Abstract
GeneLynx is a meta-database providing an extensive collection of hyperlinks to human gene-specific information in diverse databases available on the Internet. The GeneLynx project is based on the simple notion that given any gene-specific identifier (accession number, gene name, text, or sequence), scientists should be able to access a single location that provides a set of links to all the publicly available information pertinent to the specified human gene. GeneLynx was implemented as an extensible relational database with an intuitive and user-friendly Web interface. The data are automatically extracted from more than 40 external resources, using appropriate approaches to maximize coverage of the available data. Construction and curation of the system is mediated by a custom set of software tools. An indexing utility is provided to facilitate the establishment of hyperlinks in external databases. A unique feature of the GeneLynx system is a communal curation system for user-aided annotation. GeneLynx can be accessed freely at http://www.genelynx.org.
The sequencing and analysis of the human genome (International Human Genome Sequencing Consortium 2001) marks a climax of the international genome project. Despite this great success, data describing individual human genes and their encoded protein products continue to accumulate haphazardly, filling widely distributed databases accessible via diverse and often idiosyncratic Web interfaces. The collection of available information on any particular gene remains difficult and time consuming for most researchers. Useful data often remain buried in resources outside the knowledge of biologists. An ideal system would enable a researcher to submit a single query to rapidly access all the available gene-specific information about their gene of interest. Through persistent long-term efforts, such systems are available for several model organisms such as Drosophila (FlyBase Consortium 1999), Caenorhabditis elegans (Stein et al. 2001), and Saccharomyces cerevisiae (Cherry et al. 1998; Costanzo et al. 2001). The need for a similar human resource is growing daily with the increased application of parallel, high-throughput gene analysis methods, which often place a spotlight on genes outside the specialized background of research teams.
The need for integration of biological databases has been discussed widely, motivating the increased presence of cross-references between databases. However, the cross-referencing features between databases typically provide access to only a small subset of the available gene-specific information. Currently, there are several excellent resources, such as SWISS-PROT (Bairoch and Apweiler 2000), GeneCards SWISS-PROT (Rebhan et al. 1998), and LocusLink (Pruitt et al. 2000), providing valuable links to external information. Despite the quality of these efforts, there remains an unmet need for a database dedicated to connecting users with a comprehensive range of resources.
In this report we present GeneLynx, a Web-based system consisting of a comprehensive and easily extensible meta-database of hyperlinks organized around the set of human genes. The database is accompanied by an intuitive and simple user interface, efficient text, and sequence search engines, as well as a set of tools to facilitate regular updates and biologically meaningful database curation. The ultimate mission of GeneLynx is, given any reasonable gene identifier (a name, keyword, sequence identifier, or sequence), to provide links to all the information available for that gene. GeneLynx points to an extensive set of Web resources, ranging from nucleotide and protein sequence collections to summary pages and disease-related resources. For such a comprehensive resource to remain both current and accurate, it was designed with convenient procedures for updating and extending the collection of hyperlinks. In addition to the curation and quality control tools, we have equipped it with a unique system for the submission of gene-based user comments and corrections, which will be reviewed and incorporated by GeneLynx curators. It is our hope that the latter service will enable GeneLynx to become a high-quality, communally curated, comprehensive resource for studying human genes.
RESULTS
Database Contents: Gene-Based Clusters and Linked Resources
The current version (August 1, 2001) of the GeneLynx database contains 31,992 gene-based clusters (Table 1). Within the collection, a subset of 19,342 clusters contains cDNA sequences. The remaining records are based on expressed sequence tag (EST)-only clusters from UniGene (with restrictions described in Methods). Note that definitions for what constitutes a gene vary widely. For the construction of GeneLynx, we intended to consolidate cDNA and assembled EST sequences into single entries when subsegments of the processed transcripts were derived from transcription of the same chromosomal sequence in the same direction. Thus, alternative splice forms and the use of alternative promoters would generate differing transcripts for the same gene. Although the total number of GeneLynx entries approaches the number of genes predicted to be present in the human genome (International Human Genome Sequencing Consortium 2001; Venter 2001), there are likely to be many genes that are not represented in the public cDNA and EST sequence databases. At the time of submission, GeneLynx provided links to 47 categories of data from 38 external resources. The total number of links is more than half a million (not including links to EST sequences), with most summarized in Table 2.
Table 1.
Number of Links in Current GeneLynx Release (v0.9.)
| GeneLynx records with multiple cDNAs | 13044 |
| GeneLynx records with a single cDNA | 6338 |
| EST-only GeneLynx records | 12610 |
| Total number of GeneLynx records | 31992 |
Table 2.
Number of Links per Resource* in Current GeneLynx Release (0.9.1, August 1, 2001)
| Resource | Number of items linked to GeneLynx | Number of linked GeneLynx records |
|---|---|---|
| UniGene | 30379 | 30348 |
| GeneCards | 13863 | 13867 |
| cDNAs (GB/EMBL/DDBJ) | 70704 | 19432 |
| HumanPSD (Proteome) | 9240 | 9309 |
| KEGG (genes) | 14850 | 15002 |
| KEGG (pathways) | 77 | 1246 |
| MIPS | 5998 | 5127 |
| Genomic sequences | 13225 | 6106 |
| GDB | 9145 | 9265 |
| LocusLink | 14930 | 15099 |
| EGAD | 6667 | 5490 |
| Ensembl (genes) | 10487 | 10784 |
| Ensembl (transcripts) | 13603 | 10784 |
| RefSeq | 13653 | 13055 |
| HGBASE | 16082 | 4216 |
| SWISS-PROT | 6521 | 6346 |
| TrEMBL | 16719 | 10250 |
| PIR | 7333 | 5577 |
| GenPept collection | 42653 | 15438 |
| PDB | 1898 | 527 |
| HSSP | 836 | 2982 |
| InterPro | 1864 | 9900 |
| PRINTS | 896 | 3792 |
| BLOCKS/PRODOM | 2501 | 3355 |
| SUPERFAMILY | 13603 | 10784 |
| PFAM | 1370 | 6537 |
| SBASE | 25519 | 4355 |
| PROSITE | 1056 | 6879 |
| ENZYME DB/WIT/Brenda | 616 | 1382 |
| MEROPS | 287 | 286 |
| OMIM | 7857 | 7577 |
| GeneClinics | 159 | 128 |
| MGD | 4856 | 5120 |
| HumanPSD (Proteome) | 9240 | 9509 |
| Homologs (UniGene) | 13830 | 6890 |
| Homologs (LocusLink) | 5412 | 4730 |
| Homologs (nucleotide seqs) | 14681 | 7010 |
| Homologs (protein seqs) | 14531 | 12349 |
| RZPD CloneCards | 2022309 | 29039 |
| STACK | 43462 | 23244 |
| ESTs | 2290885 | 29283 |
A few resources accessible from GeneLynx are not listed in the table, due to an indeterminate number of linked items. For details, see http://www.genelynx.org/TECHNICAL/ .
For a restricted group of exceptional genes, notably those of the immune system that undergo genomic rearrangements, the selected sequence clustering algorithm was inadequate. To address these cases, we formed five major groups (immunoglobulins, major histocompatibility complex [MHC] class I antigens, MHC class 2 antigens, T-cell receptors, and natural killer cell receptors) and defined as super clusters within GeneLynx. Currently, we have not attempted to organize the extensive information available on these genes. Ultimately, a new set of specifically designed clustering methods and curation tools will be required.
User Interface
Excluding the home page and documentation, the GeneLynx interface consists of only five page types:
- 1.
The Text Search page is used for the composition of a query against the GeneLynx Index (quick search) or specified fields in GeneLynx database (advanced search). Quick Search is a fast method for locating keywords in the GeneLynx Index. Advanced Search enables the user to search specific fields and execute complex queries using Boolean operators. The details of the search procedures are described below, but the Quick Search function is likely to provide sufficient performance for most users.
- 2.
The BLAST search page enables users to submit protein or nucleotide sequences for comparison against the set of all cDNAs and assembled ESTs within GeneLynx. The user currently can specify a threshold E-value, which is more than adequate for the identification of related genes. No alignments are displayed to encourage users to perform routine BLAST searches on more computationally powerful servers.
- 3.
The Hits page displays the results from either text or BLAST searches. Text search hits are ordered by scores calculated from the number of matched words weighted relative to the frequency of the words within the index. BLAST search hits are listed in order of increasing E-value, displaying the GenBank accession number of the most significant match for each GeneLynx cluster.
- 4.
The GeneLynx Record page (the central page of GeneLynx) contains the gene name, description and locus position, and the categorized list of hyperlinks (Fig. 1).
- 5.
The User Comments page contains comment submission forms and a list of existing user comments, including those reviewed by curators, as well as those pending review.
Figure 1.
A screenshot of the GeneLynx record Web page, containing a comprehensive set of links for the given gene (hypoxanthine phosphoribosyltransferase 1 is shown).
In addition to the basic interactive set of pages, auxiliary pages include the GeneLynx Guide and specialized interfaces related to the batch generation of hyperlinks (as described below).
Search Engine
GeneLynx supports two types of text searches. For quick searches, a universal table of information is analyzed. The GeneLynx Index is an indexed two-column table in the relational database that includes all collected keywords, accession numbers, and database identifiers in one column matched to their corresponding GeneLynx identifier in the second column. Query results are processed according to search parameters defined by the user. This search feature brings GeneLynx closer to its core mission: accessing a single comprehensive set of links given any gene-specific identifier. The search time (in single-user mode) is <0.1 second on a Pentium III/700 test system running Linux. Thus, it is adequate for most text-based searches and enables processing of a high number of queries per unit time. The advanced search allows for a more precise query formulation and operates on resource-specific tables within the relational database. In the latter case, users have control over which resources are queried. External resources at their remote locations are not accessed directly by either type of query; database identifiers are stored locally for that purpose, and in most cases it is the only data from the external resources stored within the GeneLynx database. Both query systems have proven to be fast and efficient in leading users to desired gene information.
For BLAST searches, the database queried is a fixed, GeneLynx-specific collection of nucleotide sequences consisting of human cDNAs associated with GeneLynx records and the assembled sequence contigs for EST-only records. The restriction to this database enables direct and unequivocal mapping of BLAST hits to GeneLynx records.
Communal Curation Interface
An important issue in implementing a meta-database, such as GeneLynx, is the maintenance of coverage and accuracy as new data are introduced in external databases. As manual curation of all records is not feasible because of the volume and breadth of data and the inconvenient limitation of 24 hours in each day, we implemented a system through which users can submit comments, corrections, and additional information for any gene (see Fig. 2). Curators review user comments and either introduce the appropriate changes to the database or post the comment directly to the gene page. The curators' primary responsibility is to eliminate postings that do not specifically address the associated gene. Such restricted curation means that incorrect comments will be posted occasionally, and users are well advised to use their judgement in assessing the views and knowledge of their peers. If the human research community contributes, the data quality and coverage will improve with time.
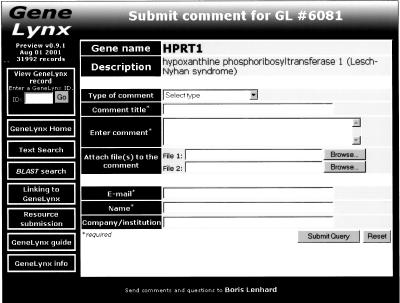
Figure 2.
A screenshot of the GeneLynx user comment submission interface. The submitted comment, together with the curator's response, is available for users to consider.
Batch Assignment and Linking to GeneLynx
An interface was provided for batch analysis of lists of gene identifiers to support the integration of GeneLynx with external databases. As a result of the breadth of coverage, GeneLynx supports the use of identifiers from more than 20 different biological databases. The output of the batch analysis is a list of those identifiers with associated GeneLynx numbers, in either HTML or a plain-text format. This service will ease the interpretation of results of microarray experiments and other high-throughput methods producing large amounts of gene-associated information. For instance, research groups may wish to obtain hyperlinks for all of the human genes represented as spots on a microarray.
Addition of New Resources to GeneLynx
A simple system is provided to the community for the generation of hyperlinks between new resources and GeneLynx. Through a Web interface, any user may submit an association list (a two-column text file containing the identifiers from the new resource and associated identifiers of a resource already represented in GeneLynx), a rule to define the Web address of each entry and an association. For example, if an external database curator submits a two-column list of their database identifiers and the corresponding SWISS-PROT identifiers, the submitted list is processed and stored to a temporary table. A random sample of up to 20 links is presented to the submitter on the submission confirmation page, to allow confirmation that the associations are correct and that the hyperlinks to external databases are functional. After review, a GeneLynx curator approves incorporation of the new data into the GeneLynx system. If the new identifiers are alphanumeric, they are added to the GeneLynx Index and become accessible to the quick search routine. The inclusion itself is still curator moderated, but nevertheless submissions by this method will be rapid and hopefully reduce the mistakes made when the association is performed by a GeneLynx curator insufficiently familiar with the new resource.
DISCUSSION
The motivation for GeneLynx is simple: Scientists working on human genes repeatedly find it difficult or impossible to rapidly access the available database information about their genes of interest using the existing and freely available Web resources. We believe that many users desire a system with a simple and intuitive user interface similar to those found on common Internet portals, to make access to gene-specific data one click away. The GeneLynx system attempts to satisfy this demand.
There are several existing Internet resources that overlap with the mission of GeneLynx (see Table 2), for example:
- 1.
GeneCards (Rebhan et al. 1998) provides a page of summary information (a card) for each human gene, with links to related information resources. As such, GeneCards is an excellent resource for quick, text-based access to basic information on characterized genes.
- 2.
SWISS-PROT (Bairoch and Apweiler 2000) offers a rich collection of curated hyperlinks for each protein entry. The database is protein-based, with a focus on hyperlinks relevant to protein structure and function. Given SWISS-PROT's admirable commitment to expert curation and high data quality, the incorporation of new sequences and data is quick, but not instantaneous.
- 3.
LocusLink (Pruitt et al. 2000) is a comprehensive resource of curated information on genetic loci for human and several other eukaryotes. Its collection of links preferentially addresses resources at the National Institutes of Health, with links to external systems limited but growing.
The strengths of GeneLynx lie in the breadth of coverage of external resources, along with additional features that are either novel or outside the scope of related systems. A tabular comparison of GeneLynx to related resources is maintained at http://www.genelynx.org/TECHNICAL/. GeneLynx serves a specific niche: quick and intuitive access to a set of links that will deliver the users to gene-specific information in databases on the Internet. Initial beta testers of the system have complimented the data coverage and ease of use. A small set of inaccuracies were detected, which could be addressed easily by the community curation feature. It was noted by users that the GeneLynx record page serves an instant educational role, by bringing new and underutilized bioinformatics resources to the attention of users. Given the limited awareness of major public data collections, (see press release from Wellcome Trust pertaining to Ensembl, at http://www.ensembl.org/News/010426.html) GeneLynx offers a convenient interface for biologists to the broad array of bioinformatics initiatives.
Although we believe the choice is justified to use a relational database system as the foundation for the GeneLynx engine, other researchers may hold different viewpoints. In particular, a case could be made for the use of the popular Sequence Retrieval System (SRS) (Etzold et al. 1996). We find the SQL (structured query language)-based database system more easily maintained and the relational aspects more intuitive than the management of flat files in SRS. SRS is particularly useful for its intended purpose in the management and retrieval of biopolymer sequences, both of which are outside the scope of the GeneLynx system. Although we suspect that SRS' cross-referencing functions could be applied successfully in a GeneLynx-like system, we prefer SQL-based relational databases for the management of gene-centric data (as opposed to sequence centric).
There are several directions in which GeneLynx could be expanded. One need to address is that of a reliable and categorized collection of literature links for each gene. Several resources have begun to address text-to-gene associations, including HumanPSD (http://www.proteome.com/) and PubGene (Jenssen et al. 2001). In addition, there are many reliable references within the GenBank records. Fully automated approaches are likely to generate a substantial proportion of irrelevant hits, which would be contrary to the desired accuracy of GeneLynx data. The current solution is that users themselves can contribute relevant literature links using the GeneLynx comment submission protocol.
The completeness and accuracy of information will be among the most important characteristics by which GeneLynx will be judged. We strive to achieve high coverage of external resources, while maintaining data accuracy. However, because of the semiautomatic nature of the database construction, some users will encounter cases of either missing or wrongly assigned data. Although such problems can be difficult to address in other systems, users are encouraged to identify flaws in the system and report them through the communal curation services in GeneLynx.
The optimistic and potentially naive concept of streamlined communal curation of GeneLynx derives from the tremendous success of similar projects for model organisms (Cherry et al. 1998; FlyBase Consortium 1999; Stein et al. 2001). Here, we suggest an organized way of joining the expertise of many researchers to make GeneLynx a complete and reliable resource for the scientific community. By simplifying and standardizing the procedure for adding links to new (or missed) resources, GeneLynx should respond quickly to its users demands and become ever more accurate and complete as we learn about the human genome.
METHODS
Database Organization
The GeneLynx database consists of a set of relational database tables that connect GeneLynx IDs to each resource, storing the identifiers required to construct hyperlinks to the target database. Because of the nature of the data and a certain amount of assignment error present in most databases, it is not possible to design a normalized database with enforced referential integrity. Instead, we developed a set of tools that performs cascade updates and deletes on the database (see below).
Clustering of cDNAs and Formation of GeneLynx Records
To form initial gene-based clusters, we used the set of human cDNA sequences available in GenBank. The sequences were classified into gene-based clusters as follows: first, the initial set is constructed by comparing pairs of sequences using BLAST (Altschul et al. 1990). A strong match was defined by two criteria: (1) BLAST comparison of two sequences should produce at least one high scoring pair (HSP) of length ≥200 nucleotides, and (2) ≥80% of the overlap region should be covered by HSPs of ≥100 nucleotides and ≥96% identity. The overlap region is defined as the length of sequence containing all HSPs and any unaligned flanking sequences extended in both the 5′ and 3′ directions to the closest transcript edges. A weak match is defined as the match that has at least one high scoring pair of length ≥200 nucleotides, but the HSP coverage within the overlap region is less than 80%.
EST-Only Clusters
Clusters containing only ESTs with a minimum of five sequences are taken directly from the most recent release of UniGene (Wheeler et al. 2001). A contiguous sequence is assembled from each cluster using the program phrap (Green 1996). The assembled sequences are compared against existing cDNAs using the described clustering criteria. The unmatched clusters are checked for repeats using Repeat- Masker (Smit and Green 1997), and those containing >30% repeats are discarded. More than 3000 clusters from Unigene build 134 (May 2001) were discarded with this criteria. Although a few of those clusters may represent legitimate transcripts, we judge that the loss of these true genes is much less serious than the level of database contamination likely to result from the inclusion of repeat-rich ESTs.
Linking External Resources
We seek to link each GeneLynx record (i.e., each gene) to as complete a set of resources as possible. To that end, a set of Perl programs with methods for extracting data from each of the linked resources was developed. Where available, distributed flat files were used for analysis. In a few cases, we resorted to direct parsing of Web pages. Parsed data are stored to relational database (RDB) tables, and tables that associate GeneLynx IDs with identifiers of external resources are (automatically) filled in several stages: (1) First, for those resources that are cross-referenced to cDNA or EST accession numbers, the link is made by a simple cross-table query. (2) For those resources that have no direct cDNA association, but are cross-referenced to one or several resources that have been linked to GeneLynx records in the first stage, the links to GeneLynx tables are formed via the first stage associations. (3) Finally, those difficult resources that are not amenable to the aforementioned linking procedures are handled with directed approaches. For instance, direct TBLASTN comparisons were required for the incorporation of a few protein sequences in GenPept (NCBI's collection of translated nucleotide coding sequences).
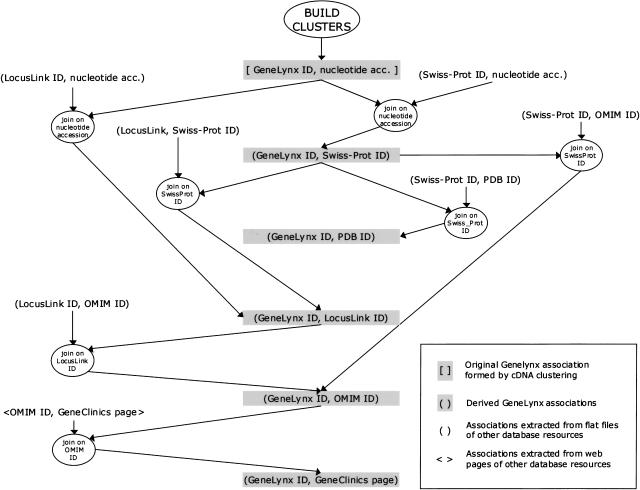
As an example, a small portion of the database-building scheme is shown in Figure 3. The entire schema is much more complex and difficult to represent without clutter in two dimensions. An up-to-date representation of the complete schema is available at http://www.genelynx.org/TECHNICAL/.
Figure 3.
A scheme of a subset of the associations-building procedure used for the construction of GeneLynx database. The central (shaded) items are the associations between GeneLynx and other resources.
Conundrum Resolution and Improvement of Data Quality by Semiautomated Curation
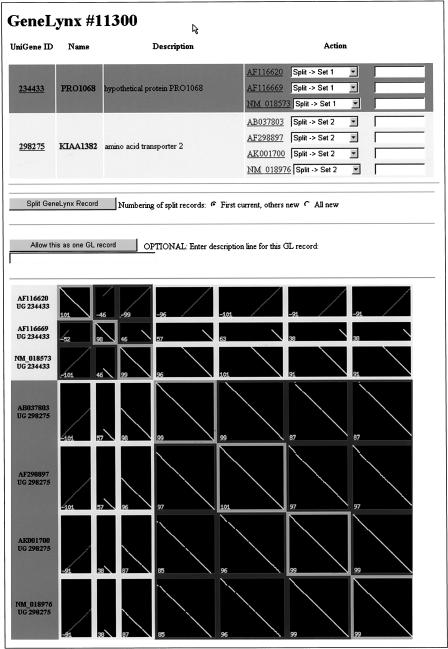
No clustering algorithm is perfect. To maximize the quality of GeneLynx records and data therein, we performed a check of GeneLynx records' relation to those resources where it was reasonable to assume that the correspondence should be of the type one-to-one, for example, UniGene and SWISS-PROT. Using a set of software tools we developed especially for this purpose (Fig. 4), we identified the cases where this assumption was violated and either rearranged GeneLynx records or explicitly allowed the conflict with external databases when we judged that the clustering in the external resource was erroneous or deliberately outside the one-to-one rule. This ‘resolver’ software generates an array of dotplots for all possible pairings of submitted cDNA sequences. Within the array, cDNAs are grouped appropriately to determine whether to accept the GeneLynx cluster or that provided by the external database. It enables easy visual inspection and, if necessary, reassignment of the sequences. We find it to be a unique and valuable curation tool.
Figure 4.
A screenshot of the Resolver curation tool for resolving ambiguous associations. The one-to-many relationship between a GeneLynx cDNA cluster and associated UniGene clusters is resolved by inspecting an array of dot plots. The curator can, if necessary, consult NCBI records for each cDNA and UniGene cluster, which are accessed via provided hyperlinks.
Platform and Availability
All programs were developed in Perl 5.005_03 (Wall et al. 1996) with extensions in C for time-critical parts on Intel Pentium III platforms running Linux 2.2 and Compaq Alpha platforms running Tru64 Unix. The programs intensively use BioPerl (http://bio.perl.org) and CGI.pm modules. Currently, the underlying database system is MySQL 3.23.27 (http://www.mysql.com), but programs access it via the DBI interface, which makes it easily portable to most relational database systems.
For BLAST searches, we currently use National Center for Biotechnology Information (NCBI) BLAST 2.0.14 (available at ftp.ncbi.nlm.nih.gov), using BLASTALL (BLASTN or TBLASTN) with default parameters and a user-defined E-value threshold. For EST contig assembly, we used phrap version 0.990329.
GeneLynx is freely available at http://www.genelynx.org for academic and nonprofit use. Information about the availability of database contents, as well as about possible mirroring, can be obtained at http://www.genelynx.org/info.html. A flat file containing a de-normalized database dump is available on request from the authors.
Acknowledgments
We are particularly indebted to James W. Fickett for his advice and vision on the needs within the biological community for a gene-centric resource . In addition, we acknowledge the contributions of the biologists at the Center for Genomics and Bioinformatics for their feedback on the design of the interface. GeneLynx would not be possible without the tremendous contribution of the wealth of database efforts providing gene-specific information on the Internet. This project was supported by funds from the Karolinska Institute and the Pharmacia Corporation.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL wyeth.wasserman@cgr.ki.se; FAX: 46-8-33-74-12.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.199801.
REFERENCES
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Bairoch A, Apweiler R. The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res. 2000;28:45–48. doi: 10.1093/nar/28.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry JM, Adler C, Ball C, Chervitz SA, Dwight SS, Hester ET, Jia Y, Juvik G, Roe T, Schroeder M, et al. SGD: Saccharomyces Genome Database. Nucleic Acids Res. 1998;26:73–79. doi: 10.1093/nar/26.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo MC, Crawford ME, Hirschman JE, Kranz JE, Olsen P, Robertson LS, Skrzypek MS, Braun BR, Hopkins KL, Kondu P, et al. YPD, PombePD and WormPD: Model organism volumes of the BioKnowledge library, an integrated resource for protein information. Nucleic Acids Res. 2001;29:75–79. doi: 10.1093/nar/29.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etzold T, Ulyanov A, Argos P. SRS: Information retrieval system for molecular biology data banks. Methods Enzymol. 1996;266:114–128. doi: 10.1016/s0076-6879(96)66010-8. [DOI] [PubMed] [Google Scholar]
- FlyBase Consortium. The FlyBase database of the Drosophila Genome Projects and community literature. Nucleic Acids Res. 1999;27:85–88. doi: 10.1093/nar/27.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green, P. 1996. PHRAP documentation at http://bozeman.mbt.washington.edu
- International Human Genome Sequencing Consortium. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Jenssen TK, Laergreid A, Komorowski J, Hovig E. A literature network of human genes for high-throughput analysis of gene expression. Nat Genet. 2001;28:21–28. doi: 10.1038/ng0501-21. [DOI] [PubMed] [Google Scholar]
- Pruitt KD, Katz KS, Sicotte H, Maglott DR. Introducing RefSeq and LocusLink: Curated human genome resources at the NCBI. Trends Genet. 2000;16:44–47. doi: 10.1016/s0168-9525(99)01882-x. [DOI] [PubMed] [Google Scholar]
- Rebhan M, Chalifa-Caspi V, Prilusky J, Lancet D. GeneCards: A novel functional genomics compendium with automated data mining and query reformulation support. Bioinformatics. 1998;14:656–664. doi: 10.1093/bioinformatics/14.8.656. [DOI] [PubMed] [Google Scholar]
- Smit, A.F.A. and Green, P. 1997. RepeatMasker at http://ftp.genome.washington.edu/RM/RepeatMasker.html
- Stein L, Sternberg P, Durbin R, Thierry-Mieg J, Spieth J. WormBase: Network access to the genome and biology of Caenorhabditis elegans. Nucleic Acids Res. 2001;29:82–86. doi: 10.1093/nar/29.1.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- Wall L, Christiansen T, Schwarz R. Programming Perl. 2d ed. Sebastopol, CA: O'Reilly & Associates, Inc.; 1996. [Google Scholar]
- Wheeler DL, Church DM, Lash AE, Leipe DD, Madden TL, Pontius JU, Schuler GD, Schriml LM, Tatusova TA, Wagner L, et al. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2001;29:11–16. doi: 10.1093/nar/29.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]