Abstract
Rice (Oryza sativa L.) will be the first major crop, as well as the first monocot plant species, to be completely sequenced. Integration of DNA sequence-based maps with cytological maps will be essential to fully characterize the rice genome. We have isolated a set of 24 chromosomal arm-specific bacterial artificial chromosomes to facilitate rice chromosome identification. A standardized rice karyotype was constructed using meiotic pachytene chromosomes of O. sativa spp. japonica rice var. Nipponbare. This karyotype is anchored by centromere-specific and chromosomal arm-specific cytological landmarks and is fully integrated with the most saturated rice genetic linkage maps in which Nipponbare was used as one of the mapping parents. An ideogram depicting the distribution of heterochromatin in the rice genome was developed based on the patterns of 4',6-diamidino-2-phenylindole staining of the Nipponbare pachytene chromosomes. The majority of the heterochromatin is distributed in the pericentric regions with some rice chromosomes containing a significantly higher proportion of heterochromatin than other chromosomes. We showed that pachytene chromosome-based fluorescence in situ hybridization analysis is the most effective approach to integrate DNA sequences with euchromatic and heterochromatic features.
Rice is one of the most important crops in the world, providing food for more than half the world's population. Compared to other cereal crops, rice has the smallest genome, 4.3 × 108 bp (Arumuganathan and Earle 1991) and is relatively easy to transform. The genetic synteny and microcolinearity between the rice genome and other grass genomes has been well demonstrated (Ahn and Tanksley 1993; Ahn et al. 1993; Chen et al. 1997, 1998). All these characteristics make rice a model monocot species for molecular biology research (Goff 1999).
The rice genome has been one of the most intensively studied plant genomes during the last decade. The restriction fragment length polymorphism (RFLP)-based genetic linkage maps of rice (Causse et al. 1994; Harushima et al. 1998) are among the most saturated maps in plants. A yeast artificial chromosome (YAC)-based physical map of rice covers more than half of the rice genome (Kurata et al. 1997). Bacterial artificial chromosome (BAC) libraries based on both HindIII and EcoRI restriction enzymes were developed in O. sativa spp. japonica rice var. Nipponbare. Collectively, these libraries contain 92,160 clones and cover 25 haploid equivalents of the rice genome (R. Wing, unpubl.). Fingerprints of 64,307 clones from the Nipponbare BAC libraries have been assembled into 1021 contigs that collectively span 436 megabases (Mb) of DNA, and 110,438 BAC end sequences have been deposited into the GenBank (R. Wing, unpubl.), providing a major resource for map-based cloning and high-throughput genome sequencing. Functional genomics using T-DNA insertional mutagenesis has been shown in rice (Jeon et al. 2000). An international rice genome sequencing project has been launched (Sasaki and Burr 2000). This is complemented by the endeavors of two private companies, Monsanto and Syngenta, to sequence the rice genome (Pennisi 2000; Davenport 2001).
In contrast to the rapid progress of molecular analyses of the rice genome, only limited success has been achieved toward a cytological characterization of the rice genome. Very few heterochromatic or euchromatic features in the rice genome have been defined at the DNA-sequence level (Cheng et al. 2001a). To date, the YAC- and BAC-based physical maps are not integrated with cytological maps. Several laboratories have used fluorescence in situ hybridization (FISH) to map DNA sequences on rice chromosomes (Fukui et al. 1994; Jiang et al. 1995; Ohmido et al. 1998). However, the majority of these reports involved mapping DNA sequences on mitotic metaphase chromosomes. FISH analyses using somatic metaphase chromosomes provide low mapping resolution and limited details regarding the cytological structure of the rice genome. Meiotic pachytene chromosome-based karyotypes have been reported previously, but none of these karyotypes are fully integrated with the currently available genetic linkage maps.
In this report, we isolated a set of 24 BAC clones that hybridize specifically to the 24 chromosomal arms of rice. Using these arm-specific BAC markers and a rice centromere-specific DNA probe, we constructed a standardized rice karyotype that is fully integrated with the most saturated genetic linkage map of rice developed by Harushima et al. (1998). An ideogram depicting the distribution of heterochromatin in the rice genome was developed based on the patterns of 4',6-diamidino-2-phenylindole (DAPI) staining of the pachytene chromosomes. The karyotype and heterochromatin distribution patterns reported in this study provide a foundation toward cytological characterization of the rice genome.
RESULTS
Development of Chromosomal Arm-Specific Cytological Markers in Rice
Cytogenetic studies rely on accurate and consistent chromosome identification, which is always a challenge in plant species with small chromosomes. To develop a reliable system for chromosome identification in rice, we isolated a set of 24 BAC clones that hybridized to each of the 24 rice chromosomal arms. RFLP markers that were single-copy in the rice genome and had been mapped to both sides of the 12 rice centromeres were selected to screen a BAC library (http://www.genome.clemson.edu/orders/Product.html). Positive BACs were used as FISH probes to hybridize to rice chromosomes in both mitotic and meiotic cells. BACs isolated by several of the RFLP markers were not useful as cytological markers because these clones either hybridized to more than one chromosome or contained extensive repetitive DNA sequences and did not generate distinct FISH signals. Only BACs that consistently produced strong and unambiguous signals were selected as chromosomal arm-specific cytological markers. Table 1 summarizes the genetic and cytological locations of the 24 chromosomal arm-specific BACs.
Table 1.
Genetic and Physical Locations of Rice Chromosomal Arm-Specific BAC Clones
| Chromosomal arm | RFLP marker | Genetic position (cM) | Relative genetic position1 | Selected BAC | Physical location2 | n3 |
|---|---|---|---|---|---|---|
| 1S | C146 | 12.3 | 6.77 | a0059H02 | 5.87 ± 0.66 | 10 |
| 1L | G370 | 140.8 | 77.49 | a0026F14 | 78.64 ± 0.83 | 10 |
| 2S | S10889 | 11.5 | 7.43 | a0095P12 | 8.00 ± 0.81 | 10 |
| 2L | C747 | 107.7 | 69.62 | a0064F10 | 76.11 ± 2.22 | 10 |
| 3S | C725 | 13.3 | 7.95 | a0076A03 | 5.95 ± 0.46 | 10 |
| 3L | G1015 | 142.8 | 85.41 | a0007K12 | 86.39 ± 1.40 | 10 |
| 4S | C820 | 17.4 | 13.44 | a0065E01 | 15.85 ± 0.87 | 10 |
| 4L | G177 | 111.2–111.7 | 85.02–86.10 | a0080P07 | 85.24 ± 1.94 | 10 |
| 5S | R3166 | 18.8 | 15.73 | a0096I15 | 6.90 ± 0.90 | 10 |
| 5L | C1230 | 119.5 | 100 | a0036L10 | 97.89 ± 0.25 | 10 |
| 6S | G200 | 33.5 | 26.80 | a0089N04 | 23.79 ± 1.70 | 10 |
| 6L | R1167 | 123.1 | 98.48 | a0088F21 | 100 ± 0.00 | 10 |
| 7S | C1057 | 11.0 | 9.40 | a0078O19 | 7.87 ± 1.27 | 10 |
| 7L | R1245 | 83.0 | 70.94 | a0085M21 | 78.73 ± 1.25 | 10 |
| 8S | C166 | 26.2 | 22.04 | a0005O22 | 11.33 ± 1.70 | 10 |
| 8L | R1963 | 118.1 | 99.34 | a0085C09 | 82.61 ± 2.05 | 10 |
| 9S | G36 | 0.8 | 0.83 | a0052I16 | 9.10 ± 0.78 | 10 |
| 9L | G293 | 94.7 | 98.54 | a0061K19 | 97.89 ± 2.50 | 10 |
| 10S | S10620 | 1.2 | 1.43 | a0045D16 | 1.54 ± 0.12 | 8 |
| 10L | C405 | 83.7 | 100 | a0056G17 | 100 ± 0.00 | 8 |
| 11S | R2918 | 10.2 | 8.60 | a0040B10 | 9.59 ± 0.73 | 10 |
| 11L | C1172 | 79.9 | 67.37 | a0071H11 | 67.57 ± 0.54 | 10 |
| 12S | R1957 | 10.1 | 9.17 | a0095C23 | 5.98 ± 1.25 | 10 |
| 12L | C901 | 110.1 | 100 | a0011E08 | 100 ± 0.00 | 10 |
Relative genetic position is calculated as (cmX ÷ cmT) × 100, where cmX = the cM value of the RFLP marker on the linkage map, cmT = the total cM value of the same linkage map (Harushima et al. 1998). For example, C146 was mapped to 12.3 cM on linkage map 1 that totals 181.7 cM. The relative genetic position of C146 is (12.3/181.7) × 100 = 6.77.
Physical location is calculated as (S ÷ T) × 100, where S = the distance (in μm) from the FISH hybridization site to the end of the short arm of the chromosome, and T = the total length of the chromosome in μm.
The number of measurements.
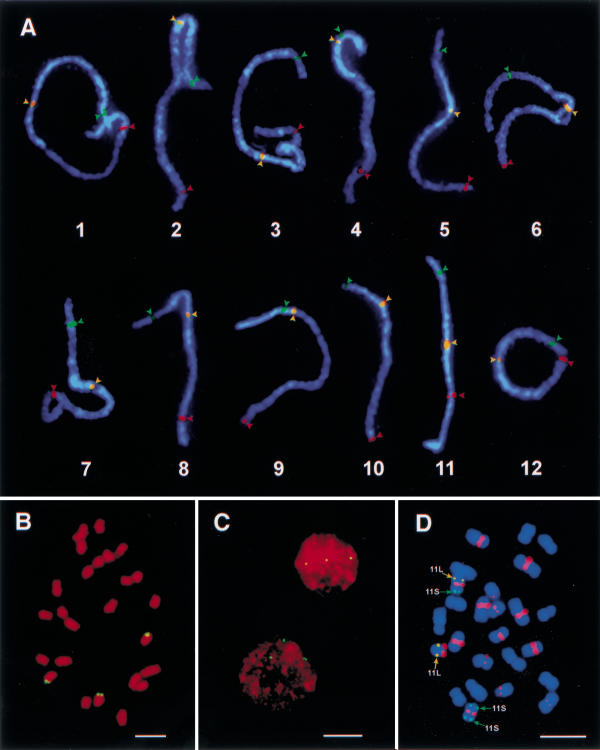
Two strategies were used to verify that the selected BACs were truly located in the same positions as the anchoring RFLP markers. First, the two BAC clones selected using RFLP markers mapped to both sides of a particular linkage group were used in the same FISH experiment together with the rice centromere-specific DNA probe pRCS2. We found that each of the 12 pairs of BACs mapped to both sides of a specific chromosome (Fig. 1A). Second, the physical locations of the selected BACs were determined on pachytene chromosomes by measuring the distances from the FISH signals to the telomeres of the short arms. The pachytene chromosome positions of all 24 selected BACs correlated with the relative genetic locations of the corresponding RFLP markers (Table 1). These data strongly supported that the 24 BACs are chromosomal arm-specific and are physically located at the same positions as their anchoring RFLP markers.
Figure 1.
Chromosomal arm-specific bacterial artificial chromosome (BAC) markers and rice chromosome identification. (A) The morphology and DAPI-staining patterns of the 12 rice pachytene bivalents were identified by hybridization to the 12 short-arm-specific BAC clones (green arrowheads; clone names are listed in Table 1), centromere-specific probe pRCS2 (yellow arrowheads), and the 12 long-arm-specific BAC clones (red arrowheads). The three probes on each pachytene bivalent were sequentially probed on the same pachytene cells. Grey-scale images of the FISH signals were pseudocolored as green, yellow, and red, respectively, and merged with the chromosomal image. (B) A somatic metaphase cell of rice primary trisomic 11 (2n = 24 + 11S·11L) probed with BAC a0071H11 that is specific to the long arm of rice chromosome 11. FISH signals are observed on three chromosomes. Bar, 5 μm. (C) Interphase nuclei from root tip cells of rice primary trisomic 11 hybridized to BAC a0071H11. Three hybridization spots are observed in each nucleus, indicating three copies of chromosome 11 in this plant. Bar, 10 μm. (D) A somatic metaphase cell of a variant derived from a primary trisomic 11 plant probed with the 11S-specific BAC a0040B10 (green signals), 11L-specific BAC a0071H11 (yellow signals), and centromere-specific clone pRCS2 (red signals). This plant contains one normal chromosome 11 (11S·11L), one telocentric chromosome derived from the long arm of 11 (11L·), and one isochromosome derived from the short arm of 11 (11S·11S). Image process is the same as that in Fig. 1A. Bar, 5 μm.
These chromosomal arm-specific BACs serve as reliable cytological markers to unambiguously identify rice chromosomes in both mitotic and meiotic cells. Figure 1B shows a somatic metaphase cell of rice primary trisomic 11 (2n = 24+ 11S·11L) probed with 11S-specific BAC a0071H11. Three chromosomes hybridize to this BAC, confirming three copies of chromosome 11 in this plant. The trisomic 11 constitution of this plant can also be determined by interphase FISH analysis (Fig. 1C). We recovered a new aneuploid plant from the progenies of trisomic 11. This new plant has 25 chromosomes and has a different morphology from euploid and trisomic 11 plants. We were not able to characterize the chromosomal constitution of this plant using the traditional pachytene chromosome analysis. When metaphase cells of this plant were sequentially probed with the 11S- and 11L-specific BAC clones and the rice centromere-specific DNA probe pRCS2, we detected one isochromosome derived from 11S and one 11L telocentric chromosome (Fig. 1D). Thus, this aneuploid plant was readily identified as 2n = 23+ 11S·11S + 11L· using chromosomal arm-specific BAC markers.
A Standardized Rice Karyotype Anchored by Centromere- and Chromosomal Arm-Specific Cytological Markers
Rice karyotypes based on meiotic pachytene chromosomes were previously constructed by several laboratories (Shastry et al. 1960; Kurata et al. 1981; Khush et al. 1984, 1996; Khush and Kinoshita 1991; Chung and Wu 1987; Cheng and Gu 1994). The differential staining of the centromeric regions in rice pachytene chromosomes are not as distinct as those in the pachytene chromosomes of maize and tomato. Most rice pachytene chromosomes show multiple lightly stained domains in the pericentric heterochromatin regions (Fig. 2A–D). FISH results using the centromere-specific probe pRCS2 showed that the centromeres of several rice pachytene chromosomes are not associated with any lightly staining chromatin domains. Thus, the centromeres of rice pachytene chromosomes are difficult to locate using traditional acetocarmine staining methods. Misidentification of centromeres can result in major errors of karyotyping and discrepancies in the numbering systems in karyotypes constructed by different researchers. Our goal was to develop a standardized rice karyotype that is fully integrated with the molecular linkage maps using the rice centromere-specific DNA marker pRCS2 (Dong et al. 1998) and the 24 chromosomal arm-specific BAC markers.
Figure 2.
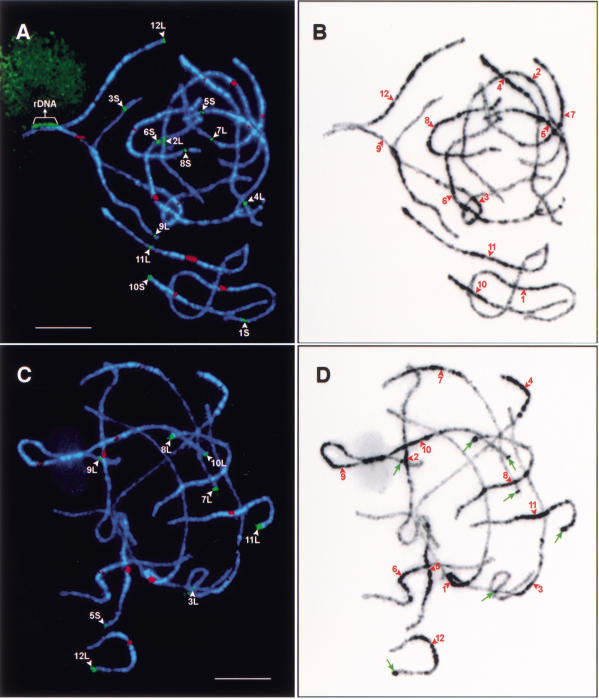
Karyotyping of rice pachytene chromosomes and mapping of euchromatin and heterochromatin in the rice genome. (A) Chromosomes in a pachytene cell of japonica rice Nipponbare were hybridized with 12 bacterial artificial chromosome (BAC) clones (green signals) that hybridize specifically to each of the 12 rice chromosomes and the centromeric probe pRCS2 (red signals) that hybridizes to all 12 rice centromeres. The 18S·26S rRNA genes are detected on chromosome 9. Bar, 10 μm. (B) DAPI-stained chromosomes in Fig. 2A were converted to a black-and-white image to enhance the visualization of distribution of euchromatin and heterochromatin along the pachytene chromosomes. Note that the centromeres—indicated by red arrowheads—are not always located within a lightly stained chromatin domain. (C) Chromosomes in a pachytene cell of indica rice Zhongxian 3037 were hybridized to the 355-bp tandem repeat Os48 (green signals) and the centromere-specific probe pRCS2 (red signals). Eight Os48 loci (denoted by white arrowheads) were detected and all eight loci, with the exception of the locus on the long arm of chromosome 9, were located on distal regions of the chromosomes. Bar, 5 μm. (D) DAPI-stained chromosomes in Fig. 2C were converted to a black-and-white image to enhance the visualization of distribution of euchromatin and heterochromatin along the pachytene chromosomes. The Os48 loci on chromosomes 3, 7, 8, 9, 10, 11, and 12 are correlated with conspicuous heterochromatin knobs (pointed to by green arrows). The centromeres are indicated by red arrowheads. Note that the heterochromatin distribution patterns in Nipponbare and Zhongxian 3037 are highly similar.
The japonica rice variety Nipponbare was selected for genome sequencing by the International Rice Genome Sequencing Project (Sasaki and Burr 2000). Thus, we used this variety for karyotype construction. To unambiguously identify every chromosome in meiotic pachytene cells we used one BAC marker for each of the 12 rice chromosomes. Multiple rounds of multicolor FISH analysis were sequentially conducted on the best pachytene preparations with each round including only two or three probes. Together with the pRCS2 probe and an rDNA probe, pTa71, the 12 chromosome-specific BAC markers were mapped to specific chromosomes after five rounds of FISH experiments. Figure 2A shows that the 12 identified bivalents in a pachytene cell with each bivalent showing FISH signals derived from pRCS2 and a BAC marker. The signal derived from pTa71 is located on chromosome 9.
The length of each pachytene bivalent was measured in 50 complete pachytene cells using IPLab Spectrum software on digital images (Table 2). The longest chromosome is chromosome 1, which has an absolute length of 61.12 μm and a relative length of 13.24%. The shortest chromosome is chromosome 10, which has an absolute length of 24.74 μm and a relative length of 5.36%. The descending order of chromosome length is 1, 3, 2, 6, 4, 5, 7, 8, 11, 9 (not including the rDNA section of the chromosome), 12, and 10. Chromosomes 3 and 6 have an arm ratio less than 1.0 (Table 2), suggesting that the current arm orientations of these two chromosomes are incorrect and the current orientation of linkage groups 3 and 6 (Harushima et al. 1998) should be reversed.
Table 2.
Length and Arm Ratio of the Pachytene Chromosomes in Nipponbare Rice
| Chr.no. | Short arm (μm) | n | Long arm (μm) | n | rDNA (μm) | Total length (μm) | Relative length (%) | DNA content (Mb)2 | Arm ratio |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 23.39 ± 3.64 | 50 | 37.73 ± 5.44 | 50 | — | 61.12 ± 8.23 | 13.24 ± 0.91 | 56.9 ± 3.9 | 1.63 ± 0.22 |
| 2 | 19.70 ± 3.30 | 50 | 32.75 ± 4.75 | 50 | — | 52.45 ± 7.35 | 11.36 ± 0.77 | 48.8 ± 3.3 | 1.68 ± 0.26 |
| 3 | 29.87 ± 4.28 | 50 | 26.54 ± 4.10 | 50 | — | 56.41 ± 7.56 | 12.21 ± 0.91 | 52.5 ± 3.9 | 0.89 ± 0.083 |
| 4 | 6.23 ± 0.83 | 50 | 31.91 ± 5.18 | 50 | — | 38.14 ± 5.80 | 8.26 ± 0.70 | 35.5 ± 3.0 | 5.13 ± 0.60 |
| 5 | 12.46 ± 1.76 | 50 | 24.62 ± 3.58 | 50 | — | 37.08 ± 4.91 | 8.05 ± 0.68 | 34.6 ± 2.9 | 1.99 ± 0.23 |
| 6 | 19.93 ± 3.13 | 50 | 18.44 ± 2.91 | 50 | — | 38.37 ± 5.96 | 8.30 ± 0.64 | 35.7 ± 2.8 | 0.93 ± 0.054 |
| 7 | 12.53 ± 2.06 | 50 | 22.16 ± 3.77 | 50 | — | 34.69 ± 5.33 | 7.51 ± 0.69 | 32.3 ± 3.0 | 1.78 ± 0.25 |
| 8 | 13.58 ± 1.99 | 50 | 18.60 ± 3.11 | 50 | — | 32.18 ± 4.54 | 6.98 ± 0.59 | 30.0 ± 2.5 | 1.38 ± 0.20 |
| 9 | 3.88 ± 0.83 | 50 | 24.72 ± 3.56 | 50 | 3.93 ± 0.78 | 28.60 ± 4.161 | 6.21 ± 0.681 | 26.7 ± 2.91 | 6.54 ± 1.061 |
| 10 | 6.14 ± 0.94 | 50 | 18.60 ± 2.70 | 50 | — | 24.74 ± 3.32 | 5.36 ± 0.38 | 23.0 ± 1.6 | 3.07 ± 0.50 |
| 11 | 13.37 ± 2.08 | 50 | 17.57 ± 2.81 | 50 | — | 30.94 ± 4.45 | 6.70 ± 0.54 | 28.8 ± 2.3 | 1.32 ± 0.18 |
| 12 | 11.65 ± 2.06 | 50 | 15.28 ± 2.36 | 50 | — | 26.93 ± 4.12 | 5.83 ± 0.55 | 25.1 ± 2.4 | 1.32 ± 0.15 |
The rDNA section of chromosome 9 was not included in the measurements.
Estimation of the DNA content of each chromosome is based on the assumption that (1) the haploid genome size of rice is 430 Mb; and (2) the euchromatic and heterochromatic regions contain equal amount of DNA (Mb/μm). Thus, the DNA content will be underestimated for relatively heterochromatic chromosomes and overestimated for relatively euchromatic chromosomes.
The arm ratio of chromosome 3 is 1.13 ± 0.12 if the current short and long arms are reversed.
The arm ratio of chromosome 6 is 1.08 ± 0.07 if the current short and long arms are reversed.
Distribution of Euchromatic and Heterochromatic Regions in Rice Pachytene Chromosomes
DAPI binds preferentially to AT-rich regions (Kapuscinski 1995) and is the most common dye to stain heterochromatin. The DAPI-staining patterns of the 12 rice pachytene bivalents are consistent in different cells and are similar to those obtained by the traditional acetocarmine staining method. In general, most of the heterochromatin, which is brightly stained with DAPI, is distributed in the pericentric regions. Chromosome 4 has the most distinct pattern, in which about one-third of the chromosome, including the entire short arm and part of the long arm, is highly heterochromatinized (Fig. 2A,B). Chromosome 10 has a heterochromatin distribution pattern similar to chromosome 4. However, the intensity of DAPI staining in the heterochromatic regions on chromosome 10 is not as bright as that of chromosome 4 (Fig. 2A,B). The heterochromatin distributions in the pachytene chromosomes of the indica variety Zhongxian 3037 are similar to those in Nipponbare (Fig. 2B,D).
Brightly DAPI-stained and knob-like heterochromatin structures were observed at the distal ends of several chromosomes in Zhongxian 3037 (Fig. 2D) but not on the same chromosomal regions in Nipponbare (Fig. 2B). We recently found that these distal knobs are associated with a 355-bp tandemly repeated DNA element, Os48 (Cheng et al. 2001a). The Os48 loci were assigned to 5L and 6S in Nipponbare, and to 3L, 5S, 7L, 8L, 9L, 10L, 11L, and 12L in Zhongxian 3037 (Fig. 2C), respectively. Only the Os48 locus on 9L is well-separated from the telomere. Figure 2, C and D, shows the heterochromatin distribution in indica rice variety Zhongxian 3037 and the relationship between the Os48 sequence and the distal heterochromatic knobs.
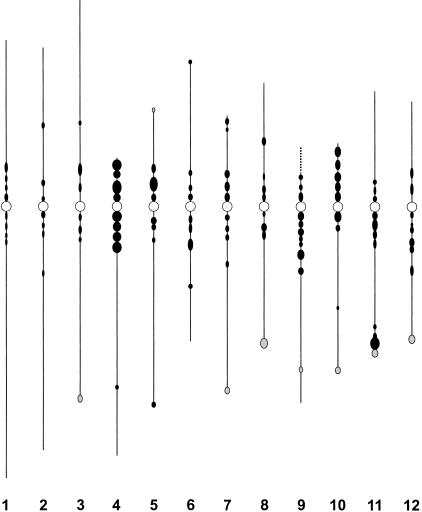
To depict the distribution of heterochromatin in rice genome, an ideogram based on the DAPI-staining patterns of the Nipponbare pachytene chromosomes was developed. The ideogram was generalized based on observations in the same 50 pachytene cells used for karyotype construction. Each distinct DAPI-bright region, which is consistently observed in a majority of the pachytene cells, is represented by a solid circle (Fig. 3). The length (from south to north) of the circle represents the size of the DAPI-bright region on the pachytene bivalent, and the width (from east to west) of the circle represents the relative intensity of DAPI staining. The numbers of distinct DAPI-staining regions on each pachytene bivalent is dependent on early or late stages of pachytenema, as some closely adjacent regions tend to fuse during late pachytenema. The ideogram is constructed based on typical DAPI-staining patterns of late pachytene chromosomes. The short arm and the pericentric region on the long arm of chromosome 4 is the most heterochromatinized region in the rice genome based on its brightest DAPI-staining pattern. Other notable heterochromatic regions were observed in the pericentric region of chromosome 5, the distal region on the long arm of chromosome 11, and the short arm and pericentric region on the long arm of chromosome 10 (Fig. 3). The three longest chromosomes (1, 2, and 3) are among the most euchromatic chromosomes and show relatively low DAPI-staining intensities along the entire length of the chromosomes (Fig. 3).
Figure 3.
Ideogram of the distribution of DAPI-bright regions on rice pachytene chromosomes. The ideogram was generalized based on observations in 50 pachytene cells. Each circle represents a distinct DAPI-bright region that is consistently observed in the majority of the pachytene chromosomes. Shaded circles represent the DAPI-bright regions observed in indica rice Zhongxian 3037 but not in japonica rice Nipponbare. Open circles represent the location of the centromeres. The dotted line on the top of chromosome 9 represents the rDNA section. The relative length of each chromosome and the arm ratio were drawn based on Table 2 data.
DISCUSSION
Rice Chromosome Identification
Cytogenetic studies of higher eukaryotic genomes rely on reliable and easy-to-use techniques for chromosome identification. The somatic metaphase chromosomes of rice are small (1–3 μm) and similar in size. Thus, routine and unambiguous identification of individual rice chromosomes based on their morphology is almost impossible. Rice pachytene chromosomes provide more cytological features for identification. However, pachytene chromosome preparation is an elaborate process. Several pairs of rice pachytene chromosomes, including 1 and 2, 5 and 7, 6 and 8, 9 and 10, and 11 and 12, share similar size, arm ratio, and heterochromatin distribution patterns and thus are difficult to distinguish. A new methodology for chromosome identification using chromosome-specific BAC clones was recently demonstrated in potato by Dong et al. (2000). FISH signals derived from chromosome-specific BACs can be used as reliable cytological markers for chromosome identification. This methodology is particularly useful for plant species with a large number of small chromosomes. In this study, we isolated a set of chromosomal arm-specific BACs in rice. We showed that these BACs are excellent cytological markers for chromosome identification in both mitotic and meiotic cells. Such cytological markers are powerful tools to characterize rice aneuploids with extra and/or modified chromosomes, which can hardly be characterized using conventional cytogenetic techniques (Fig. 1B–D).
Rice Karyotype
Numerous rice karyotypes based on mitotic prometaphase chromosomes or meiotic pachytene chromosomes have been reported since the identification of the rice chromosome number as 2n = 24 (Kuwada 1910). However, the accuracy of the measurements of chromosome sizes and arm ratios in these previous efforts has been disputed because of the potential misidentification of the centromeric locations in pachytene chromosomes and the large variation of the measurements. In addition, different researchers used their own nomenclature systems in karyotyping. Thus, significant discrepancies exist among the previously published karyotypes. More importantly, none of these karyotypes are fully integrated with the two most saturated rice genetic linkage maps constructed by Causse et al. (1994) and by Harushima et al. (1998). To avoid the limitations associated with the previously published karyotypes, we developed 24 chromosomal arm-specific BAC clones using genetically mapped RFLP markers. These BACs not only allowed the correct identification of individual chromosomal arms, but also enabled us to completely integrate our karyotype with the genetic linkage map of rice constructed by Harushima et al. (1998). We also avoided misidentification of centromeric locations by using a rice centromere-specific probe during karyotype construction. Computer-based measurements were conducted on every chromosome in 50 pachytene cells. The number of measurements from each chromosome was significantly more than any previous karyotyping effort and resulted in the smallest variation reported to date.
The relationships between the 12 linkage groups and the 12 rice chromosomes were established previously by DNA dosage analysis of genetically mapped RFLP markers using rice trisomics (McCouch et al. 1988; Khush et al. 1996; Singh et al. 1996; Harushima et al. 1998). Our results show that the length of the pachytene bivalents in the current numbering system is not in the same descending order as for the linkage groups. The current descending order of the pachytene bivalent length is 1, 3, 2, 6, 4, 5, 7, 8, 11, 9 (not including the rDNA section), 12, and 10. In addition, according to the current nomenclature system, the short arms of chromosomes 3 and 6 are longer than their long arms. These unusual characteristics of the current karyotype are due to inaccurate cytological characterization of the rice trisomics. Cytological identification of rice trisomics was done mainly according to the size and the morphology of the trivalent involving the extra chromosome at the pachytene stage. Compared to the corresponding bivalents, the morphology of the trivalents is often altered significantly because of the synapsis of multiple chromosomes. Thus, it is difficult to accurately measure the length of such multivalents. In addition, conventional acetocarmine staining does not clearly visualize the centromeres of rice pachytene chromosomes. Thus, misidentification of the centromeric location may also contribute to the inaccurate characterization of the rice trisomics.
Integration of DNA Sequence-Based Maps with Cytological Maps in Rice
A complete sequence of the rice genome will soon be publicly available (Sasaki and Burr 2000, http://rgp.dna.affrc.go.jp/cgi-bin/statusdb/seqcollab.pl). Identification of euchromatin and heterochromatin at both the cytological and DNA sequence level will be a critical component for full characterization of the rice genome. The power and importance of integration of molecular and cytological data was distinctly illustrated by the recent characterization of a heterochromatic structure on the short arm of Arabidopsis thaliana chromosome 4 (Fransz et al. 2000; McCombie et al. 2000). Based on the DAPI-staining patterns of pachytene chromosomes, the proportion of heterochromatin in the rice genome is significantly larger than in the A. thaliana genome. In A. thaliana, the majority of the cytologically defined heterochromatin is located in the centromeric regions (Fransz et al. 1998). However, DAPI-bright regions cover significant parts of the pericentric regions in the majority of the rice chromosomes (Figs. 2, 3). Telomeric and interstitial heterochromatin features are also present in most pachytene chromosomes (Figs. 2, 3). FISH-based analyses will be critical to integrate DNA sequence information with different types of chromatin structures. We recently showed that several heterochromatic knobs in rice pachytene chromosomes are associated with a 355-bp tandem repeat Os48. The Os48 sequences are heavily methylated and physically located very close to the telomeres of rice chromosomes (Cheng et al. 2001a).
Pachytenema is the best meiotic stage to distinguish euchromatin and heterochromatin. Thus, pachytene chromosome-based FISH mapping is the most effective approach to integrate DNA sequence information with cytological features (Fransz et al. 2000; Cheng et al. 2001a). Pachytene chromosomes are 10 times longer than somatic metaphase chromosomes. Therefore, pachytene FISH also provides for a high resolution for mapping DNA probes to specific chromosomal regions. BAC clones separated by ∼100 kb can be well-resolved on rice pachytene chromosomes (Cheng et al. 2001b). Pachytene FISH of BACs anchored by genetically mapped RFLP markers has proven to be a powerful approach in revealing the genetic and physical relationships in specific chromosomal regions (Cheng et al. 2001b). The genetic linkage map of rice reported by Harushima et al. (1998) contains more markers per cM than any other plant genetic map. However, it still contains 39 gaps that span >5 cM. It is not known if these linkage gaps represent recombination hot spots or large physical gaps caused by the lack of markers in these regions. We previously showed that pachytene FISH of BACs flanking these linkage gaps is an effective strategy to reveal the physical nature of such gaps (Cheng et al. 2001b). Comprehensive physical characterization of the large linkage gaps will be of critical importance in the acceleration of the International Rice Genome Sequencing Project.
METHODS
Materials
O. sativa spp. japonica rice var. Nipponbare and O. sativa spp. indica rice var. Zhongxian 3037 were used for cytological studies. The rice aneuploids used in this study were also developed from Zhongxian 3037 (Cheng et al. 2001c). All BAC clones used for FISH mapping were identified by screening a Nipponbare BAC library (http://www.genome.clemson.edu/orders/Product.html) using RFLP makers previously mapped on different chromosomes (Harushima et al. 1998). Several other DNA clones were used in our FISH analyses, including pRCS2, which contains a repetitive sequence specific to rice centromeres (Dong et al. 1998); pOs48, which contains a 355-bp subtelomeric tandem repeat of rice (Wu and Wu 1987); and pTa71, which contains the coding sequences for the 18S·26S ribosomal RNA genes of wheat (Gerlach and Bedbrook 1979).
Chromosome Preparation
Rice root tips were harvested from germinated seeds or plants growing in the field, pretreated in 0.002M 8-hydroxyquinoline at 20°C for 2 h to accumulate prometaphase and metaphase cells, and fixed in methanol:acetic acid (3:1). Root tips were macerated in 2% cellulose and 1% pectolyase at 37°C for 1.5 h and squashes were made in the same fixative. Young panicles of rice were harvested and fixed in 3:1 (100% ethanol:glacial acetic acid) Carnoy's solution. Microsporocytes at the pachytene stage were squashed in acetocarmine solution according to Wu (1967). Slides were stored at −80°C until use. After removing the coverslips, slides were dehydrated through an ethanol series (70%, 90%, and 100%) prior to use in FISH.
FISH
The FISH procedure applied to both mitotic and meiotic chromosomes was essentially the same as previously published protocols (Jiang et al. 1995). Rice Cot-1 DNA was included in the hybridization mixtures to reduce hybridization background for some BAC probes. High quality pachytene chromosome preparations were used for repeated probing. After the first round of probing and image capture, the slides were soaked in a 1× PBS (phosphate-buffered saline) solution to remove the coverslips. The slides were then dehydrated in an ethanol series (70%, 90%, and 100%, 5 min each), denatured again in 70% formamide at 80°C for 2 min, dehydrated in a second ethanol series, and incubated with a different FISH probe. This procedure was repeated up to five rounds. Biotin-labeled and digoxigenin-labeled probes were detected using a fluorescein isothiocyanate (FITC)-conjugated antibiotin antibody (Vector Laboratories) and a rhodamine-conjugated antidigoxigenin antibody (Roche Diagnostics), respectively. In repeated probing experiments, no more than three probes were used for each round to unambiguously identify signals from the individual probes. When three probes were used in a FISH experiment, the first and second probes were labeled by biotin and digoxigenin; the third probe was labeled by 50% biotin and 50% digoxigenin, which resulted in a yellow detection color. Chromosomes were counterstained with DAPI in an antifade solution (Vector Laboratories).
Cytological Measurements and Analysis
All images were captured digitally using a SenSys CCD (charge coupled device) camera (Roper Scientific) attached to an Olympus BX60 epifluorescence microscope. The CCD camera was controlled using IPLab Spectrum v3.1 software (Signal Analytics) on a Macintosh computer. Grey-scale images were captured for each color channel and then merged. Measurements were made on the digital images of the FISH signals and chromosomes within IPLab Spectrum software and final image adjustments were done with Adobe Photoshop v5.1. At least eight data points were collected to map BACs on pachytene chromosomes. For karyotype construction, chromosomes in 50 complete pachytene cells were measured and standard deviations were calculated.
Acknowledgments
This research was supported by a grant from the Consortium for Plant Biotechnology Research, Inc.; Novartis Seeds, Inc.; and Dow AgroSciences to J.J. Funding support to C.R.B. includes USDA-CSREES grant 99-35317-8275, NSF grant DBI998282, and DOE grant DE-FG02-99ER20357. Funding support to R.A.W. includes USDA grant 99-35317-8505 and NSF grant DBI-9982594.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL jjiang1@facstaff.wisc.edu; FAX 608-262-4743.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.194601.
REFERENCES
- Ahn S, Tanksley SD. Comparative linkage maps of rice and maize genomes. Proc Natl Acad Sci. 1993;90:7980–7984. doi: 10.1073/pnas.90.17.7980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn S, Anderson JA, Sorrells ME, Tanksley SD. Homoeologous relationships of rice, wheat and maize chromosomes. Mol Gen Genet. 1993;241:483–490. doi: 10.1007/BF00279889. [DOI] [PubMed] [Google Scholar]
- Arumuganathan K, Earle ED. Nuclear DNA content of some important plant species. Plant Mol Biol Rep. 1991;9:208–218. [Google Scholar]
- Causse MA, Fulton TM, Cho YG, Ahn SN, Chunwongse J, Wu KS, Xiao JH, Yu ZH, Ronald PC, Harrington SE, et al. Saturated molecular map of the rice genome based on an interspecific backcross population. Genetics. 1994;138:1251–1274. doi: 10.1093/genetics/138.4.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, SanMiguel P, de Oliveira AC, Woo S-S, Zhang H, Wing RA, Bennetzen JL. Microcolinearity in the sh2-homologous regions of maize, rice, and sorghum genomes. Proc Natl Acad Sci. 1997;94:3431–3435. doi: 10.1073/pnas.94.7.3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, SanMiguel P, Bennetzen JL. Sequence organization and conservation in sh2/a1-homologous regions of sorghum and rice. Genetics. 1998;148:435–443. doi: 10.1093/genetics/148.1.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng ZK, Gu MH. Karyotype analysis for pachytene chromosome of indica, japonica rice and their hybrid. Chinese J Genet. 1994;21:182–187. [Google Scholar]
- Cheng ZK, Stupar RM, Gu MH, Jiang J. A tandemly repeated DNA sequence is associated with both knob-like heterochromatin and a highly decondensed structure in the meiotic pachytene chromosomes of rice. Chromosoma. 2001a;110:24–31. doi: 10.1007/s004120000126. [DOI] [PubMed] [Google Scholar]
- Cheng ZK, Presting GG, Buell CR, Wing RA, Jiang J. High resolution pachytene chromosome mapping of bacterial artificial chromosomes anchored by genetic markers reveals the centromere location and the distribution of genetic recombination along chromosome 10 of rice. Genetics. 2001b;157:1749–1757. doi: 10.1093/genetics/157.4.1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng ZK, Yan H, Yu H, Tang S, Jiang J, Gu MH, Zhu L. Development and applications of a complete set of rice telotrisomics. Genetics. 2001c;157:361–368. doi: 10.1093/genetics/157.1.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung MC, Wu HK. Karyotype analysis of ‘IR36’ and two trisomic lines of rice. Bot Bull Acad Sin. 1987;28:289–304. [Google Scholar]
- Davenport RJ. Syngenta finishes, consortium goes on. Science. 2001;291:807. doi: 10.1126/science.291.5505.807a. [DOI] [PubMed] [Google Scholar]
- Dong F, Miller JT, Jackson SA, Wang GL, Ronald PC, Jiang J. Rice (Oryza sativa) centromeric regions consist of complex DNA. Proc Natl Acad Sci. 1998;95:8135–8140. doi: 10.1073/pnas.95.14.8135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong F, Song J, Naess SK, Helgeson JP, Gebhardt C, Jiang J. Development and applications of a set of chromosome-specific cytogenetic DNA markers in potato. Theor Appl Genet. 2000;101:1001–1007. [Google Scholar]
- Fransz PF, Armstrong S, Alonso-Blanco C, Fischer TC, Torres-Ruiz RA, Jones J. Cytogenetics for the model system Arabidopsis thaliana. Plant J. 1998;13:867–876. doi: 10.1046/j.1365-313x.1998.00086.x. [DOI] [PubMed] [Google Scholar]
- Fransz PF, Armstrong S, de Jong JH, Parnell LD, van Drunen C, Dean C, Zabel P, Bisseling T, Jones GH. Integrated cytogenetic map of chromosome arm 4S of A. thaliana: Structural organization of heterochromatic knob and centromere region. Cell. 2000;100:367–376. doi: 10.1016/s0092-8674(00)80672-8. [DOI] [PubMed] [Google Scholar]
- Fukui K, Ohmido N, Khush GS. Variability in rDNA loci in the genus Oryza detected through fluorescence in situ hybridization. Theor Appl Genet. 1994;87:893–899. doi: 10.1007/BF00225782. [DOI] [PubMed] [Google Scholar]
- Gerlach WL, Bedbrook JR. Cloning and characterization of ribosomal RNA genes from wheat and barley. Nucleic Acids Res. 1979;7:1869–1885. doi: 10.1093/nar/7.7.1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff SA. Rice as a model for cereal genomes. Curr Opin Plant Biol. 1999;2:86–89. doi: 10.1016/S1369-5266(99)80018-1. [DOI] [PubMed] [Google Scholar]
- Harushima Y, Yano M, Shomura A, Sato M, Shimano T, Kuboki Y, Yamamoto T, Lin SY, Antonio BA, Parco A, et al. A high-density rice genetic linkage map with 2275 markers using a single F2 population. Genetics. 1998;148:479–494. doi: 10.1093/genetics/148.1.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon J-S, Lee S, Jung K-H, Jun S-H, Jeong D-H, Lee J, Kim C, Jang S, Lee S, Yang J, et al. T-DNA insertional mutagenesis for functional genomics in rice. Plant J. 2000;22:561–570. doi: 10.1046/j.1365-313x.2000.00767.x. [DOI] [PubMed] [Google Scholar]
- Jiang J, Gill BS, Wang GL, Ronald PC, Ward DC. Metaphase and interphase fluorescence in situ hybridization mapping of the rice genome with bacterial artificial chromosomes. Proc Natl Acad Sci. 1995;92:4487–4491. doi: 10.1073/pnas.92.10.4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapuscinski J. DAPI: A DNA-specific fluorescent probe. Biotechnic Histochem. 1995;70:220–233. doi: 10.3109/10520299509108199. [DOI] [PubMed] [Google Scholar]
- Khush GS, Kinoshita T. Rice karyotype, marker genes, and linkage groups. In: Khush GS, Toenniessen GH, editors. Rice Biotechnology. Wallingford, UK: CAB International; 1991. pp. 83–108. [Google Scholar]
- Khush GS, Singh RJ, Sur SC, Librojo AL. Primary trisomics of rice: origin, morphology, cytology, and use in linkage mapping. Genetics. 1984;107:141–163. doi: 10.1093/genetics/107.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khush GS, Singh K, Ishii T, Parco A, Huang N, Brar DS, Multani DS. Centromere mapping and orientation of the cytological, classical, and molecular linkage maps of rice. In: Khush GS, editor. Rice Genetics III. Proceedings of the Third International Rice Genetics Symposium. Manila, Philippines: International Rice Research Institute; 1996. pp. 57–75. [Google Scholar]
- Kurata N, Omura T, Iwata N. Studies on centromere, chromomere and nucleolus in pachytene nuclei of rice, Oryza sativa, microsporocytes. Cytologia. 1981;46:791–800. [Google Scholar]
- Kurata N, Umehara Y, Tanoue H, Sasaki T. Physical mapping of the rice genome with YAC clones. Plant Mol Biol. 1997;35:101–113. [PubMed] [Google Scholar]
- Kuwada Y. A cytological study of Oryza sativa L. Shokubutsugaku Zasshi. 1910;24:267–281. [Google Scholar]
- McCombie WR, de la Bastide M, Habermann K, Parnell L, Dedhia N, Gnoj L, Schutz K, Huang E, Spiegel L, Yordan C, et al. The complete sequence of a heterochromatic island from a higher eukaryote. Cell. 2000;100:377–386. [PubMed] [Google Scholar]
- McCouch SR, Kochert G, Yu ZH, Wang ZY, Khush GS, Coffman WR, Tanksley SD. Molecular mapping of rice chromosomes. Theor Appl Genet. 1988;76:815–829. doi: 10.1007/BF00273666. [DOI] [PubMed] [Google Scholar]
- Ohmido N, Akiyama Y, Fukui K. Physical mapping of unique nucleotide sequences on identified rice chromosomes. Plant Mol Biol. 1998;38:1043–1052. doi: 10.1023/a:1006062905742. [DOI] [PubMed] [Google Scholar]
- Pennisi E. Stealth genome rocks rice researchers. Science. 2000;288:239–241. doi: 10.1126/science.288.5464.239. [DOI] [PubMed] [Google Scholar]
- Sasaki T, Burr B. International Rice Genome Sequencing Project: The effort to completely sequence the rice genome. Curr Opin Plant Biol. 2000;3:138–141. doi: 10.1016/s1369-5266(99)00047-3. [DOI] [PubMed] [Google Scholar]
- Shastry SVS, Rangao Rao DR, Mistra RN. Pachytene analysis in Oryza. I. Chromosome morphology in Oryza sativa L. Indian J Genet Breed. 1960;20:15–21. [Google Scholar]
- Singh K, Ishii T, Parco A, Huang N, Brar DS, Khush GS. Centromere mapping and orientation of the molecular linkage map of rice (Oryza sativa L.) Proc Natl Acad Sci. 1996;93:6163–6168. doi: 10.1073/pnas.93.12.6163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu HK. Note on preparing of pachytene chromosomes by double mordant. Sci Agric. 1967;15:40–44. [Google Scholar]
- Wu T, Wu R. A new rice repetitive DNA sequence shows homology to both 5s RNA and tRNA. Nucl Acids Res. 1987;15:5913–5923. doi: 10.1093/nar/15.15.5913. [DOI] [PMC free article] [PubMed] [Google Scholar]