aggarwal AK. DNA synthesis across an abasic lesion by human DNA polymerase iota. Structure. 2009;17:530–537. doi: 10.1016/j.str.2009.02.015. •• The structures presented here describe a unique strategy for nucleotide incorporation opposite an abasic site. Rather than employing hydrogen bonds, pol ι imposes van der Waals interactions between the incoming nucleotide and abasic site.
- 34.Kunkel TA. DNA replication fidelity. J Biol Chem. 2004;279:16895–16898. doi: 10.1074/jbc.R400006200. [DOI] [PubMed] [Google Scholar]
- 35.Hsu GW, Ober M, Carell T, Beese LS. Error-prone replication of oxidatively damaged DNA by a high-fidelity DNA polymerase. Nature. 2004;431:217–221. doi: 10.1038/nature02908. [DOI] [PubMed] [Google Scholar]
- 36.Brieba LG, Kokoska RJ, Bebenek K, Kunkel TA, Ellenberger T. A lysine residue in the fingers subdomain of T7 DNA polymerase modulates the miscoding potential of 8-oxo-7,8-dihydroguanosine. Structure. 2005;13:1653–1659. doi: 10.1016/j.str.2005.07.020. [DOI] [PubMed] [Google Scholar]
- 37.Hogg M, Rudnicki J, Midkiff J, Reha-Krantz L, Doublié S, Wallace SS. Kinetics of mismatch formation opposite lesions by the replicative DNA polymerase from bacteriophage RB69. Biochemistry. 2010;49:2317–2325. doi: 10.1021/bi901488d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beckman J, Wang M, Blaha G, Wang J, Konigsberg WH. Substitution of Ala for Tyr567 in RB69 DNA polymerase allows dAMP to be inserted opposite 7,8-dihydro-8-oxoguanine. Biochemistry. 2010;49:4116–4125. doi: 10.1021/bi100102s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shibutani S, Takeshita M, Grollman AP. Insertion of specific bases during DNA synthesis past the oxidation-damaged base 8-oxodG. Nature. 1991;349:431–434. doi: 10.1038/349431a0. [DOI] [PubMed] [Google Scholar]
- 40.Krahn JM, Beard WA, Miller H, Grollman AP, Wilson SH. Structure of DNA polymerase beta with the mutagenic DNA lesion 8-oxodeoxyguanine reveals structural insights into its coding potential. Structure. 2003;11:121–127. doi: 10.1016/s0969-2126(02)00930-9. [DOI] [PubMed] [Google Scholar]
- 41.Batra VK, Beard WA, Hou EW, Pedersen LC, Prasad R, Wilson SH. Mutagenic conformation of 8-oxo-7,8-dihydro-2'-dGTP in the confines of a DNA polymerase active site. Nat Struct Mol Biol. 2010;17:889–890. doi: 10.1038/nsmb.1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Johnson RE, Prakash L, Prakash S. Distinct mechanisms of cis-syn thymine dimer bypass by Dpo4 and DNA polymerase eta. Proc Natl Acad Sci U S A. 2005;102:12359–12364. doi: 10.1073/pnas.0504380102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Masutani C, Kusumoto R, Yamada A, Dohmae N, Yokoi M, Yuasa M, Araki M, Iwai S, Takio K, Hanaoka F. The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase eta. Nature. 1999;399:700–704. doi: 10.1038/21447. [DOI] [PubMed] [Google Scholar]
- 44.Johnson RE, Kondratick CM, Prakash S, Prakash L. hRAD30 mutations in the variant form of xeroderma pigmentosum. Science. 1999;285:263–265. doi: 10.1126/science.285.5425.263. [DOI] [PubMed] [Google Scholar]
- 45.Hogg M, Wallace SS, Doublié S. Bumps in the road: how replicative DNA polymerases see DNA damage. Curr Opin Struct Biol. 2005;15:86–93. doi: 10.1016/j.sbi.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 46.Zang H, Irimia A, Choi JY, Angel KC, Loukachevitch LV, Egli M, Guengerich FP. Efficient and high fidelity incorporation of dCTP opposite 7,8-dihydro-8-oxodeoxyguanosine by Sulfolobus solfataricus DNA polymerase Dpo4. J Biol Chem. 2006;281:2358–2372. doi: 10.1074/jbc.M510889200. [DOI] [PubMed] [Google Scholar]
- 47. Rechkoblit O, Malinina L, Cheng Y, Kuryavyi V, Broyde S, Geacintov NE, Patel DJ. Stepwise translocation of Dpo4 polymerase during error-free bypass of an oxoG lesion. PLoS Biol. 2006;4:e11. doi: 10.1371/journal.pbio.0040011. • Some of the first structures of Dpo4 replicating past 8-oxoG are unveiled in this work.
- 48. Eoff RL, Irimia A, Angel KC, Egli M, Guengerich FP. Hydrogen bonding of 7,8-dihydro-8-oxodeoxyguanosine with a charged residue in the little finger domain determines miscoding events in Sulfolobus solfataricus DNA polymerase Dpo4. J Biol Chem. 2007;282:19831–19843. doi: 10.1074/jbc.M702290200. • A rigorous effort is applied here to characterize the role of the PAD of Dpo4 in TLS.
- 49. Biertumpfel C, Zhao Y, Kondo Y, Ramon-Maiques S, Gregory M, Lee JY, Masutani C, Lehmann AR, Hanaoka F, Yang W. Structure and mechanism of human DNA polymerase eta. Nature. 2010;465:1044–1048. doi: 10.1038/nature09196. •• Pol η is the single polymerase of human origin which, when defective, is associated with cancer. Crystallization of this enzyme in complex with a thymine dimer required substantial effort in terms of protein expression and selection of the proper polymerase variant, which was the key to this story of error-free bypass of UV damage in DNA.
- 50. Silverstein TD, Jain R, Johnson RE, Prakash L, Prakash S, Aggarwal AK. Structural basis for error-free replication of oxidatively damaged DNA by yeast DNA polymerase eta. Structure. 2010;18:1463–1470. doi: 10.1016/j.str.2010.08.019. •• This manuscript reveals the first structure of eukaryotic pol η with 8-oxoG, which represents an important development in the polymerase field, where this process had previously been modeled with Dpo4, an enzyme of archaeal origin.
- 51.Silverstein TD, Johnson RE, Jain R, Prakash L, Prakash S, Aggarwal AK. Structural basis for the suppression of skin cancers by DNA polymerase eta. Nature. 2010;465:1039–1043. doi: 10.1038/nature09104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Trincao J, Johnson RE, Escalante CR, Prakash S, Prakash L, Aggarwal AK. Structure of the catalytic core of S. cerevisiae DNA polymerase eta: implications for translesion DNA synthesis. Mol Cell. 2001;8:417–426. doi: 10.1016/s1097-2765(01)00306-9. [DOI] [PubMed] [Google Scholar]
- 53. Vasquez-Del Carpio R, Silverstein TD, Lone S, Swan MK, Choudhury JR, Johnson RE, Prakash S, Prakash L, Aggarwal AK. Structure of human DNA polymerase kappa inserting dATP opposite an 8-OxoG DNA lesion. PLoS One. 2009;4:e5766. doi: 10.1371/journal.pone.0005766. • Pol κ inserts dAMP opposite 8-oxoG, relying on a methionine residue for full activity.
- 54. Irimia A, Eoff RL, Guengerich FP, Egli M. Structural and functional elucidation of the mechanism promoting error-prone synthesis by human DNA polymerase kappa opposite the 7,8-dihydro-8-oxo-2'-deoxyguanosine adduct. J Biol Chem. 2009;284:22467–22480. doi: 10.1074/jbc.M109.003905. • Although pol κ prefers to insert dAMP opposite 8-oxoG, the PAD can be mutated to elevate error-free bypass.
- 55. Kirouac KN, Ling H. Unique active site promotes error-free replication opposite an 8-oxo-guanine lesion by human DNA polymerase iota. Proc Natl Acad Sci U S A. 2011 doi: 10.1073/pnas.1013909108. •• Pol ι was crystallized with all four nucleotides opposite 8-oxoG in order to describe the propensity of this enzyme to incorporate dCMP correctly.
- 56.Li Y, Kong Y, Korolev S, Waksman G. Crystal structures of the Klenow fragment of Thermus aquaticus DNA polymerase I complexed with deoxyribonucleoside triphosphates. Protein Sci. 1998;7:1116–1123. doi: 10.1002/pro.5560070505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Irimia A, Loukachevitch LV, Eoff RL, Guengerich FP, Egli M. Metal-ion dependence of the active-site conformation of the translesion DNA polymerase Dpo4 from Sulfolobus solfataricus. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2010;66:1013–1018. doi: 10.1107/S1744309110029374. [DOI] [PMC free article] [PubMed] [Google Scholar]