Abstract
The human pseudoautosomal region 1 (PAR1) is essential for meiotic pairing and recombination, and its deletion causes male sterility. Comparative studies of human and mouse pseudoautosomal genes are valuable in charting the evolution of this interesting region, but have been limited by the paucity of genes conserved between the two species. We have cloned a novel human PAR1 gene, DHRSXY, encoding an oxidoreductase of the short-chain dehydrogenase/reductase family, and isolated a mouse ortholog Dhrsxy. We also searched for mouse homologs of recently reported PGPL and TRAMP genes that flank it within PAR1. We recovered a highly conserved mouse ortholog of PGPL by cross-hybridization, but found no mouse homolog of TRAMP. Like Csf2ra and Il3ra, both mouse homologs are autosomal; Pgpl on chromosome 5, and Dhrsxy subtelomeric on chromosome 4. TRAMP, like the human genes within or near PAR1, is probably very divergent or absent in the mouse genome. We interpret the rapid divergence and loss of pseudoautosomal genes in terms of a model of selection for the concentration of repetitive recombinogenic sequences that predispose to high recombination and translocation.
[The sequence data described in this paper have been submitted to the EMBL data library under accession nos. AJ293620, AJ296079, and AJ293619.]
The human sex chromosomes, heteromorphic in both size and gene content, are fossils of an ancestral, homologous chromosome pair (Ohno 1967). Over the 200 million years of mammalian evolution, the X and Y chromosomes lost homology as the Y chromosome was progressively degraded as the result of drift in nonrecombining regions and/or selection at linked loci on the Y (Charlesworth 1990). However, pseudoautosomal regions 1 and 2 (PAR1 and PAR2) at the termini of the short and long arms of X and Y chromosomes still recombine during male meiosis, ensuring X-Y nucleotide sequence identity that is necessary for normal male meiosis (Rappold 1993). In this paper, we will consider only the short-arm PAR1.
Between-species comparisons of the PAR and neighboring X-specific regions have aided our understanding of the genesis of this region, but also have presented mysteries. Cloning and mapping the marsupial homologs of genes within and near the human PAR1 revealed that this region was added to the X and Y since the divergence of marsupials and eutherian mammals 130 million years ago (Toder and Graves 1998). Comparisons with carnivores and artiodactyls showed that the human PAR1 has been recently reduced from a larger homologous region that included the steroid sulfatase (STS) gene, which lies just outside the PAR on the human X chromosome, and detects an inactive copy on the long arm of the human Y (Toder et al. 1997).
Comparison with mouse genes is harder to interpret. Human and mouse pseudoautosomal regions appear to have distinct evolutionary origin, as there are no genes that are pseudoautosomal in both species. Of the 10 cloned genes assigned to the human PAR1, eight appear to have no mouse homologs. The two that do recognize mouse homologs, Csfgmra (the homolog of CSF2RA) and Il3ra (the homolog of IL3RA), both have diverged considerably from their human homologs (Hara and Miyajima 1992; Park et al. 1992) and both have been assigned to mouse autosomes (chromosomes 19 and 14, respectively) (Disteche et al. 1992; Ellison et al. 1996). The only gene assigned to the PAR of the mouse is steroid sulfatase (Sts), which is pseudoautosomal in other eutherians, but not human. Mouse Sts is very divergent from human STS (63% identity at nucleotide and 59% at amino-acid levels [Salido et al. 1996]).
Spanning the boundary of the mouse PAR is the gene Fxy, which codes for a RING finger protein. The human homolog FXY is located on Xp22.3, near but not within PAR1 (Perry et al. 1998). The 5′ region of the mouse gene (exons 1–3, including a significant portion of the coding region) has no Y homolog, while the 3′ region (exons 4–10) lies within PAR1 and therefore is present both on the X and Y chromosomes. Human FXY and mouse Fxy shows a very high sequence similarity at the DNA and protein levels, but the X-Y shared 3′ region is much less conserved (estimated at 170-fold higher for synonymous sites) than the 5′ portion (Perry and Ashworth 1999). It is hard to explain why genes in the pseudoautosomal region should diverge more rapidly than genes in the differentiated region of the X chromosome or the autosomes.
It is not clear, from the few human genes with mouse homologs, how general this observation is. It would be a great advantage to analyze other human PAR1 genes with mouse homologs. To further explore the relationship between human and mouse PARs, we have isolated a novel pseudoautosomal human gene called DHRSXY and undertaken comparative studies of this and two other genes that we have recently isolated: PGPL and TRAMP. We found a highly conserved mouse ortholog of PGPL and a less conserved mouse ortholog of DHRSXY, both of which are autosomal. There appears to be no mouse TRAMP ortholog. We discuss the implications of this finding in understanding the evolution of the pseudoautosomal region.
RESULTS
Cloning of a Novel Pseudoautosomal Gene
We previously isolated two human PAR1 genes, PGPL and TRAMP (Gianfrancesco et al. 1998; Esposito et al. 1999). To identify additional transcripts, we started from X-linked expressed sequence tags (ESTs) of the Human Transcript Map (http://www.ncbi.nlm.nih.gov/genemap99/map.cgi?CHR=X). We used the IMAGE clone 447388 (AA702323) belonging to the most telomeric UniGene cluster (Hs.16129) of the map. A PCR product of 262 bp made from this EST was used to screen a cDNA library derived from a human uninduced male teratocarcinoma cell line, NT2/D1 (Skowronski et al. 1988). A cDNA clone of 2554 bp was isolated, sequenced, and submitted to the European Molecular Biology Laboratory (EMBL) with accession number AJ293620 (Fig. 1).
Figure 1.
The nucleotide sequence of the human DHRSXY gene together with the conceptual translation of the open reading frame. Gray boxes denote SDR protein-like domains. Solid lines indicate the primers used in reverse transcriptase-PCR assay.
Sequence analysis of this cDNA clone revealed an open reading frame (ORF) of 993 bp encoding a putative protein of 330 amino acids. Using this protein as a query, the BLAST algorithm revealed homology with numerous proteins known as members of the short-chain dehydrogenase/reductase (SDR) family. Consistent with the recommendations of the Human Gene Organization nomenclature committee, we have named it DHRSXY (dehydrogenase/reductase of SDR family on X and Y chromosomes). The SDR family encompasses a wide variety of enzymes, most of which are known to be NAD- or NADP-dependent oxidoreductases of average-size 250 to 300 amino-acid residues. Although the similarity between the different SDR proteins can be as low as about 30%, two domains are conserved: the first binding the coenzyme, often NAD (GXXXGXG), and the second binding the substrate (YXXXK) (Jornvall et al. 1995). This latter domain determines the substrate specificity and contains amino acids involved in catalysis. Although DHRSXY is much larger than the average SDR enzymes, it exhibits the sequence features and distances between conserved motifs that are characteristic of SDR enzymes. In fact, DHRSXY amino-acid sequence analysis identified both the coenzyme, binding site GGTDGIG at position 50–56 and the potential substrate binding site YAQSK at positions 208–212. At this stage, it is difficult to predict the role that DHRSXY may play in the metabolism of the cell, but its unique features make it an interesting target for further investigation.
Fine Localization and Expression of DHRSXY
Because the EST sequence used to clone human DHSRXY came from the most distal portion of the X transcript map, DHRSXY must lie within PAR1. To map DHRSXY more precisely, YAC clones covering the PAR1 were screened by PCR using sequence from the putative gene to design primers (Ried et al. 1995). Two YAC clones (H1130 and D1116) were positive. We also analyzed the cosmids covering these two YACs (unpubl.), finding three (LLNOYCO3′M‘43C10, LLNOYCO3′M‘16C12 and LLNOYCO3′M‘21E1) that were positive for DHRSXY. These cosmids lie distal to ASMT. One (LLNOYCO3′M‘21E1) overlaps with the ASMT-containing cosmid LLNOYCO3′M‘52D2, so DHRSXY must be physically adjacent to ASMT (Fig. 2).
Figure 2.
Localization of the DHRSXY gene in the human pseudoautosomal region.
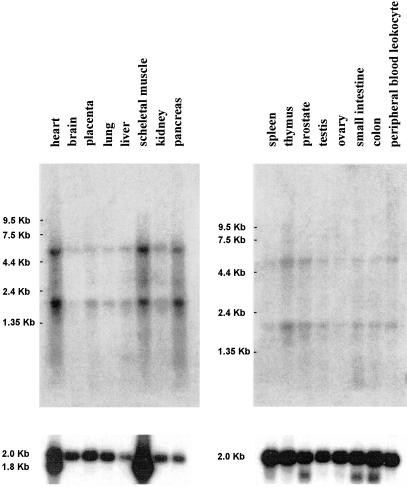
The PCR product used for cDNA library screening was used as a probe to examine the levels of expression in various tissue types. Northern analysis detected two RNA species of about 1800 and 5000 bp in all human adult tissues tested (Fig. 3). Expression studies by reverse transcriptase (RT) and PCR using various somatic cell hybrid lines (Esposito et al. 1997) showed that the RT-PCR products were present in lines containing an active X, inactive X, or Y chromosome (data not shown). Thus, this novel gene, like other pseudoautosomal genes, escapes X inactivation and has a functional copy on the Y chromosome.
Figure 3.
Gene expression of DHRSXY in human tissues. The bars on the left indicate the position of migration of RNA markers. Reprobing of the same filter with a β-actin probe is shown in the bottom panel.
Isolation of Mouse Homologs of Human DHRSXY and PGPL
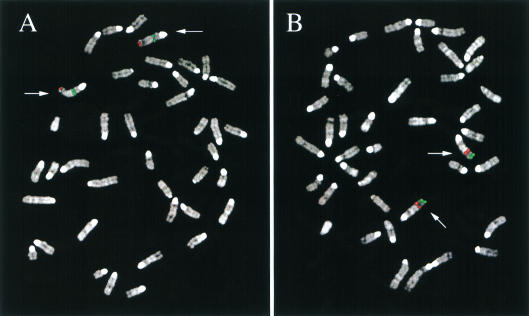
To isolate the murine ortholog of the human DHRSXY gene, we performed an EST database search to identify ESTs of the mouse-related gene. A variety of mouse ESTs were detected, but only a few showed significant homology at the amino acid levels using the TBLASTN program. The EST (AI466831) with the highest similarity with the putative DHRSXY protein was analyzed. The sequence of this IMAGE clone 716487 of 852 bp (named Dhrsxy) was deposited in the EMBL (accession no. AJ296079). This clone does not contain the full-length sequence, as the termination codon was lacking. Comparison of human DHRSXY to mouse Dhrsxy by “BLAST2 sequences” revealed a 64% identity at nucleotide and a 59% identity (84% similarity) at protein level (data not shown). Using primer pairs based on the mouse cDNA sequence, a genomic DNA of about 3 kb, containing a portion of this gene, was amplified by PCR to obtain a probe for in situ localization. Fluorescence in situ hybridization (FISH) was performed and a specific hybridization signal on chromosome 4E was detected (Fig. 4A).
Figure 4.
Chromosomal localization of the mouse orthologs as determined by fluorescence in situ hybridization analysis. (A) Localization of the Dhrsxy gene on chromosome 4E (red signal). The green signal is the result of the PAC 372I17 mapping at 4B. (B) Localization of the Pgpl gene (red signal) on chromosome 5, band E, cohybridized with PAC 426A18 (green signal) mapping at 5G.
To isolate the mouse ortholog of the recently reported PGPL gene (Gianfrancesco et al. 1998), its cDNA was used to screen a 10.5-d embryonic mouse cDNA library (Stratagene). A cDNA of almost 1300 bp was isolated, subcloned, and fully sequenced (EMBL accession no. AJ293619). Comparison of mouse Pgpl and human PGPL revealed 71% identity at the nucleotide level (data not shown) and 68% identity and 87% similarity at the protein level (data not shown). Two mouse multiple-tissue Northern blots were hybridized with the full-length mouse cDNA. Low expression of a transcript of 1800 bp was detected in heart, brain, spleen, lung, liver, kidney, and testis, but not in skeletal muscle. The same transcript also was detected in fetal tissues (data not shown). A genomic clone obtained by screening a mouse λ library (129 strain) was used for FISH to chromosomes of a mouse cell line. Pgpl was localized to chromosome 5, portion 5E (Fig. 4B).
Recently, we reported the cloning of a novel human PAR1 gene called TRAMP, having some characteristics of an ancient transposable element (Esposito et al. 1999). To isolate the murine homolog, we used the human full-length cDNA to probe a mouse genomic library (129 strain). Even at reduced stringency, the human probes failed to detect any cross-hybridizing signal. Moreover, we found no sequences in a mouse EST database that showed significant homology to human TRAMP either at nucleotide or protein level. FISH, using the human TRAMP clone at low stringency, failed to detect any hybridization signal on mouse chromosomes. The results suggest that the TRAMP rodent gene, if it exists, is highly divergent from its human counterpart.
DISCUSSION
The pseudoautosomal region, as the last vestige of the autosome pair that differentiated into the sex chromosomes, might be expected to maintain the characteristics of autosomal regions. Whereas the differential region of the Y chromosome is expected to have a high mutation and attrition rate, there was no reason to suppose that the pseudoautosomal region would be subject to any special evolutionary forces and should therefore be as conserved in gene content and sequence as any autosomal region (Charlesworth 1991).
Our results with previous data (Blaschke and Rappold 1997) demonstrate that this clearly is not the case. Only one human PAR gene (PGPL) is conserved in the mouse genome. The other three human PAR genes with mouse homologs (DHRSXY, CSF2RA, and IL3RA) and one that maps to the original larger eutherian PAR (STS) are poorly conserved in mouse and could be isolated only by functional strategies rather than by cross-hybridization experiments. FXY, whose mouse counterpart straddles the pseudoautosomal boundary, is much more divergent on the PAR than the differentiated side. Moreover, several human PAR genes (TRAMP, SHOX, XE7, ANT3, ASMT, MIC2, PBDX/Y), as well as several genes that were in the ancient eutherian PAR but differentiated recently in primates (KAL, PRKY, ARSD, ARSE), cannot be detected at all in mouse, implying that they have diverged beyond recognition, or have been lost from the genome (Ellison et al. 1996; Smith and Goodfellow 1994). The PAR also seems to have been peculiarly subject to rearrangement in mouse, given that the mouse homologs of four human PAR genes (PGPL, DHRSXY, CSF2RA, and IL3RA) are located on four different mouse autosomes 5, 4, 19, and 14, respectively), implying four different translocation events within less than a megabase.
Overlapping sets of genes within and near the PARs in different eutherian species, and correspondence with the conserved X chromosome, suggest that they all represent relics of a larger ancestral PAR that once extended from Xp22 to Xpter. It included human genes shared between the X and Y chromosome such as ZFX/Y, STS/STSP, as well as the present human PAR genes (Graves 1995; Graves et al. 1998; Lahn and Page 1999). Gradually, parts of the PAR have been rearranged or lost independently in primates and rodents. Comparative mapping in marsupials shows that this PAR was a part of a larger autosomal region translocated to the X and Y in an ancestral eutherian, as it includes CSF2RA, as well as STS and several genes from human Xp (Toder and Graves 1998). The difference in gene content between the human and mouse PAR therefore must represent losses from the mouse PAR, rather than gains to the primate PAR.
Why is the pseudoautosomal region undergoing this rapid evolution? An intriguing hypothesis involves the recombination rate. Recombination within the human PAR is 10 times more frequent during male meiosis than female meiosis (Rappold 1993). This is because there is an obligate crossing over event within this 2.6-Mb region in males that is required for correct segregation of the X and Y chromosomes. This high frequency of recombination may render the PAR more prone to mistakes and repair, creating sequence divergence between the two species. A higher recombination rate also may predispose the region to interchromosomal exchanges, making translocation to autosomes more frequent.
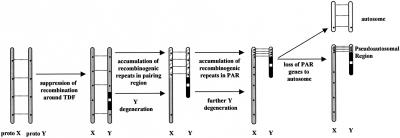
Why and how did the PAR evolve this high frequency of recombination? Heteromorphic sex chromosomes evolved originally from an autosomal pair, driven initially by differences at a sex-determining locus. Ohno (1967) suggested that subsequent differentiation of this pair to the sex pair was accomplished exclusively at the expense of one member of the pair (the Y chromosome in mammals) that was selected to accumulate sex-specific functions. Acquisition of a sex-determining function by locus on the proto-Y was the starting point for Y attrition (Fig. 5). As well as attrition, comparative mapping in marsupial and monotreme mammals demonstrates the addition of a large autosomal region to an ancient PAR early in eutherian divergence (Graves 1995). The process of attrition continued along this added region, ultimately leaving the present human PAR as a relic of this addition (Graves et al. 1998). The independent movement of the pseudoautosomal boundary contributed to the human/mouse differentiation. Thus the PAR is a product of two competing processes: addition of autosomal regions to the X and Y, and progressive attrition of the Y.
Figure 5.
Evolution of sticky pseudoautosomal region that is rich in repetitive recombinogenic sequences and shows high recombination in males. Evolutionary steps are indicated by color code: grey, homologous region; white, SRY gene; black, unpaired region in testis-determining factor region; hatched grey, male-specific recombinogenic region.
Attrition of the Y appears to be inexorable. Its end-point is likely to be complete loss of homology between the X and Y. This appears to have occurred in akodont rodents and in all marsupials, which do not undergo homologous pairing and recombination (Ashley et al. 1989; Sharp 1982; Toder et al. 2000), and must presumably have evolved alternate means of controlling chromosome segregation and avoiding nondisjunction.
We propose that to avoid this scenario, a fierce selection is working to accumulate and concentrate repetitive recombinogenic sequences on the sex chromosomes, for more robust pairing and/or increased recombination, which are essential (at least in mouse and man) for male fertility (Fig. 5). Only some of these sites may be active during female meiosis, in which the two X chromosomes may pair along their full length. The mouse PAR is rich in repetitive sequences and the sole surviving PAR gene Sts has a highly diverged sequence to the extent that it is not recognizable by hybridization with human STS (Salido et al. 1996). This may represent the penultimate stage, in which selection is for conservation of a pairing function even at the expense of gene activity. Studies of a proviral insertion in the mouse PAR (Harbers et al. 1986) suggested that repetitive flanking regions are rearranged and duplicated frequently, perhaps by unequal recombination. The accumulation of these repetitive recombinogenic sequences also may act to promote translocations of pseudoautosomal genes to autosomes.
METHODS
cDNAs and Sequence Analysis
The cDNA library for the isolation of DHRSXY cDNA was from a human uninduced male teratocarcinoma cell line, NT2/D1 (Skowronski et al. 1988). Mouse PGPL-related cDNA clone was isolated from a mouse 11 days embryo cDNA library (Clontech). The cDNAs were subcloned into pGEM-4Z vector (Promega-Biotech) and analyzed by Dye-Terminator cycle sequencing on an Applied Biosystem 377 automated sequencer. The Pgpl genomic clone, used for the FISH assay, was isolated from a mouse genomic library (Stratagene) using the Pgpl cDNA as a probe.
Northern Blot Analysis
Two human and two mouse multiple-tissue Northern blots (Clontech) were hybridized with the PCR product of DHRSXY gene and with a Pgpl cDNA, respectively. The Northern blots were prehybridized, hybridized, and washed according to the manufacturer’s directions (Clontech).
Yeast Artificial Chromosome and Cosmid Screening
For the localization of the DHRSXY gene, YACs from the PAR1 were screened (Ried et al. 1995). Cosmids identified by PCR with the DHRSXY primers were derived from the ICRF X chromosome-specific cosmid library and the Lawrence Livermore X chromosome-specific cosmid library.
Cell Lines
The panel of somatic-cell hybrid lines used in RT-PCR assays comprises two hybrids retaining the active human X chromosome; three hybrids retaining an inactive human X chromosome; a hybrid retaining two inactive human X chromosomes; and two hybrids retaining the human Y chromosome (Esposito et al. 1997).
Reverse Transcriptase-PCR
The RT-PCR for X inactivation experiments of DHRSXY were carried out using 100 ng of RNA and 5 ng of cDNA as the template in a 10 μL PCR reaction containing 1× TNK 100 buffer (Blanchard et al. 1993), 0.2 mM dNTPs, 0.35 units of AmpliTaq polymerase (Boehringer Mannheim), and 0.5 μM each of the primer sequences derived from cDNA. Using a DNA Thermal Cycler MJR (M.J. Research Inc.), we carried out 35 cycles of amplification using: 1 min at 94°C; 2 min at 56°C; and 2 min at 72°C. The DHRSXY primers used in RT-PCR assay are 5′ ATA GTC AGT GGG ATT TTG TC 3′ and 5′ AAA ACC TCA CAC AGC CAA AG 3′.
FISH Analysis
Mouse spreads were prepared from an EM mouse cell line, which is mosaic for a Robertsonian fusion between chromosomes 2 and 15. Chromosome identification was facilitated by co-hybridizing Dhrsxy and Pgpl probes with PACs 372I17 and 426A18 (de Jong RP-21 library) mapping to chromosomes 4, band B, and chromosome 5, band G, respectively (http://www.biologia.uniba.it/rmc/9-MOUSE/-MOUSE.html). Digital images were obtained using a Leica DMRXA epifluorescence microscope equipped with a cooled CCD camera (Princeton Instruments). Cy3 (red), FluorX (green), and DAPI (blue) fluorescence signals were detected using specific filters and recorded separately as gray-scale images. Pseudocoloring and merging of images were performed using Adobe Photoshop software.
Acknowledgments
We gratefully acknowledge M. D'Esposito and V. Ursini for critical reading of the manuscript. This work was supported by Telethon Italy to M.D., Ricerca Avanzata prot.18899.2000/University of Modena to A.F., the financial support cofin98-MURST to N.A., and Deutsche Forschungsgemeinschaft to G.R.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL F.Gianfrancesco@igm.ss.cnr.it; FAX 39 079 946714.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.197001.
REFERENCES
- Ashley T, Jaarola M, Fredga K. The behavior during pachynema of a normal and an inverted Y chromosome in Microtus agrestis. Hereditas. 1989;111:281–294. doi: 10.1111/j.1601-5223.1990.tb00407.x. [DOI] [PubMed] [Google Scholar]
- Blanchard MM, Taillon-Miller P, Nowotny V. PCR buffer optimization with uniform temperature regimen to facilitate automation. PCR Methods Appl. 1993;2:234–240. doi: 10.1101/gr.2.3.234. [DOI] [PubMed] [Google Scholar]
- Blaschke R, Rappold GA. Man to mouse — lessons learned from the distal end of the human X chromosome. Genome Res. 1997;7:1114–1117. doi: 10.1101/gr.7.12.1114. [DOI] [PubMed] [Google Scholar]
- Charlesworth B. Mutation-selection balance and the evolutionary advantage of sex and recombination. Genet Res. 1990;55:199–221. doi: 10.1017/s0016672300025532. [DOI] [PubMed] [Google Scholar]
- ————— The evolution of sex chromosomes. Science. 1991;251:1030–1033. doi: 10.1126/science.1998119. [DOI] [PubMed] [Google Scholar]
- Disteche CM, Brannon CI, Larsen A, Adler D, Schorderet DF, Gearing D, Copeland NG, Jenkins NA, Park LS. The human pseudoautosomal GM-CSF receptor alpha subunit gene is autosomal in mouse. Nat Genet. 1992;1:333–336. doi: 10.1038/ng0892-333. [DOI] [PubMed] [Google Scholar]
- Ellison JW, Li X, Francke U. Rapid evolution of human pseudoautosomal genes and their mouse homologs. Mamm Genome. 1996;7:25–30. doi: 10.1007/s003359900007. [DOI] [PubMed] [Google Scholar]
- Esposito T, Gianfrancesco F, Ciccodicola A, D'Esposito M, Nagaraja R, Mazzarella R, D'Urso M, Forabosco A. Escape from X inactivation of two new genes associated with DXS6974E and DXS7020E. Genomics. 1997;15:183–190. doi: 10.1006/geno.1997.4797. [DOI] [PubMed] [Google Scholar]
- Esposito T, Gianfrancesco F, Ciccodicola A, Montanini L, Mumm S, D'Urso M, Forabosco A. A novel pseudoautosomal human gene encodes a putative protein similar to Ac-like transposases. Hum Mol Genet. 1999;8:61–67. doi: 10.1093/hmg/8.1.61. [DOI] [PubMed] [Google Scholar]
- Gianfrancesco F, Esposito T, Montanini L, Ciccodicola A, Mumm S, Mazzarella R, Rao E, Giglio S, Rappold G, Forabosco A. A novel pseudoautosomal gene encoding a putative GTP-binding protein resides in the vicinity of the Xp/Yp-telomere. Hum Mol Genet. 1998;7:407–414. doi: 10.1093/hmg/7.3.407. [DOI] [PubMed] [Google Scholar]
- Graves JA. The origin and function of the mammalian Y chromosome and Y-borne genes — an evolving understanding. BioEssays. 1995;17:311–320. doi: 10.1002/bies.950170407. [DOI] [PubMed] [Google Scholar]
- Graves JAM, Wakefield MJ, Toder R. Evolution of the pseudoautosomal region of mammalian sex chromosomes. Hum Molec Genet. 1998;7:1991–1996. doi: 10.1093/hmg/7.13.1991. [DOI] [PubMed] [Google Scholar]
- Hara T, Miyajima A. Two distinct functional high affinity receptors for interleukin-3 (IL-3) EMBO J. 1992;11:1875–1884. doi: 10.1002/j.1460-2075.1992.tb05239.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbers K, Soriano P, Muller U, Jaensich R. High frequency of unequal recombination in pseudoautosomal region shown by proviral insertion in transgenic mouse. Nature. 1986;324:682–685. doi: 10.1038/324682a0. [DOI] [PubMed] [Google Scholar]
- Jornvall H, Persson B, Krook M, Atrian S, Gonzalez-Duarte R, Jeffery J, Ghosh D. Short-chain dehydrogenases/reductases (SDR) Biochemistry. 1995;34:6003–6013. doi: 10.1021/bi00018a001. [DOI] [PubMed] [Google Scholar]
- Lahn BT, Page DC. Four evolutionary strata on the human X chromosome. Science. 1999;286:964–967. doi: 10.1126/science.286.5441.964. [DOI] [PubMed] [Google Scholar]
- Ohno S. Sex Chromosomes and Sex-Linked Genes. Berlin: Springer-Verlag; 1967. [Google Scholar]
- Park LS, Martin U, Sorenson R, Luhr S, Morrissey PJ, Cosman D, Larsen A. Cloning of the low-affinity murine granulocyte-macrophage colony stimulating factor receptor and reconstitution of a high-affinity receptor complex. Proc Natl Acad Sci. 1992;89:4295–4299. doi: 10.1073/pnas.89.10.4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry J, Feather S, Smith A, Palmer S, Ashworth A. The human FXY gene is located within Xp22.3: Implications for evolution of the mammalian X chromosome. Hum Mol Genet. 1998;7:299–305. doi: 10.1093/hmg/7.2.299. [DOI] [PubMed] [Google Scholar]
- Perry J, Ashworth A. Evolutionary rate of a gene affected by chromosomal position. Curr Biol. 1999;9:987–989. doi: 10.1016/s0960-9822(99)80430-8. [DOI] [PubMed] [Google Scholar]
- Rappold GA. The pseudoautosomal regions of the human sex chromosomes. Hum Genet. 1993;92:315–324. doi: 10.1007/BF01247327. [DOI] [PubMed] [Google Scholar]
- Ried K, Mertz A, Nagaraja R, Trusgnich M, Riley JH, Anand R, Lehrach H, Page D, Ellison JW, Rappold G. Characterization of a YAC contig spanning the pseudoautosomal region. Genomics. 1995;29:787–792. doi: 10.1006/geno.1995.9933. [DOI] [PubMed] [Google Scholar]
- Salido EC, Li XM, Yen PH, Martin N, Mohandas TK, Shapiro LJ. Cloning and expression of the mouse pseudoautosomal steroid sulphatase gene (Sts) Nat Genet. 1996;13:83–86. doi: 10.1038/ng0596-83. [DOI] [PubMed] [Google Scholar]
- Sharp P. Sex chromosome pairing during male meiosis in marsupials. Chromosoma. 1982;86:27–47. doi: 10.1007/BF00330728. [DOI] [PubMed] [Google Scholar]
- Skowronski J, Fanning TG, Singer M F. Unit-length line-1 transcripts in human teratocarcinoma cells. Mol Cell Biol. 1988;8:1385–1397. doi: 10.1128/mcb.8.4.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MJ, Goodfellow PN. MIC2R: A transcribed MIC2-related sequence associated with a CpG island in the human pseudoautosomal region. Hum Mol Genet. 1994;3:1575–1582. doi: 10.1093/hmg/3.9.1575. [DOI] [PubMed] [Google Scholar]
- Toder R, Glaser B, Schiebel K, Wilcox SA, Rappold G, Graves JAM, Schempp W. Genes located in and near the human pseudoautosomal region are located in the X-Y pairing region in dog and sheep. Chromosome Res. 1997;5:1–6. doi: 10.1023/B:CHRO.0000038760.84605.0d. [DOI] [PubMed] [Google Scholar]
- Toder R, Graves JAM. CSF2RA, ANT3 and STS are autosomal in marsupials: Implications for the origin and evolution of the pseudoautosomal region of mammalian sex chromosomes. Mamm Genome. 1998;9:373–376. doi: 10.1007/s003359900772. [DOI] [PubMed] [Google Scholar]
- Toder R, Wakefield M, Graves JAM. The minimal mammalian Y chromosome — the marsupial Y as a model system. Cytogenet Cell Genet. 2000;91:285–292. doi: 10.1159/000056858. [DOI] [PubMed] [Google Scholar]