Abstract
Background
Diffusion Tensor Imaging (DTI) infers the trajectory and location of large white matter tracts by measuring anisotropic diffusion of water. DTI data may then be analyzed and presented as tractography for visualization of the tracts in three dimensions. Despite the important information contained in tractography images, usefulness for neurosurgical planning has been limited by the inability to define which are critical structures within the mass of demonstrated fibers and to clarify their relationship to the tumor.
Objective
Our goal was to develop a method to allow the interactive querying of tractography datasets for surgical planning and to provide a working software package for the research community.
Methods
Tool was implemented within open-source software project.
Echoplanar DTI at 3T was performed on five patients, followed by tensor calculation.
Technical Development
Software was developed allowing placement of dynamic seedpoint for local selection of fibers, and for fiber display around a segmented structure - both with tunable parameters. A neurosurgeon was trained in use of software in less than one hour and used to review cases.
Results
Tracts near tumor and critical structures were interactively visualized in three dimensions to determine spatial relationships to lesion. Tracts were selected using 3 methods: (1) anatomical and fMRI-defined regions of interest (ROIs), (2) distance from the segmented tumor volume, (3) dynamic seedpoint spheres.
Conclusion
Interactive tractography successfully enabled inspection of white matter structures that were in proximity to lesions, critical structures, and functional cortical areas allowing the surgeon to explore the relationships between them.
Keywords: diffusion tensor imaging, magnetic resonance imaging, neurosurgery, surgical planning, tractography
For most patients with brain tumors gross total resection increases time to progression, lengthens survival, reduces mass effect and associated neurologic deficits, provides significant cytoreduction, and may be curative for some tumor types.1–4 Tumors located near critical cortical regions or functionally significant white matter (WM) fiber tracts are difficult to resect maximally while avoiding postoperative neurological deficits.5 Defining the limits of the tumor relative to key cortical areas and associated white matter tracts can be difficult due to the inability to distinguish tumor from brain and to identify those key structures intra-operatively. Functional brain mapping using a variety of techniques (functional MRI, Positron Emission Tomography, Magnetic source imaging) now allows pre-operative and non-invasive demonstration of critical cortical areas.6–8 Defining critical WM anatomy and its relationship to cortical areas has been more problematic as WM tracts cannot be visualized on conventional imaging and there is no reliable way to test for their presence. Moreover, in patients with brain tumors, variable displacement, disruption, or infiltration of white matter tracts may occur5, 9 and functional WM tracts have even been found within tumor boundaries.10 If injury to critical white matter tracts occurs, the patient will incur a new neurologic deficit even if the eloquent cortex has been respected.11 Subcortical stimulation mapping of the white matter is the clinical gold standard and the only functional method for identifying the motor12 and language13 pathways. However, this technique does not reveal the full 3D extent of the tracts,13–15 is not in general use, and is not available pre-operatively. If the surgeon could have a pre-operative understanding of the WM anatomy of the patient, he or she would be able to assess risks of surgery, and the likelihood of complete resection, and could counsel patients accordingly. Such pre-operative information could also guide the deployment of intra-operative subcortical mapping when available, thereby making it more efficient and possibly validating DTI and intra-operative subcortical WM stimulation against one another.
Diffusion Tensor Imaging (DTI) is an emerging MRI-based technique that can demonstrate white matter anatomy by measuring the directional anisotropy of water. Diffusion MRI is the first noninvasive technique for measuring white matter fiber structure in vivo.16 In DTI analysis, a tensor model is used to represent the orientation of white matter fibers. In voxels where one white matter fiber population is predominant, the principal diffusion direction is aligned with the white matter fiber tract direction.17 By following principal directions of diffusion, a process called tractography18, 19 estimates the trajectories of white matter fiber tracts. These reconstructions may then be displayed in three dimensions providing a detailed map of the configuration of the tracts and their relationship to other structures. Numerous studies have used DTI tractography to define white matter anatomy in healthy subjects and in patients with neurological and neurosurgical diseases. In particular, in patients with brain tumors, DTI can demonstrate displacement, interruption, or infiltration of white matter tracts by the tumor.9, 20–29 Tractography to demonstrate WM anatomy in neurosurgical patients has been incorporated into neuronavigation systems30, 31 and even acquired intraoperatively.32
Despite the important information contained in these images, usefulness for neurosurgical planning has been limited by the inability to define which are critical structures within the mass of fibers and to clearly demonstrate the relationship of tracts to the tumor. Our goal was to develop a method to allow the interactive querying of tractography datasets for surgical planning in order to define clinically important WM tracts and the relationship to the surgical lesion in order to provide the neurosurgeon with an optimal understanding of the functional brain anatomy for individual patients. This effort is aided by recent progress in computational power making such an interactive approach feasible to run on a laptop computer.
In order to isolate white matter structures of interest, the standard method is to seed tractography trajectories ("fibers") using manually identified regions of interest (ROIs), such as edited contours, spheres, or boxes.33, 34 To further select fibers of interest, additional ROIs are used to limit the tractography to a specific structure.20, 35 However, space-occupying lesions are known to have many pathological effects on white matter tracts, including disruption, destruction, infiltration, displacement, and production of edema.9, 36 Due to these and other pathological changes throughout the brain, the manual identification of ROIs based on known neuroanatomy is often not straightforward in tumor patients. In order to define the relevant anatomy, it would be useful to identify those fibers that pass within a certain (variable) distance of the tumor or which run through the tumor, as well as those associated with particular, patient-specific, cortical areas such as fMRI activations or MEG findings. This can provide a pre-operative functional brain map defining both the critical cortical functional areas and the WM tracts leading to and from these areas. By interactively querying the data, the surgeon should be able to build a mental representation of the relevant anatomy. Moreover, such an approach could be used intra-operatively to select and display critical white matter tracts based on intra-operative findings, particularly the determination of eloquent cortex in the region of the lesion using electrocortical stimulation testing (see Elhawary et al., Neurosurgery accepted 2010 for more information).
With these goals in mind we set out to develop a software module which could display WM tracts selectively and interactively. We wanted a tool that would show all the tracts within a certain distance of the tumor boundary and that could interactively change that distance in order to progressively view the layers of tracts around the tumor. The second tool was designed to allow the placement of a dynamic seedpoint of variable size which would seed all the tracts passing through that area and which could be moved around the 3D dataset to demonstrate tracts “on-the-fly”. We also wished to be able to use certain regions as seed regions, for example fMRI activations or anatomic structures. The tools were developed with the goal of being disseminated widely for investigational use by clinicians. The tools were developed in 3D Slicer, version 3 (www.slicer.org), a freely available anatomic visualization software project based at Brigham and Women’s Hospital.37
METHODS
Subjects
All subjects were recruited in accordance with the policies of the Partners Healthcare Institutional Review Board for human subjects. Written informed consent was obtained from all subjects. All patients underwent functional magnetic resonance imaging (fMRI) and DTI pre-operatively. As directed by clinical considerations and tumor location, fMRI was used to define motor cortex, somato-sensory cortex, visual cortex, and/or language areas. DTI imaging was acquired over the whole brain during the same scanning session. For purposes of developing the tool, we applied the method in patients with newly diagnosed or recurrent primary glial tumors or metastatic brain tumors.
Patient 1
Patient 1 is a 19 year old male who presented with personality changes and new onset of seizures who was found to have a large non-enhancing lesion in the left parietal lobe. DTI imaging was acquired with 1 baseline and 31 gradient directions. In addition, the patient underwent functional MRI during motor, sensory and visual tasks. Using the tools described, tractography was performed to delineate the principal motor and somatosensory tracts by seeding from fMRI activations and dynamically from known anatomic landmarks. The patient underwent surgery for subtotal resection of the tumor. Pathology demonstrated WHO grade III astrocytoma.
Patient 2
Patient 2 is 31-year-old right handed female with a large right hemisphere lesion who noted visual changes a month prior to presentation. MRI revealed a very large non-enhancing mass involving occipital, temporal, and parietal lobes. She underwent stereotactic biopsy at an outside institution demonstrating infiltrating oligoastrocytoma, WHO grade II. DTI imaging was acquired with 1 baseline and 55 gradient directions. fMRI was performed for left motor mapping (foot, hand and face) and visual field mapping. She underwent subtotal resection under local anesthesia with intraoperative electrocortical determination of somatosensory and visual cortical areas. The patient had slight worsening of her right sided visual field cut post-operatively.
Patient 3
Patient 3 is a 30 year old ambidextrous bilingual (English and Portuguese) female with a very large T2 intense non-contrast enhancing lesion involving the left frontal and temporal lobes. She underwent pre-operative fMRI during language tasks, face and right hand-clenching motor tasks, and DTI with 5 baselines and 31 gradient directions. Surgical resection of the lesion was performed under local anesthesia with language mapping in both English and Portuguese. Electrocortical stimulation testing demonstrated two regions in which stimulation at low current reliably interfered with language task performance in both languages: one directly superficial to the tumor in the posterior inferior frontal lobe and one directly superficial to the tumor in the superior temporal gyrus. Intra-operative findings were congruent with fMRI-demonstrated language associated activations. The resection was performed with continuous monitoring of language function and pre-operatively visualized areas of preserved white matter tracts as well as areas subjacent to positive cortical stimulation points were spared during the resection. There were no post-operative language changes.
Patient 4
Patient 4 is a 28 year old right handed female with right posterior temporal WHO Grade II mixed glioma (oligoastrocytoma) presenting with a probable seizure. Pre-surgical MRI scans were taken for three fMRI tasks (left hand clenching, lip pursing, and visual field stimulation), as well as DTI (31 directions, 1 baseline). She underwent resection of the lesion without new neurologic deficit.
Patient 5
Patient 5 is a 55 year old right-handed female with metastatic adenocarcinoma of the lung who presented with an asymptomatic contrast enhancing lesion in the superior left temporal lobe. She underwent fMRI (three language tasks, right hand clenching, lip pursing) and DTI (31 directions; 5 baselines) prior to surgical resection of the lesion performed under local anesthesia with language mapping. The patient did not have any language changes associated with the procedure.
Image acquisition
MR images were obtained using a 3.0 Tesla scanner (EXCITE Signa scanner, GE Medical System, Milwaukee, WI, USA) with Excite 14.0, using an 8-channel head coil and ASSET. As a first step, a high resolution whole brain T1-weighted axial 3D SPGR (TR=7500ms, TE=30ms, matrix=256×256, FOV=25.6cm, FA=20°; imaging 120–182 slices of 1mm thickness) was acquired. Next, DWI was acquired with a multi-slice single shot diffusion weighted echo-planar-imaging sequence (TR=14000ms, TE=30ms) consisting of 55 or 31 gradient directions with a b-value of 1000 s/mm2, and 1 or 5 baseline T2 images. The FOV was 25.6cm. Imaging matrix was 128×128 with slice thickness of 2.6mm. A single-shot gradient-echo echo-planar imaging (EPI) was used to acquire BOLD functional images (TR = 2000 ms, TE = 40 ms, flip angle = 90°, FOV = 24 cm, acquisition matrix = 64 × 64, slice gap = 0 mm, voxel size = 3.75 × 3.75 × 5 mm3). In each image volume, 28 axial slices were acquired using ascending interleaved scanning sequence. Behavioral paradigms were selected based on clinical considerations and included: visual alternating whole field checkerboard; self-paced motor tasks for hand, foot and mouth; somatosensory stimulation; and visually presented antonym generation and noun categorization language tasks.
High-resolution T2-weighted gradient-echo MR images (TR = 8000 ms, TE = 98 ms, flip angle = 90°, matrix = 512 × 512, 93 slices, voxel size = 0.5 × 0.5 × 1.5 mm3) were acquired in order to demonstrate the surgical pathology.
Image analyses
For each subject, a diffusion tensor volume derived from the Diffusion Weighted Image volume was calculated using the Teem (http://teem.sf.net) library through 3D Slicer. As implemented in 3D Slicer, the white matter tract trajectories are estimated using a single tensor model. The standard streamline tractography method (as in Basser et al.38) repeatedly steps in the principal diffusion direction defined by the tensor at each location.
Functional MRI data were realigned, motion corrected, and analyzed using SPM2 (Wellcome Department of Cognitive Neurology, London, UK).
Since images may be acquired in multiple sessions, and since patients often move during the course of an imaging session, image-based registration techniques are used to align the structural, DTI, and fMRI volumes. Our software relies on the Insight Toolkit (ITK, http://www.itk.org39) to provide image registration using a wide range of linear and non-linear algorithms. Care must be taken when reviewing the alignment of these registrations since MRI scans in general, and in particular the EPI sequences used for fMRI and DTI, often trade improved signal at the cost of increased geometric distortion.40 A full characterization of these distortions is beyond the scope of the current discussion, as the complexity of these non-linear distortions is difficult to capture using current tools. In practice we currently rely on an a process of manual registration to rough fit, followed by automatic affine registration (linear-only compensation for variations in scale, translation, rotation and shear; ITK-implemented algorithm39), followed by manual inspection and correction as necessary.
Hardware requirements
Data analyses and interactive tractography were performed on a Dell workstation with Intel Core 2 Duo 6300 processor at 1.86 Ghz, 2 Gb of RAM, 7200 RPM SATA hard-drive, nVidia Quadro FX3450 graphics accelerator with 256 Mb dedicated memory, and Linux operating system kernel running proprietary nVidia drivers. For case 3, with a 256×256, 38 axial-slice Diffusion-Weighted volume with 5 baseline scans and 31 gradient directions, estimation of diffusion tensors in 3D Slicer required approximately 1 minute. Interactive seedpoint tractography runs at near-realtime on this system using default parameters and seeding regions up to 6mm radius, with increasing update delays as the seeding region is enlarged.
Software environment
The software discussed in this report has been implemented in the 3D Slicer (version 3) open source software platform (www.slicer.org). 3D Slicer is an open source research software package, facilitating rapid improvement of image analysis techniques. A suite of software tools known collectively as the NA-MIC Kit support the core functionality needed in the implementation of the custom tools described here.41 All components of the NA-MIC Kit have all been developed according to the Open Source model of software development (http://opensource.org) meaning that they are available for download, free of charge and restrictions, by any groups wishing to replicate or extend our results. A complete inventory of the software is available at the NA-MIC Web Site; for the work described in this paper modifications were made primarily to the Teem library (http://teem.sf.net) and the 3D Slicer application software. 3D Slicer includes modules for the selection of structures of interest including tumor segmentation, ROI analysis, and interactive DTI.
Tract Selection Tools
A primary goal of this research is to provide the surgeon with a detailed mental model of the pre-operative condition of the tumor and surrounding anatomy. Our techniques rely on interactive 3D graphics and allow exploration of the local white matter anatomy using structural or functionally-defined regions of interest as input. In addition, novel techniques allow a controlled exploration of clinically relevant portions of the diffusion dataset. Tract selection and display was performed 3 ways:
ROI from fMRI
Using seed points in the WM adjacent to cortical areas associated with functional activation can suggest the pathways associated with the eloquent cortex that should be preserved during tumor removal. Tractography is performed using the coregistered (to DTI) functional activation as the seeding input voxels. Seedpoints are selected by software at either fixed-distance intervals or a randomly distributed grid within the activation volume.
Distance seeding from tumor segmentation
Tracts generated from seed points at the boundary of the tumor provide an indication of the white matter pathways in the immediate vicinity of the tumor. Segmentation of tumors was performed using a combination of sub-volume thresholding, threshold levelset selection, and manual adjustments using labelmap drawing tools. The software allows generation of tracts from an isocontour shell inward or outward from the segmentation boundary, allowing exploration of the local white matter anatomy. For example, by seeding near the tumor, the presence of preserved tracts within or near an infiltrating tumor can be demonstrated (see Figure 4d and video supplement). This technique can also be applied to any structure in the scan, including normal or pathological tissue that can be delineated and segmented. The method pre-computes a distance transform around a segmented model of the tumor boundary (or other tissue). Isocontour surfaces of the distance transform represent all points at specific distances from the tumor margin. Tractography is updated with each distance change, interactively demonstrating fiber tracts that pass within the specified distance of the tumor (within the limits of the tractography method). To allow interactive updates as the distance is varied, the number of seeds per update is set to a user-defined maximum to minimize latency (varies with dataset and computer configuration). In practice, 200–500 seedpoints provide a sufficient overview of regional tractography. This method is usually followed by use of dynamic seedpoints for specific tract selection.
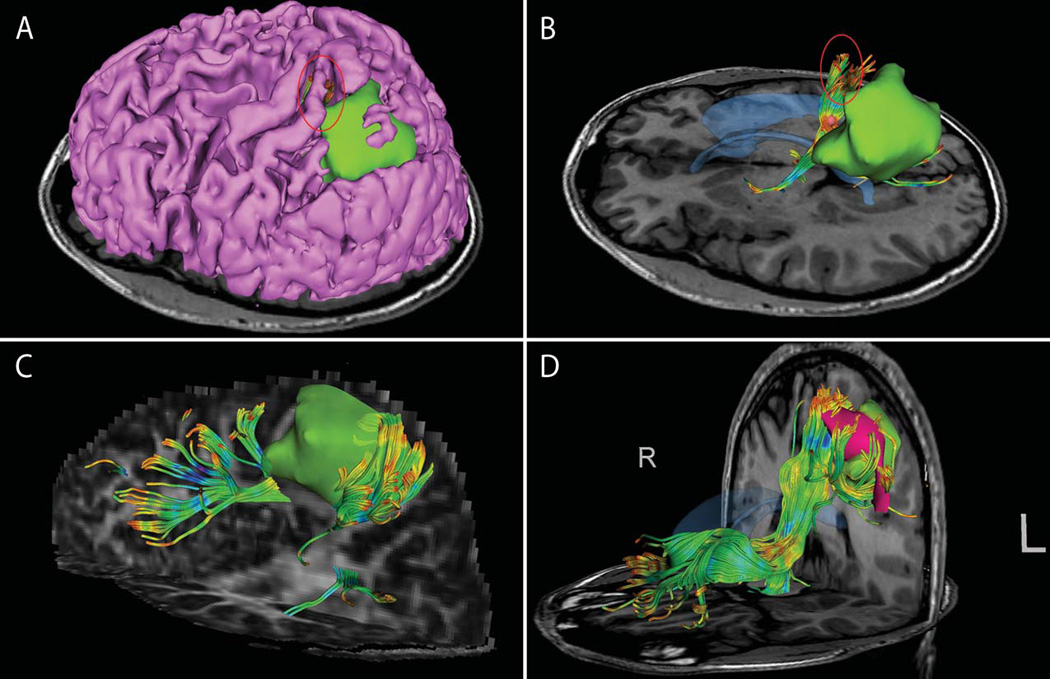
Figure 4. Images from patient 4 with right posterior temporal tumor.
(A) Case overview with segmented cortical surface (pink), segmented tumor (green), and whole-field Vision fMRI (posterior yellow area) (B) Identification using dynamic seedpoints of optic radiations (blue) as a subset of tracts (red) seeded from whole-field Vision fMRI (orange). (C) ROI seeding demonstrates tracts passing through medial and anterior portions of tumor. (D) Distance-map seeding shell (5 mm) demonstrates tracts at the tumor margins.
Dynamic seedpoints
Our software allows the clinician to select arbitrary 3D points (dynamic seedpoints) to define a cluster of seed points within an adjustable sphere around the selected center point. The location of the seedpoint can be interactively moved while the tractography is updated in real time superimposed with the multi-modal image display. The user specifies the spherical seeding region size (mm radius around dynamic seedpoint) and intra-sphere seeding step size (mm). This mode of exploration allows for precise movement of the seeds with respect to the anatomy and allows the surgeon to build an intuitive understanding of areas in the peritumoral volume that may contain critical white matter tracts (see figures 1A–C and figure 3D). Because only a small volume is used as a seed, this tool reduces the visual clutter generated by larger seed regions and can be used for precise probing of specific areas of interest.
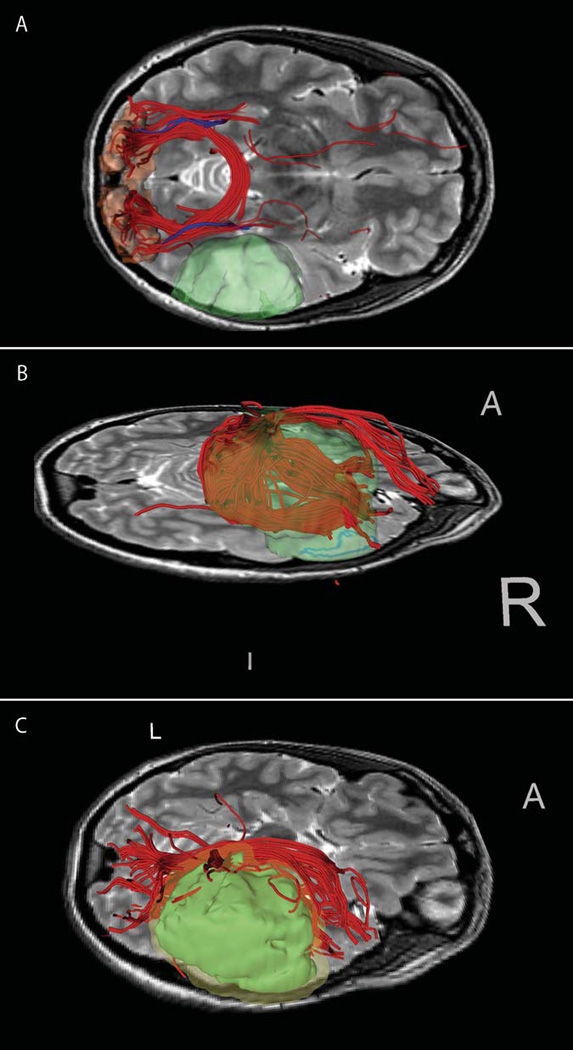
Figure 1. Images from patient 1 with left fronto-parietal tumor.
(A–B) segmented tumor (green) with relative location of CST (red ellipse), with and without cortical surface rendering (pink). Tract was defined by manual seeding exploration near tumor. (C) Posterior dynamic seeding shows parietal tracts in tumor vicinity against FA map (D) ROI seeding from Right Hand fMRI activation area (magenta) shows diverse fiber projections.
Figure 3. Images from patient 3 with left fronto-temporal tumor.
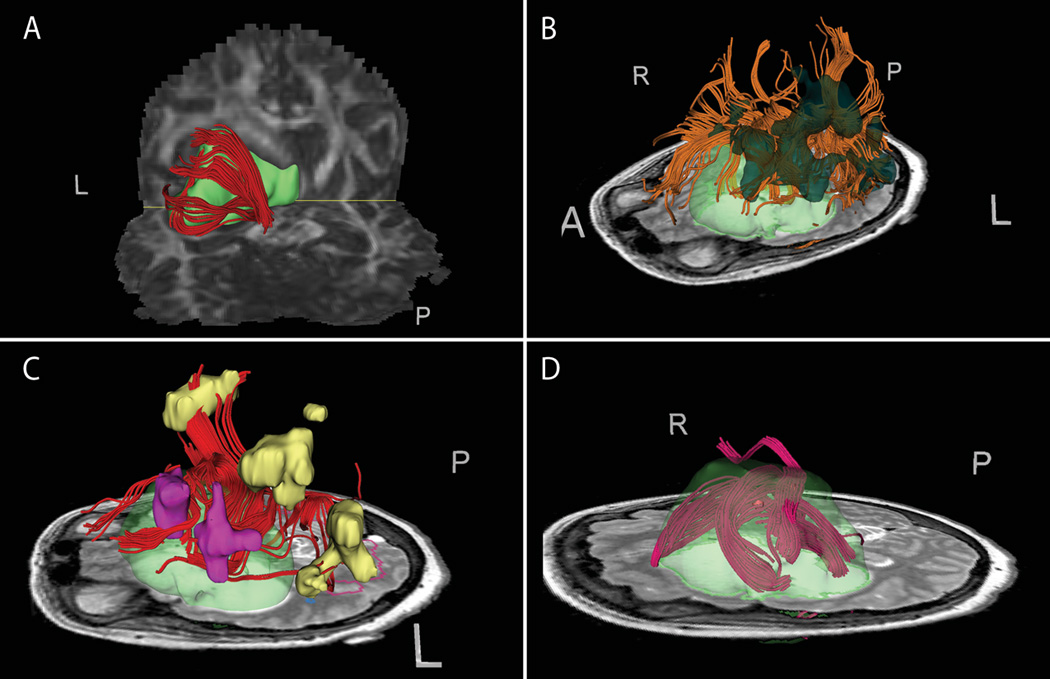
(A) Dynamically seeded tracts wrapping along the posterior tumor margin displayed against DTI FA map. (B) Seeding from Portuguese antonym fMRI task activation. (C) Seeding from a near-tumor subset of fMRI activation for the English noun task. The seeded area (pink) yields tracts connecting to unseeded areas within the same task activation (yellow). (D) Dynamic seedpoint exploration demonstrates tracts of concern in the superior margin of the tumor against T2 with segmented 3D tumor model (green semi-transparent).
Parameters
The methods detailed above provide a number of parameters to control the tractography algorithm. Fractional Anisotropy (FA) is a derived measure of the orientation of water diffusion on a per-voxel basis. FA ranges from 0 (isotropic) to 1 (fully anisotropic), with the minimum for tractography computation generally set around 0.2 (patient, scanner, and anatomy dependent). Stopping Tract curvature (deg/mm) controls the maximum radius of curvature of a tractography path: the tractography algorithm will not return paths which exceed this value (anatomy and algorithm dependent). Path length range restricts the display of fibers to those within the given range and is useful to reduce the visual clutter associated with very short tracts, or to specifically select only certain tracts. Seedpoint spacing determines the minimum distance between starts of tractography computation. Integration step length sets the distance between successive calculations of principle diffusion direction. For a detailed explanation of these parameters and the general principles of Diffusion Tensor Imaging please see: 16, 17, 19, 21, 34, 36, 38
RESULTS
The Structural, fMRI and DTI data were loaded into 3D Slicer for interactive tractography. The clinician (AG) was able to use the tool immediately with assistance from the software developers regarding workflow. The use of the software tool with five patients whose images were retrospectively reviewed is discussed, with illustrations for patients 1–4 and Supplemental Video (available online) for patient 5.
Figure 1 shows images from the investigation of patient 1. In this case the tumor (green model) is adjacent to the central sulcus and particular interest was given to the location of the corticospinal tract and its relationship to the tumor. Seeding was performed with dynamic seedpoints to explore the corticospinal tract (A with cortex, B without) and parietal tracts peripheral to the tumor (C, displayed with FA map). For comparison, ROI seeding from right hand motor fMRI activations (magenta: anterior and lateral to the tumor) shows a similar set of tracts demonstrated adjacent to the tumor (D).
Figure 2 shows images from patient 2 showing tracts both displaced by and within the tumor. (A) shows segmentation of high-intensity tumor regions displayed with cortical outline. ROI-based tractography was used for exploratory seeding within the brightest region of T2 (B), showing antero-superior tracts consistent with displacement rather than infiltration. Dynamically-seeded tracts wrapping around the tumor are displayed against the FA map (C) demonstrating in particular a tract running through the T2-bright area of the lesion. ROI-based seeding from the cerebral peduncle was used to demonstrate the corticospinal tract (D). Intraoperative electrocortical stimulation points were recorded and used for retrospective tract seeding (E–F), giving results consistent with the integrity of the wrapped tract in (C), and supporting the location of the CST in (D).
Figure 2. Images from patient 2 with large R fronto-parieto-occipital tumor.
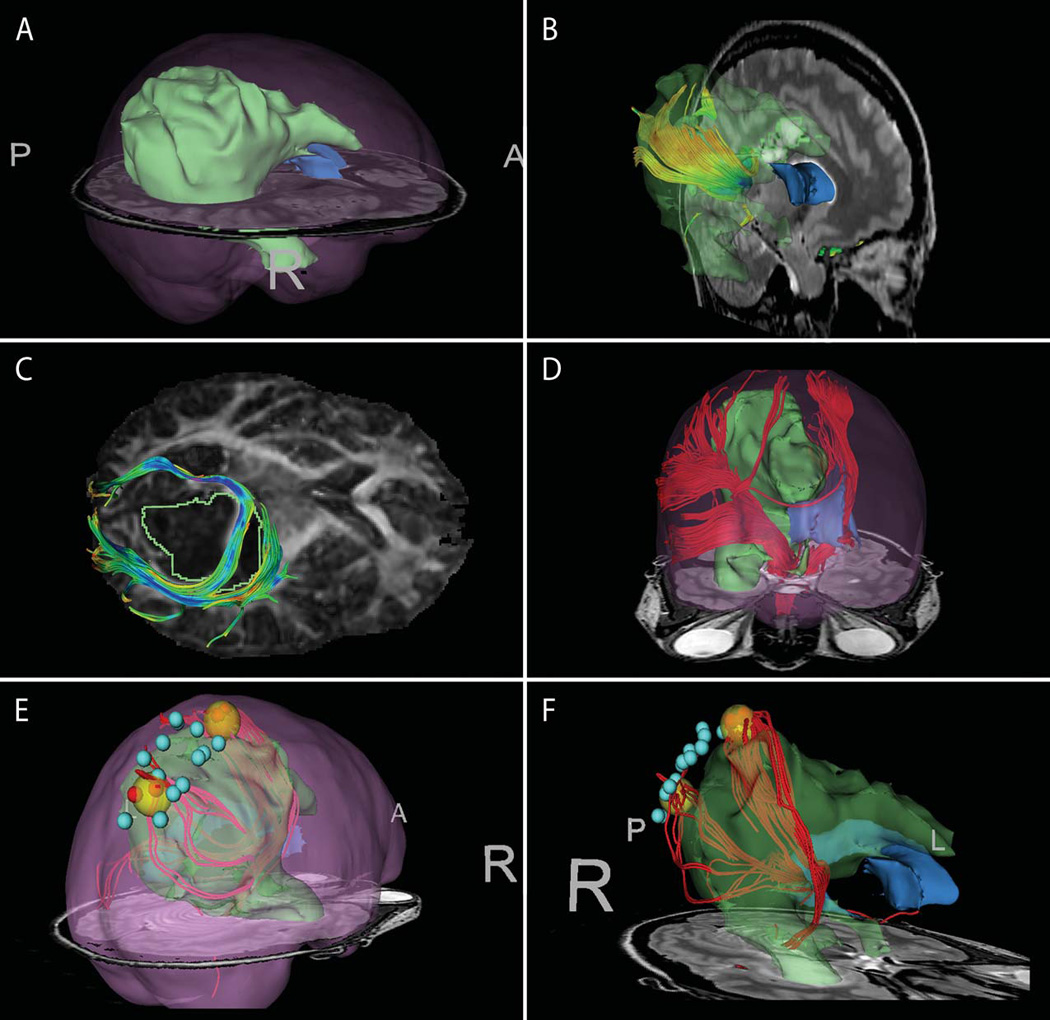
(A) tumor segmentation (green) with brain outline rendering (pink) (B) Seeding within high intensity T2-bright area demonstrates tracts within tumor region. (C) Manually-seeded infiltrating and displaced tracts against the DTI FA map (D) ROI seeding from cerebral peduncle to identify CST. Seeding region was outlined on several T2 slices, and tractography was seeded within this volume.(E–F) offline, post-operative tract seeding from intra-operative cortical stimulation locations (red spheres positive). The yellow spheres represent enlarged seeding area around positive sites in order to seed DTI tracts that do not reach the cortical surface.
Figure 3 shows tractography including fMRI seeding results for patient 3. Dynamically-seeded tracts are observed (A) wrapped or displaced posterior to the tumor, with display against the FA map. Tractography was seeded from the Portuguese antonym activation (B). Seeding from a tumor-proximate subset of the English noun categorization task demonstrated tractography connections to several unseeded activation areas from the same task (C). Figure (D) demonstrates intra-tumor seeding with manual dynamic seedpoints to explore tracts in the superior portion of the tumor near the margin (based on T2 intensity) which could be at risk during resection.
Figure 4 shows images from patient 4 using seeding from fMRI, dynamic seedpoints, and from a shell around the lesion. (A) ROI seeding was performed based on visual task fMRI (orange), which resulted in many tracts (red tracts) in addition to the optic radiation being displayed, thus limiting direct utility of this approach in this case. The optic radiations (blue tracts) were manually delineated using dynamic seedpoints from the LGN, showing correlation with fMRI findings. Using the dynamic seedpoints was helpful in selecting limited tracts of interest from among those obtained by seeding from the fMRI analogous to dissection in cadaveric study.42 Dynamic seedpoint exploration was used (B) to locate tracts wrapping the medial and superior margins of the tumor. The dynamic seeding shell method was used (C) to show tracts at the medial tumor margin.
A short video (available on website) shows the interactive tractography being performed in patient 5. The video demonstrates rotation of the volume and use of the dynamic seedpoint method to develop appreciation for the inherently 3-dimensional qualities of these tools. The initial clip introduces the scene, with cortical surface and lesion segmentations displayed prominently. For context, the next scene demonstrates the result of whole-brain seeding combined with spectral clustering of tracts using a distance-affinity measure.43 Next, the distance map function is used to demonstrate the creation of a seeding shell around the tumor in order to explore the bulk organization of tracts near the tumor. Finally, the cortico-spinal tract is located using manually-placed dynamic seedpoints, and the tumor periphery is explored interactively to locate the putative arcuate fasciculus.
DISCUSSION
We have demonstrated the use of a novel software tool which allows the interactive querying of DTI data in order to define the location and trajectory of white matter tracts around the tumor and critical areas of the brain. Knowledge of the anatomy of the white matter and the relationship to the tumor can be very helpful in planning and carrying out surgical resections in the region of key cortical areas and associated white matter tracts31 Presence of eloquent WM tracts in the vicinity of the tumor is strong predictor of residual tumor.44, 45 Clinically it is critically important to distinguish between cases where the lesion infiltrates the white matter and cases where the fibers are displaced by the lesion as this determines the extent of resectability of the lesion. To the extent that DTI is able to resolve tracts given the limitations imposed by resolution and anisotropy thresholds, this tool allows the clinician to explore areas of potential infiltration with precise seedpoint placement. However, this tool requires some understanding of the underlying DTI methods and inherent limitations. For basic validation, we recommend frequent comparison of the tracts displayed by our selection tools against both the FA map (for example, figure 2-C) and the tensor direction maps available in Slicer. Moreover, as with any imaging method DTI tractography must be used to supplement fundamental clinical understanding and considerations.
Whereas the information provided by DTI is a great advance over previous understanding of the white matter which was based on rough extrapolations from normal anatomical knowledge, making sense of the results is challenging even for neurosurgeons accustomed to thinking in three dimensions and knowledgeable about WM anatomy in general. We have developed this method in response to several questions that have arisen repeatedly when looking at DTI data. 1) Does a tract run in the lesion or just beyond it? When viewing 3D tractography datasets this can be difficult to appreciate. 2) How far is a given tract from the edge of the lesion? The user can define the edge however they feel is most clinically relevant. 3) Which tracts are associated with a particular area of cortex. Cortical areas may be defined based on a priori knowledge or any functional mapping technique such as fMRI or MEG. 4) At the margins of the lesion what are the various WM structures all around? The user can interactively move a dynamic seedpoint around to query any region in detail.
Our method of distance transform isosurfacing is conceptually complementary to previous work based on hulls around fiber pathways by Enders et al.46 In order to better visualize the relationship between fibers and tumor, that method creates a set of tractography paths which are visually simplified to a single hull of elliptical cross-section, with a radial size that is varied to represent a user-specified percentile of enclosed paths. In our method, rather than varying the size of the geometric representation of the fiber pathway, we vary the size of the seeding geometry around the tumor to capture fibers which traverse the region. A related example of interactive DTI analysis is given by Sherbondy et al.47 That method pre-computes DTI tractography paths which may then be selected using interactively defined Volumes of Interest. Our method computes tractography on request, allowing for fine-tuned tract selection using dynamic seedpoints and facilitating the distance-based tract selection method.
This interactive visualization approach can be used with any whole brain tractography acquisition and analysis methods thus taking advantages of technical and theoretical improvements as they are developed. For example, two-tensor models of DTI have been found to resolve crossing fibers better than the single tensor model.48 This can be particularly relevant for demonstrating the lateral projections of the CST to the hand and face regions of the motor cortex49 Thus, as advances in tractography make DTI increasingly accurate, robust, and useful, those results can be formulated and displayed optimally for surgical planning purposes by using this type of approach.
The software allows the use of fMRI to select functionally relevant tracts.50 Intact and sometimes functional WM tracts may be found within some infiltrative low grade tumors.10 fMRI-derived ROIs have been shown to perform better than anatomically-defined ROIs to delineate pyramidal tracts and the superior longitudinal fasciculus.51 Care must be taken when interpreting the output of this style of analysis since the diffusion signal becomes more isotropic as the white matter merges into gray matter and there may be little or no real geometric overlap between detectable white matter tracts and cortical activations. Two possible approaches are to seed from a region representing a dilated fMRI activation, or to place and iteratively move around dynamic seedpoints in the WM directly deep to the fMRI.
Interactive approaches and selective visualization has been employed in a variety of fields including engineering, manufacturing, architecture, and gaming.52, 53 Various approaches have been taken to visualize complex three-dimensional volumetric data such as peeling away layers to view inside the volume.54, 55 Some approaches depend on presenting the viewer with a vantage point that is not fixed thus allowing the inference of three dimensional relationships. Other common approaches include the demonstration of only selected portions of the structure. Our tool takes advantage of both these general approaches to allow the viewer to gradually form an internal representation of the complex 3D structure. The printed images included in the present report are poorly suited to fully demonstrating the advantages of such an approach due to their static nature. Thus we have included a video capture of a session in which clinical data is queried. The viewer is able to gradually form an internal representation of the data which then makes both the moving images and the static images more readily interpreted.
While this approach can serve as a useful adjunct to other DTI images, there are some potential pitfalls of this approach. The approach is prone to subjective decisions made by the person interrogating the data, and may be influenced by existing biases. Thus, for example, if the user has not considered the likely presence of a critical tract in the region, they may not visualize it using the tools. Such a subjective bias is always true of surgeon decision-making. It is reasonable, therefore, to complement this approach with other visualizations such as 2D cross sectional images with DTI glyphs which show the data in a less processed form. Nevertheless, an interactive approach such as that presented puts some of the decision making in the hands of the person doing the surgery rather than a third party who may not be aware of all the clinical facts.
Although the software developed for this research is available for download, it is not approved as a medical device by the FDA or any other authority. Investigators and institutions wishing to make use of the tools must take appropriate care to establish appropriate research protocols. In our experience, effective use of these techniques requires that the research team include one or more members with image analysis experience who are responsible for ensuring that site-specific variability is accounted for in the processing workflow. In particular, fMRI and DWI data acquisition is often the source of incompatibilities due to vendor-dependent file formats, variations in processing techniques, difficulties in image registration, and a reliance on research scanning protocols.
CONCLUSION
Allowing the clinician to interact with DTI-based WM tractography pre-operatively has the potential to provide helpful anatomic and functional information that can make neurosurgical resection safer and more effective. We have developed a tool to allow clinicians to interactively query DTI tractography data in order to define functionally important and anatomically relevant WM tracts.
Supplementary Material
Supplemental Video (available online): Video shows use of the interactive diffusion tensor tractography tool to review tracts for Patient 5 with left temporal lobe metastatic adenocarcinoma. We first demonstrate use of the seeding shell method to explore all tracts within a certain variable distance from the tumor margin. Next, the dynamic seedpoint method is used to precisely select several tracts of interest including the corticospinal tract as well as peri-tumoral tracts including the arcuate fasciculus.
Acknowledgments
Funding Disclosure
NIH 1U41RR019703-01A2
NIH P01-CA67165
Brain Science Foundation
Klarman Family Foundation
References
- 1.Berger MS, Deliganis AV, Dobbins J, Keles GE. The effect of extent of resection on recurrence in patients with low grade cerebral hemisphere gliomas. Cancer. 1994;74(6):1784–1791. doi: 10.1002/1097-0142(19940915)74:6<1784::aid-cncr2820740622>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 2.Bradley WG. Achieving gross total resection of brain tumors: intraoperative MR imaging can make a big difference. AJNR. American Journal of Neuroradiology. 2002;23(3):348–349. [PMC free article] [PubMed] [Google Scholar]
- 3.Claus EB, Horlacher A, Hsu L, et al. Survival rates in patients with low-grade glioma after intraoperative magnetic resonance image guidance. Cancer. 2005;103(6):1227–1233. doi: 10.1002/cncr.20867. [DOI] [PubMed] [Google Scholar]
- 4.Sanai N, Berger MS. Glioma extent of resection and its impact on patient outcome. Neurosurgery. 2008;62(4):753–764. doi: 10.1227/01.neu.0000318159.21731.cf. discussion 264–266. [DOI] [PubMed] [Google Scholar]
- 5.Talos I-F, Zou KH, Ohno-Machado L, et al. Supratentorial low-grade glioma resectability: statistical predictive analysis based on anatomic MR features and tumor characteristics. Radiology. 2006;239(2):506–513. doi: 10.1148/radiol.2392050661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alberstone CD, Skirboll SL, Benzel EC, et al. Magnetic source imaging and brain surgery: presurgical and intraoperative planning in 26 patients. Journal of Neurosurgery. 2000;92(1):79–90. doi: 10.3171/jns.2000.92.1.0079. [DOI] [PubMed] [Google Scholar]
- 7.Fried I, Nenov VI, Ojemann SG, Woods RP. Functional MR and PET imaging of rolandic and visual cortices for neurosurgical planning. Journal of Neurosurgery. 1995;83(5):854–861. doi: 10.3171/jns.1995.83.5.0854. [DOI] [PubMed] [Google Scholar]
- 8.Tharin S, Golby A. Functional brain mapping and its applications to neurosurgery. Neurosurgery. 2007;60(4) Suppl 2:185–201. doi: 10.1227/01.NEU.0000255386.95464.52. discussion 201–202. [DOI] [PubMed] [Google Scholar]
- 9.Witwer BP, Moftakhar R, Hasan KM, et al. Diffusion-tensor imaging of white matter tracts in patients with cerebral neoplasm. Journal of Neurosurgery. 2002;97(3):568–575. doi: 10.3171/jns.2002.97.3.0568. [DOI] [PubMed] [Google Scholar]
- 10.Skirboll SS, Ojemann GA, Berger MS, Lettich E, Winn HR. Functional cortex and subcortical white matter located within gliomas. Neurosurgery. 1996 Apr;38(4):678–684. discussion 684–675. [PubMed] [Google Scholar]
- 11.Kinoshita M, Yamada K, Hashimoto N, et al. Fiber-tracking does not accurately estimate size of fiber bundle in pathological condition: initial neurosurgical experience using neuronavigation and subcortical white matter stimulation. NeuroImage. 2005;25(2):424–429. doi: 10.1016/j.neuroimage.2004.07.076. [DOI] [PubMed] [Google Scholar]
- 12.Keles GE, Lundin DA, Lamborn KR, Chang EF, Ojemann G, Berger MS. Intraoperative subcortical stimulation mapping for hemispherical perirolandic gliomas located within or adjacent to the descending motor pathways: evaluation of morbidity and assessment of functional outcome in 294 patients. Journal of Neurosurgery. 2004;100(3):369–375. doi: 10.3171/jns.2004.100.3.0369. [DOI] [PubMed] [Google Scholar]
- 13.Duffau H, Capelle L, Sichez N, et al. Intraoperative mapping of the subcortical language pathways using direct stimulations. An anatomo-functional study. Brain: A Journal of Neurology. 2002;125(Pt 1):199–214. doi: 10.1093/brain/awf016. [DOI] [PubMed] [Google Scholar]
- 14.Duffau H, Capelle L, Denvil D, et al. Usefulness of intraoperative electrical subcortical mapping during surgery for low-grade gliomas located within eloquent brain regions: functional results in a consecutive series of 103 patients. Journal of Neurosurgery. 2003;98(4):764–778. doi: 10.3171/jns.2003.98.4.0764. [DOI] [PubMed] [Google Scholar]
- 15.Duffau H, Peggy Gatignol ST, Mandonnet E, Capelle L, Taillandier L. Intraoperative subcortical stimulation mapping of language pathways in a consecutive series of 115 patients with Grade II glioma in the left dominant hemisphere. J Neurosurg. 2008 Sep;109(3):461–471. doi: 10.3171/JNS/2008/109/9/0461. [DOI] [PubMed] [Google Scholar]
- 16.Basser PJ, Mattiello J, LeBihan D. Estimation of the effective self-diffusion tensor from the NMR spin echo. J Magn Reson B. 1994 Mar;103(3):247–254. doi: 10.1006/jmrb.1994.1037. [DOI] [PubMed] [Google Scholar]
- 17.Beaulieu C. The basis of anisotropic water diffusion in the nervous system - a technical review. NMR Biomed. 2002 Nov-Dec;15(7–8):435–455. doi: 10.1002/nbm.782. [DOI] [PubMed] [Google Scholar]
- 18.Mori S, Barker PB. Diffusion magnetic resonance imaging: its principle and applications. The Anatomical Record. 1999;257(3):102–109. doi: 10.1002/(SICI)1097-0185(19990615)257:3<102::AID-AR7>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 19.Mori S, van Zijl PC. Fiber tracking: principles and strategies - a technical review. NMR Biomed. 2002 Nov-Dec;15(7–8):468–480. doi: 10.1002/nbm.781. [DOI] [PubMed] [Google Scholar]
- 20.Bello L, Gambini A, Castellano A, et al. Motor and language DTI Fiber Tracking combined with intraoperative subcortical mapping for surgical removal of gliomas. NeuroImage. 2008;39(1):369–382. doi: 10.1016/j.neuroimage.2007.08.031. [DOI] [PubMed] [Google Scholar]
- 21.Beppu T, Inoue T, Shibata Y, et al. Measurement of fractional anisotropy using diffusion tensor MRI in supratentorial astrocytic tumors. Journal of Neuro-Oncology. 2003;63(2):109–116. doi: 10.1023/a:1023977520909. [DOI] [PubMed] [Google Scholar]
- 22.Clark CA, Barrick TR, Murphy MM, Bell BA. White matter fiber tracking in patients with space-occupying lesions of the brain: a new technique for neurosurgical planning? NeuroImage. 2003;20(3):1601–1608. doi: 10.1016/j.neuroimage.2003.07.022. [DOI] [PubMed] [Google Scholar]
- 23.Hendler T, Pianka P, Sigal M, et al. Delineating gray and white matter involvement in brain lesions: three-dimensional alignment of functional magnetic resonance and diffusion-tensor imaging. Journal of Neurosurgery. 2003;99(6):1018–1027. doi: 10.3171/jns.2003.99.6.1018. [DOI] [PubMed] [Google Scholar]
- 24.Lu S, Ahn D, Johnson G, Cha S. Peritumoral diffusion tensor imaging of high-grade gliomas and metastatic brain tumors. AJNR. American Journal of Neuroradiology. 2003;24(5):937–941. [PMC free article] [PubMed] [Google Scholar]
- 25.Price SJ, Burnet NG, Donovan T, et al. Diffusion tensor imaging of brain tumours at 3T: a potential tool for assessing white matter tract invasion? Clinical Radiology. 2003;58(6):455–462. doi: 10.1016/s0009-9260(03)00115-6. [DOI] [PubMed] [Google Scholar]
- 26.Smits M, Vernooij MW, Wielopolski PA, Vincent AJPE, Houston GC, van der Lugt A. Incorporating functional MR imaging into diffusion tensor tractography in the preoperative assessment of the corticospinal tract in patients with brain tumors. AJNR. American Journal of Neuroradiology. 2007;28(7):1354–1361. doi: 10.3174/ajnr.A0538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tummala RP, Chu RM, Liu H, Truwit CL, Hall WA. Application of diffusion tensor imaging to magnetic-resonance-guided brain tumor resection. Pediatric Neurosurgery. 2003;39(1):39–43. doi: 10.1159/000070879. [DOI] [PubMed] [Google Scholar]
- 28.Wieshmann UC, Symms MR, Parker GJ, et al. Diffusion tensor imaging demonstrates deviation of fibres in normal appearing white matter adjacent to a brain tumour. Journal of Neurology, Neurosurgery, and Psychiatry. 2000;68(4):501–503. doi: 10.1136/jnnp.68.4.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamada K, Kizu O, Mori S, et al. Brain fiber tracking with clinically feasible diffusion-tensor MR imaging: initial experience. Radiology. 2003;227(1):295–301. doi: 10.1148/radiol.2271020313. [DOI] [PubMed] [Google Scholar]
- 30.Nimsky C, Ganslandt O, Merhof D, Sorensen AG, Fahlbusch R. Intraoperative visualization of the pyramidal tract by diffusion-tensor-imaging-based fiber tracking. NeuroImage. 2006;30(4):1219–1229. doi: 10.1016/j.neuroimage.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 31.Wu JS, Zhou LF, Tang WJ, et al. Clinical evaluation and follow-up outcome of diffusion tensor imaging-based functional neuronavigation: a prospective, controlled study in patients with gliomas involving pyramidal tracts. Neurosurgery. 2007 Nov;61(5):935–948. doi: 10.1227/01.neu.0000303189.80049.ab. discussion 948–939. [DOI] [PubMed] [Google Scholar]
- 32.Nimsky C, Ganslandt O, Hastreiter P, et al. Preoperative and intraoperative diffusion tensor imaging-based fiber tracking in glioma surgery. Neurosurgery. 2007 Jul;61(1 Suppl):178–185. doi: 10.1227/01.neu.0000279214.00139.3b. discussion 186. [DOI] [PubMed] [Google Scholar]
- 33.Catani M, Howard RJ, Pajevic S, Jones DK. Virtual in vivo interactive dissection of white matter fasciculi in the human brain. Neuroimage. 2002 Sep;17(1):77–94. doi: 10.1006/nimg.2002.1136. [DOI] [PubMed] [Google Scholar]
- 34.Conturo TE, Lori NF, Cull TS, et al. Tracking neuronal fiber pathways in the living human brain. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(18):10422–10427. doi: 10.1073/pnas.96.18.10422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wakana S, Caprihan A, Panzenboeck MM, et al. Reproducibility of quantitative tractography methods applied to cerebral white matter. Neuroimage. 2007 Jul 1;36(3):630–644. doi: 10.1016/j.neuroimage.2007.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jellison BJ, Field AS, Medow J, Lazar M, Salamat MS, Alexander AL. Diffusion tensor imaging of cerebral white matter: a pictorial review of physics, fiber tract anatomy, and tumor imaging patterns. AJNR. American Journal of Neuroradiology. 2004;25(3):356–369. [PMC free article] [PubMed] [Google Scholar]
- 37.Pieper S, Halle M, Kikinis R. 3D Slicer. Biomedical Imaging: Nano to Macro, 2004. IEEE International Symposium on. 2004;Vol. 631:632–635. [Google Scholar]
- 38.Basser PJ, Pajevic S, Pierpaoli C, Duda J, Aldroubi A. In vivo fiber tractography using DT-MRI data. Magnetic Resonance in Medicine: Official Journal of the Society of Magnetic Resonance in Medicine / Society of Magnetic Resonance in Medicine. 2000;44(4):625–632. doi: 10.1002/1522-2594(200010)44:4<625::aid-mrm17>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 39.Yoo TS. Insight into Images: Principles and Practice for Segmentation, Registration, and Image Analysis. AK Peters Ltd; 2004. [Google Scholar]
- 40.Jezzard P, Balaban RS. Correction for geometric distortion in echo planar images from B0 field variations. Magn Reson Med. 1995 Jul;34(1):65–73. doi: 10.1002/mrm.1910340111. [DOI] [PubMed] [Google Scholar]
- 41.Pieper S, Lorensen B, Schroeder W, Kikinis R. The NA-MIC Kit: ITK, VTK, pipelines, grids and 3D slicer as an open platform for the medical image computing community. Biomedical Imaging: Nano to Macro, 2006. 3rd IEEE International Symposium on. 2006:698–701. [Google Scholar]
- 42.Ture U, Yasargil MG, Friedman AH, Al-Mefty O. Fiber dissection technique: lateral aspect of the brain. Neurosurgery. 2000 Aug;47(2):417–426. doi: 10.1097/00006123-200008000-00028. discussion 426–417. [DOI] [PubMed] [Google Scholar]
- 43.O'Donnell LJ, Westin C-F, Golby AJ. Tract-based morphometry. Medical Image Computing and Computer-Assisted Intervention: MICCAI … International Conference on Medical Image Computing and Computer-Assisted Intervention. 2007;10(Pt 2):161–168. [PubMed] [Google Scholar]
- 44.Mandonnet E, Jbabdi S, Taillandier L, et al. Preoperative estimation of residual volume for WHO grade II glioma resected with intraoperative functional mapping. Neuro-Oncology. 2007;9(1):63–69. doi: 10.1215/15228517-2006-015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Talos IF, Zou KH, Kikinis R, Jolesz FA. Volumetric assessment of tumor infiltration of adjacent white matter based on anatomic MRI and diffusion tensor tractography. Acad Radiol. 2007 Apr;14(4):431–436. doi: 10.1016/j.acra.2007.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Enders F, Sauber N, Merhof D, Hastreiter P, Nimsky C, Stamminger MS. Visualization of White Matter Tracts with Wrapped Streamlines; Visualization Conference, IEEE; Los Alamitos, CA, USA: IEEE Computer Society; 2005. p. 7. [Google Scholar]
- 47.Sherbondy A, Akers D, Mackenzie R, Dougherty R, Wandell B. Exploring connectivity of the brain's white matter with dynamic queries. Visualization and Computer Graphics, IEEE Transactions on. 2005;11(4):419–430. doi: 10.1109/TVCG.2005.59. [DOI] [PubMed] [Google Scholar]
- 48.Peled S, Friman O, Jolesz F, Westin C-F. Geometrically constrained two-tensor model for crossing tracts in DWI. Magnetic Resonance Imaging. 2006;24(9):1263–1270. doi: 10.1016/j.mri.2006.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Qazi AA, Radmanesh A, O'Donnell L, et al. Resolving crossings in the corticospinal tract by two-tensor streamline tractography: Method and clinical assessment using fMRI. NeuroImage. 2008 doi: 10.1016/j.neuroimage.2008.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guye M, Parker GJM, Symms M, et al. Combined functional MRI and tractography to demonstrate the connectivity of the human primary motor cortex in vivo. NeuroImage. 2003;19(4):1349–1360. doi: 10.1016/s1053-8119(03)00165-4. [DOI] [PubMed] [Google Scholar]
- 51.Schonberg T, Pianka P, Hendler T, Pasternak O, Assaf Y. Characterization of displaced white matter by brain tumors using combined DTI and fMRI. NeuroImage. 2006;30(4):1100–1111. doi: 10.1016/j.neuroimage.2005.11.015. [DOI] [PubMed] [Google Scholar]
- 52.Li W, Agrawala M, Curless B, Salesin D. ACM SIGGRAPH 2008 papers. Los Angeles, California: ACM; 2008. Automated generation of interactive 3D exploded view diagrams; pp. 1–7. [Google Scholar]
- 53.Li W, Ritter L, Agrawala M, Curless B, Salesin D. ACM SIGGRAPH 2007 papers. San Diego, California: ACM; 2007. Interactive cutaway illustrations of complex 3D models; p. 31. [Google Scholar]
- 54.Correa CD, Silver D, Chen M. Feature Aligned Volume Manipulation for Illustration and Visualization. Visualization and Computer Graphics, IEEE Transactions on. 2006;12(5):1069–1076. doi: 10.1109/TVCG.2006.144. [DOI] [PubMed] [Google Scholar]
- 55.McGuffin MJ, Tancau L, Balakrishnan R. Proceedings of the 14th IEEE Visualization 2003 (VIS'03) IEEE Computer Society; 2003. Using Deformations for Browsing Volumetric Data; p. 53. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Video (available online): Video shows use of the interactive diffusion tensor tractography tool to review tracts for Patient 5 with left temporal lobe metastatic adenocarcinoma. We first demonstrate use of the seeding shell method to explore all tracts within a certain variable distance from the tumor margin. Next, the dynamic seedpoint method is used to precisely select several tracts of interest including the corticospinal tract as well as peri-tumoral tracts including the arcuate fasciculus.


