Abstract
Aldehyde dehydrogenases (ALDHs) belong to a superfamily of NAD(P)+-dependent enzymes, which catalyze the oxidation of endogenous and exogenous aldehydes to their corresponding acids. Increased expression and/or activity of ALDHs, particularly ALDH1A1, have been reported to occur in human cancers. It is proposed that the metabolic function of ALDH1A1 confers the “stemness” properties to normal and cancer stem cells. Nevertheless, the identity of ALDH isozymes that contribute to the enhanced ALDH activity in specific types of human cancers remains to be elucidated. ALDH1B1 is a mitochondrial ALDH that metabolizes a wide range of aldehyde substrates including acetaldehyde and products of lipid peroxidation (LPO). In the present study, we immunohistochemically examined the expression profile of ALDH1A1 and ALDH1B1 in human adenocarcinomas of colon (N=40), lung (N=30), breast (N=33) and ovary (N=33) using an NIH tissue array. The immunohistochemical expression of ALDH1A1 or ALDH1B1 in tumor tissues was scored by their intensity (scale = 1–3) and extensiveness (% of total cancer cells). Herein we report a 5.6-fold higher expression score for ALDH1B1 in cancerous tissues than that for ALDH1A1. Remarkably, 39 out of 40 colonic cancer specimens were positive for ALDH1B1 with a staining intensity of 2.8 ± 0.5. Our study demonstrates that ALDH1B1 is more profoundly expressed in the adenocarcinomas examined in this study relative to ALDH1A1 and that ALDH1B1 is dramatically upregulated in human colonic adenocarcinoma, making it a potential biomarker for human colon cancer.
Keywords: ALDH1B1, epithelial cancer, colon cancer, cancer stem cell, biomarker
Introduction
The ALDH superfamily of enzymes catalyze the NAD(P)+-dependent oxidation of wide varieties of endogenous and exogenous aldehyde substrates to their corresponding acids (1). To date, twenty mouse Aldh and nineteen human ALDH genes have been reported to encode proteins; functional polymorphisms in these genes are associated with distinct phenotypes in humans and rodents (2), indicating that these ALDH enzymes play diverse but physiologically important roles. In recent years, the ALDH activity has been used as a specific marker for the selection of normal and cancer stem cells (2). It was first found that human hematopoietic stem cells (HSCs) were rich in ALDH activity (3). Through the use of a synthetic fluorescent substrate for ALDH, named BODIPY aminoacetaldehyde (BAAA), primitive HSCs were effectively isolated from human umbilical cord blood using fluorescence activated cell sorting (3). Later studies provided several lines of evidence supporting a critical role of ALDH-mediated retinoic acid (RA) signaling in regulating HSC self-renewal and differentiation (4;5). It was hypothesized that the main ALDH isozyme involved was ALDH1A1 due to its ability to convert retinol to RA (5). More recently, the functional activity of ALDH has been widely used to identify and isolate cancer stem cells (CSCs) found in the bone marrow (6), breast (7), lung (8), ovary (9), colon (10), prostate (11), and pancreas (12). In these studies, a commercially available ALDEFLUOR assay was used to sort out a subpopulation of cells possessing high ALDH activity (ALDHbri cells). The ALDHbri cells, in some cases simultaneously positive for other markers of stem cells, revealed much higher tumorigenic potential than ALDHlow cells based on in vivo tumorigenicity and ex vivo clonogenicity assays. Similarly, the expression of ALDH enzymes, mainly ALDH1A1, was found to be upregulated in tumor samples by immunohistochemical studies (9). The molecular basis of ALDH1A1, and likely other ALDHs, being selectively expressed in CSCs is not completely understood. It is proposed, however, that the metabolic activity of ALDH1A1 towards cytotoxic aldehydes arising from chemotherapies is a key determinant of cell survival and drug resistance of CSCs (8;13). Nevertheless, since the existing ALDEFLUOR assay selects only for the total ALDH activity, without specificity for individual ALDH isozymes, the identity of specific ALDH isozyme(s) in contributing to the enhanced ALDH activity in a specific type of human cancers remains to be elucidated.
ALDH1B1 is a mitochondrial ALDH isozyme and shares 65% homology in peptide sequence with cytosolic ALDH1A1 in human. We have recently characterized the biochemical properties of ALDH1B1 enzyme using human recombinant ALDH1B1 (14). Human ALDH1B1 is catalytically active towards a wide range of aldehyde substrates, including aliphatic and aromatic aldehydes and the products of LPO. Furthermore, ALDH1B1 possesses the second lowest Km (55 µM) among ALDHs for the oxidation of acetaldehyde, supporting its putative role in ethanol metabolism. By immunohistochemical analysis, human ALDH1B1 was found to be expressed at high levels in the small intestine, liver, and pancreas, and at lower levels in the lung and colon (14). At present, the physiological and/or pharmacological-toxicological role(s) of ALDH1B1 are largely unknown. To begin to understand the role of ALDH1B1 in cancer biology, we examined the expression profile of ALDH1B1 and ALDH1A1 in human adenocarcinomas of the colon, lung, breast and ovary by immunohistochemical staining. Our results demonstrated that the two enzymes are differentially expressed in each tissue examined, and the ALDH1B1 protein appears to be more highly expressed in human epithelial cancers than is ALDH1A1.
Materials and methods
Human tissues
Formalin-fixed and paraffin-embedded tissues of normal human liver, colon, lung, breast and ovary were procured by IHCtech (Aurora, CO) in accordance with relevant approvals and guidelines for use in immnohistochemical staining. Human cancer tissues of the colon, lung, breast and ovary were purchased as tissue microarray slides (T-MTA-6A) from the Cooperative Human Tissue Network and the Tissue Array Research Program (TARP) of the National Cancer Institute (NIH, Bethesda, MD). Patient diagnosis, gender, age and cancer characteristics were provided by TARP.
Cross-reactivity of ALDH antibodies
The immuno-reactivity of anti-human ALDH1A1 (BD Pharmingen, San Diego, CA) and anti-human ALDH1B1 (14) towards six ALDH isozymes were assessed by Western blotting analysis (See Supplementary Material).
Immunohistochemical staining (IHC)
IHC was performed by IHCtech according to a standard procedure (15) with minor modifications. Briefly, paraffin-embedded tissue sections (5-µm) were deparaffinized and rehydrated, followed by heat-induced epitope retrieval in Retrieval Solution (Leica Microsystems, Bannockburn, IL) at 90°C for 10 min. After incubation with Protein Blocker (Open Biosystems, Huntsville, AL), sections were incubated with monoclonal anti-human ALDH1A1 (BD Pharmingen, San Diego, CA) at 1:1000 or polyclonal anti-human ALDH1B1 (14) at 1:750 in Protein Blocker for 60 min at room temperature. Corresponding HRP-conjugated secondary antibodies (BioCare Medical, Concord, CA) were used at 1:500 for 10–20 min at room temperature. Slides were incubated with DAB (Open Biosystems, Huntsville, AL) for 10 min, followed by counterstaining with Hematoxylin. Negative control slides were processed in the same manner except that Protein Blocker was applied replacing anti-human ALDH1A1 or pre-immune serum was applied replacing anti-human ALDH1B1.
Cancer sample analysis
Tissue sections of adenocarcinoma samples of colon, lung, breast and ovary (50 samples each) were present on the tissue microarray slide and each was evaluated for the immunopositivity of ALDH1A1 and ALDH1B1 staining as described (16). Briefly, the IHC signal for each antibody in each tissue was rated semi-quantitatively in positively stained sections for intensity (I) on a scale of 1–3 (1 = faint staining, 2 = moderate staining, and 3 = strong staining) and for extensiveness (E, % of tumor cells showing an IHC signal). Samples were examined and scored by two independent observers (YC and DJO). Those samples lacking enough tumor tissue for evaluation were excluded from the final analysis. The IHC expression score (S) for each antibody in each sample was calculated by S = I × E. Data are reported as mean ± SEM.
Results
Evaluation of the cross-reactivity of anti-human ALDH1A1 and anti-human ALDH1B1
Before starting our IHC survey of tissues, the specificity of the antibodies was evaluated. The monoclonal anti-human ALDH1A1 is commercially available and has been used in numerous studies for the immuno-detection of ALDH1A1 protein (9;10;17). The polyclonal anti-human ALDH1B1 antibody was raised against a 58-peptide (amino acids 353–411) of human ALDH1B1 and purified through an antigen-bound affinity column (14). We examined the immunoreactivity of these antibodies towards six human recombinant ALDH proteins by Western immunoblotting analysis (See Supplementary Material). Under the conditions we used, the anti-ALDH1A1 antibody reacted exclusively with ALDH1A1 protein; on the other hand, the anti-ALDH1B1 antibody exhibited strongest immunoreactivity towards ALDH1B1 protein, albeit showing some degree of cross-reactivity to ALDH1A1 protein and no cross-reactivity to other ALDH proteins.
ALDH1A1 and ALDH1B1 distribution in normal human tissues
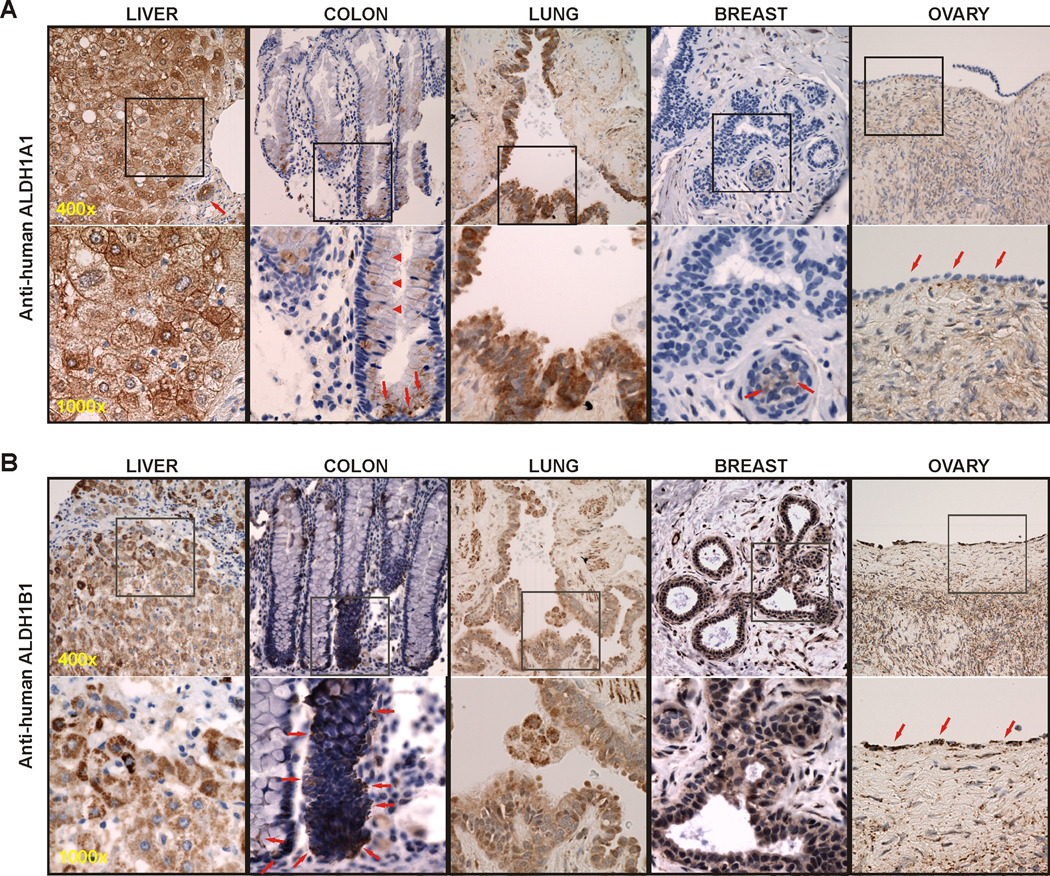
IHC staining was performed in normal human tissues of liver, colon, lung, breast and ovary for the presence of ALDH1A1 and ALDH1B1 protein, respectively (Fig. 1).
Figure 1. Distribution of ALDH1A1 and ALDH1B1 in normal human tissues by IHC.
Positive staining (brown) displayed a diffuse cytoplasmic pattern for ALDH1A1 (A), but a cytoplasmic punctate pattern for ALDH1B1 (B). Hepatocytes were strongly stained for both proteins. In the colon, cells at the crypt bottom were positively stained with both ALDH1A1 and ALDH1B1 (arrows), whereas the immunopositivity of ALDH1A1 was also noted in partially differentiated cells at higher crypt levels (arrowheads). The bronchiole epithelium and Clara cells in the lung showed immunopositivity of ALDH1A1 and to a lesser extend of ALDH1B1. In the breast, IHC-signal of ALDH1A1 was barely notable and observed only in luminal epithelial cells (arrows); in contrast, the acinar and ductal lining columnar cells and the basal cells beneath them were all positively stained for ALDH1B1. In the ovary, the surface simple cuboidal cells (arrows) stained negatively for ALDH1A1, but quite intensely for ALDH1B1. Representative images are presented at two magnifications, with the squared field in the top panel (400×) enlarged in the bottom panel (1000×).
ALDH1A1 expression (Fig. 1A)
In the liver, both hepatocytes (boxed area) and cholangiocytes (arrow) showed immunopositivity, whereas most stromal cells were negative. The staining of hepatocytes was of a “checkerboard pattern”, showing high variability from cell to cell without apparent order. Hepatocytes in the peri-portal and centrilobular portions exhibited similar quantities of ALDH1A1 staining, which showed both a cytoplasmic and a plasma membrane component. The cholangiocyte staining was more uniform and only appeared to be of the cytoplasmic variety. In the colon, we observed a cytoplasmic staining of ALDH1A1 both in cells located at the bottom of the crypts (arrows) as well as in many of the partially differentiated absorptive and secretory cells at higher crypt levels (arrowheads), whereas the fully differentiated cells were negative. A few of the stromal macrophages were slightly positive, while other stromal cells were negative. In the lung, the Clara cells showed strong cytoplasmic immunopositivity; the basal cells and ciliated cells were also positive, but to a lesser extent. Stromal elements appeared negative. In the breast, ALDH1A1 immunopositivity was sparse and included only some of the luminal epithelial cells (arrows); the rest of the epithelial structures and the stromal elements were negative. In the ovary, the surface simple cuboidal cells (arrows) or germinal epithelium was negative as was most of the stroma. A small amount of staining appeared to associate with either smooth muscle cells or fibroblasts.
ALDH1B1 expression (Fig. 1B)
In the liver, both hepatocytes and cholangiocytes stained positively for ALDH1B1, whereas stromal cells were negative. The ALDH1B1 staining exhibited a punctate intracellular morphology, consistent with being confined within well dispersed intracellular organelles. Hepatocyte staining of ALDH1B1 in the peri-portal areas was more intense than in the centrilobular portions, where the staining showed a reasonably homogeneous intensity from cell to cell. The cholangiocyte staining was restricted to the luminal half of the cell and was also of a punctate nature (not shown). In the colon, ALDH1B1 immunopositivity was noted in a small subset of undifferentiated appearing slim columnar cells predominantly near the sides of the crypt bottom (arrows) and in small numbers sporadically along the sides of the upper portion of the crypt. The differentiated cells were negative. In addition, a subpopulation of macrophages in the stroma also stained positively. In the lung, the Clara cell stained intensely for ALDH1B1; however the basal cells of the epithelium of the bronchioles as well as lung macrophages and smooth muscle cells were also positive. Ciliated cells and fibroblasts were negative. In the breast, the acinar and ductal lining columnar cells and the basal cells beneath them all stained positive for ALDH1B1, again in a punctate manner. The myoepithelial cells were negative as was most of the stroma, except for some of the smooth muscle cells. In the ovary, the surface simple cuboidal cells stained intensely for ALDH1B1 in a punctate manner. In addition, some of the endothelial cells and smooth muscle cells were positively stained, but with much less intensity. Other cell types of the stroma and follicles were negative.
ALDH1A1 and ALDH1B1 expression in human cancerous tissues
The human tissue microarray slide (T-MTA-6A) contained fifty cancer tissues each of human colon, lung, breast and ovary. All of the tumors were diagnosed as adenocarcinoma with varying histological grades. The average age in years of the patients was 70 ± 11, 63 ± 12, 56 ± 15, and 61 ± 15 for colon, lung, breast and ovarian cancers, respectively. For colon and lung cancers, female and male patients were equally represented. The intensity (I; 1–3 scale) and extensiveness (E; 1–100 scale) of IHC staining for each primary antibody in each tissue sample was assessed and the overall expression score (S; 1–300 scale) was calculated by S = I × E. Based on this scoring system, the expression profiles for ALDH1A1 and ALDH1B1 in these adenocarcinomas are summarized in Tables 1 and 2, respectively.
Table 1.
Expression of ALDH1A1 in human cancer specimens by IHC
| Tumor Features |
Colon | Lung | Breast | Ovary | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n/N1 | I2 | E3 | S4 | n/N | I | E | S | n/N | I | E | S | n/N | I | E | S | ||
| Overall | 15/41 | 1.3 ± 0.1 | 22 ± 6 | 32 ± 10 | 9/30 | 1.4 ± 0.2 | 32 ± 8 | 55 ± 18 | 1/34 | 1.0 | 25 | 25 | 9/34 | 1.4 ± 0.2 | 21 ± 6 | 36 ± 16 | |
| Differentiation | |||||||||||||||||
| well | 7/19 | 1.1 ± 0.1 | 23 ± 11 | 34 ± 22 | 1/3 | 3.0 | 75 | 225 | 0/3 | 0 | 0 | 0 | 2/5 | 2.0 ± 1.0 | 45 ± 5 | 95 ± 25 | |
| moderate | 5/12 | 1.7 ± 0.1 | 21 ± 6 | 34 ± 8 | 2/6 | 1.5 ± 0.5 | 15 ± 10 | 28 ± 9 | 1/11 | 1.0 | 25 | 25 | 3/11 | 1.0 ± 0.0 | 5 ± 0.0 | 5 ± 0.0 | |
| poor | 3/10 | 1.2 ± 0.4 | 20 ± 8 | 23 ± 14 | 6/21 | 1.2 ± 0.2 | 31 ± 9 | 31 ± 10 | 0/20 | 0 | 0 | 0 | 4/16 | 1.4 ± 0.2 | 21 ± 8.3 | 25 ± 11 | |
| Metastasis | |||||||||||||||||
| low-moderate | 8/31 | 1.4 ± 0.8 | 18 ± 9 | 32 ± 19 | 5/14 | 1.6 ± 0.4 | 34 ± 13 | 70 ± 40 | 1/9 | 1 | 25 | 25 | 5/16 | 1.4 ± 0.4 | 21 ± 10 | 35 ± 24 | |
| high | 7/10 | 1.2 ± 0.2 | 26 ± 6 | 32 ± 9 | 4/16 | 1.3 ± 0.3 | 30 ± 11 | 36 ± 12 | 0/25 | 0 | 0 | 0 | 4/14 | 1.4 ± 0.2 | 21 ± 8.3 | 37 ± 15 | |
| Stroma | 0.8 ± 0.1 | 0.1 ± 0.07 | 0.1 ± 0.05 | 0.0 | |||||||||||||
n denotes the number of cancer samples that showed immunopositivity of ALDH1A1; N denotes the total number of sample sections that have been examined.
The intensity of ALDH1A1 immunostaining in positively stained sections was scored as: 1 = faint staining, 2 = moderate staining, 3 = strong staining.
The extensiveness of ALDH1A1 immunostaining in positively stained sections was scored as percent of total cancer cells.
The overall immunopositivity score (S) of ALDH1A1 was calculated as S = I × E.
Results were expressed as mean ± s.e.m.
Table 2.
Expression of ALDH1B1 in human cancer specimens by IHC
| Tumor Features |
Colon | Lung | Breast | Ovary | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n/N1 | I2 | E3 | S4 | n/N | I | E | S | n/N | I | E | S | n/N | I | E | S | ||
| Overall | 39/40 | 2.8 ± 0.5 | 99 ± 3 | 284 ± 7 | 23/30 | 1.9 ± 0.2 | 84 ± 6 | 165 ± 20 | 20/33 | 1.9 ± 0.2 | 87 ± 5 | 171 ± 22 | 28/33 | 2.3 ± 0.1 | 94 ± 3 | 215 ± 14 | |
| Differentiation | |||||||||||||||||
| well | 18/19 | 2.9 ± 0.4 | 100 ± 0 | 272 ± 10 | 3/3 | 1.8 ± 0.7 | 100 ± 0 | 183 ± 44 | 0/2 | 0 | 0 | 0 | 3/5 | 2.3 ± 0.3 | 100 ± 0 | 233 ± 33 | |
| moderate | 12/12 | 2.9 ± 0.2 | 100 ± 0 | 292 ± 6 | 5/6 | 2.0 ± 0.4 | 80 ± 12 | 175 ± 54 | 7/11 | 1.9 ± 0.3 | 100 ± 0 | 193 ± 28 | 10/11 | 2.5 ± 0.2 | 98 ± 3 | 243 ± 19 | |
| poor | 9/9 | 2.8 ± 0.5 | 98 ± 7 | 280 ± 20 | 15/21 | 1.9 ± 0.2 | 83 ± 8 | 125 ± 24 | 13/20 | 1.8 ± 0.3 | 80 ± 8 | 148 ± 31 | 14/16 | 2.1 ± 0.2 | 89 ± 6 | 193 ± 22 | |
| Metastasis | |||||||||||||||||
| low-moderate | 30/31 | 2.9 ± 0.3 | 100 ± 0 | 292 ± 6 | 14/14 | 1.9 ± 0.2 | 79 ± 8 | 159 ± 26 | 4/7 | 2.5 ± 0.2 | 100 ± 0 | 250 ± 20 | 13/16 | 2.1 ± 0.2 | 96 ± 4 | 193 ± 23 | |
| high | 9/9 | 2.8 ± 0.5 | 99 ± 5 | 279 ± 13 | 9/16 | 1.8 ± 0.3 | 92 ± 8 | 176 ± 30 | 15/25 | 1.7 ± 0.2 | 83 ± 7 | 151 ± 27 | 12/14 | 2.5 ± 0.2 | 90 ± 6 | 223 ± 23 | |
| Stroma | 0.4 ± 0.2 | 0.6 ± 0.3 | 0.3 ± 0.3 | 0.5 ± 0.3 | |||||||||||||
n denotes the number of cancer samples that showed immunopositivity of ALDH1B1; N denotes the total number of sample sections that have been examined.
The intensity (I) of ALDH1B1 immunostaining in positively stained sections was scored as: 1 = faint staining, 2 = moderate staining, 3 = strong staining.
The extensiveness (E) of ALDH1B1 immunostaining in positively stained sections was scored as percent of total cancer cells.
The overall immunopositivity score (S) of ALDH1B1 was calculated as S = I × E.
Results were expressed as mean ± s.e.m.
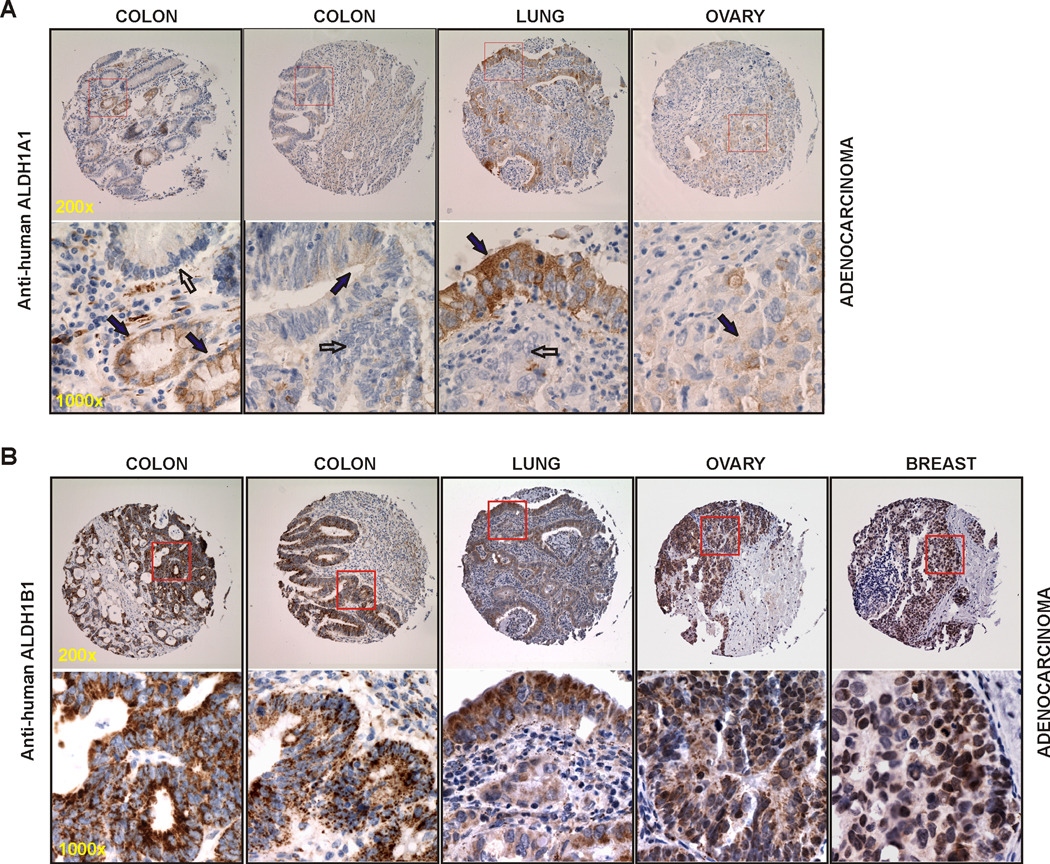
ALDH1A1 expression (Table 1 & Fig. 2A)
Figure 2. Expression of ALDH1A1 and ALDH1B1 in human adenocarcinoma by IHC.
(A) Cancer specimens stained with anti-ALDH1A1 showed positive cystoplasmic staining (brown) in ~20% cancer cells (solid arrows) and negative staining in neighboring cancer cells (open arrows). (B) Cancer specimens stained with anti-ALDH1B1 showed strong positive punctate staining (brown) in almost all cancer cells. Representative images are presented at two magnifications, with the squared field in the top panel (200×) enlarged in the bottom panel (1000×).
In colonic adenocarcinoma, 15 out of 41 (36.6%) samples were positively stained for ALDH1A1. The staining intensity was low (I = 1.3 ± 0.1) and highly variable both between different cases and within any one case (E = 22 ± 6). The overall expression score was 32 ± 10. When tumor features were analyzed, we found a high proportion (70%) of highly metastatic colon adenocarcinomas were positive for the presence of ALDH1A1. Two cases are presented in Fig. 2A. In the case on the left, we observed positive cytoplasmic staining in all of the cells of two well differentiated glands (solid arrows), yet an adjacent less differentiated gland with equal nuclear atypia was negative (open arrow); in the same specimen, a subset of the stromal histiocytes also stained positive. In the case on the right, we noted portions of two moderately differentiated glands, one with low intensity staining confined to the luminal portions of the cancerous cells (solid arrow) and the other was entirely negative (open arrow); in this specimen, the stroma showed a very weak intensity stain throughout it. In lung adenocarcinoma, 9 out of 30 (30%) of the cases were positive for ALDH1A1 IHC-staining. The staining intensity was low (I = 1.4 ± 0.2), the extensiveness was 32% and the overall expression score was 55 ± 18. ALDH1A1 expression in lung adenocarcinoma did not appear to associate with tumor histological grade or metastatic status. One case presented in Fig. 2A showed immunopositivity of ALDH1A1 in the full thickness of the moderately differentiated respiratory epithelium (solid arrow), whereas the invasive portion of the cancer (open arrow) is negative. In this specimen, the stroma and inflammatory infiltrate are both negative. In ovarian adenocarcinoma, 9 out of 34 (26.5%) samples were immunopositive for ALDH1A1. The staining was of low intensity (I = 1.4 ± 0.2) and the extensiveness was 21%, giving a total expression score of 36 ± 16. One case presented in Fig. 2A showed a poorly differentiated ovarian adenocarcinoma with a low level of immunopositivity of ALDH1A1 (solid arrow). The stroma in this specimen was negative. In breast adenocarcinoma, out of a total of 34 evaluated specimens, we only noted one positively stained sample (3%), which had a low level of 1A1 immunopositivity in about 25% of cancer cells. This one case (not shown) was a moderately differentiated ductal adenocarcinoma with moderate metastasis.
ALDH1B1 expression (Table 2 & Fig. 2B)
In colonic adenocarcinoma, 39 out of 40 (97.5%) samples were IHC-positive for ALDH1B1. The staining was intense (I = 2.8 ± 0.5) and extensive (E = 99 ± 3), giving an overall expression score of 284 ± 7. Two cases shown in Fig. 2B represented a moderately differentiated (left) and a well differentiated (right) adenocarcinoma. In both cases, ALDH1B1 stain was very intense and exhibited a punctate pattern in all the cancer cells. The stroma showed no staining (left) or some of the stromal cells including histiocytes were positive (right). In lung adenocarcinoma, 23 out of 30 (77%) cases showed immunopositivity of ALDH1B1. The staining intensity was moderate (I = 1.9 ± 0.2) but found in 84 ± 6% of cancer cells. The overall expression score was 165 ± 20. In addition, tumors that were well or moderately differentiated or less aggressive tended to have a higher incidence of ALDH1B1-positive staining. The same lung cancer specimen shown in Fig.3A is presented here in Fig. 2B, but with ALDH1B1 staining. A strong punctate ALDH1B1 immunostaining was noted in all the cancer cells of this moderately well differentiated lung adenocarcinoma. Some of the stromal cells also stained positively, but did so much less intensely. In ovarian adenocarcinoma, 28 out of 33 (85%) cases were immunopositive for ALDH1B1. The staining intensity was moderate to high (I = 2.3 ± 0.1) and quite extensive (E = 94 ± 3), giving a final expression score of 215 ± 14. One case shown in Fig. 2B was a poorly differentiated ovarian adenocarcinoma, in which all of the cancer cells showed an intense positive punctate staining. Only a few cells of the stromal compartment were positive, some of which appeared to be histiocytes. In breast adenocarcinoma, 20 out of 33 (61%) samples were found to be positively stained for ALDH1B1 with a moderate intensity (I = 1.9 ± 0.2), but present in the majority of the cancer cells (E = 87 ± 5). The overall expression score was 171 ± 22. One case presented in Fig. 2B showed a poorly differentiated breast adenocarcinoma, in which a large proportion of the cancer cells were positively stained. In this specimen, the stromal tissue and inflammatory infiltrate were negative. In these 163 adenocarcinoma samples analyzed, the overall expression score for ALDH1B1 (~208) was 5.6-fold higher than that for ALDH1A1 (~37); in addition, the ALDH1B1 staining did not show any trend of association with the tumor histological grade or metastatic level.
Discussion
ALDH1A1 and ALDH1B1 belong to the family 1 of ALDH superfamily. ALDH1A1 resides in the cytoplasm, whereas ALDH1B1 is predicted to be a mitochondrial enzyme due to a 19-residue mitochondrial signal sequence at its N-terminus (14). In recent years, enhanced expression of ALDH1A1 in cancers has been reported and it is proposed that the metabolic function of ALDH1A1 contributes to the cancer development (2). When compared to ALDH1A1, much less is known about the physiological/pathological roles of ALDH1B1. Herein, we report the expression profile of ALDH1B1 in normal tissues and 163 adenocarcinoma specimens from human colon, lung, breast and ovary. In normal tissues of lung, breast and ovary, ALDH1B1 was expressed at moderate to high levels in the parenchymal cells and at low levels in the stroma. In the colon, however, the expression of ALDH1B1 appeared to be confined to a small number of undifferentiated cells that are consistent with being the stem cell compartment. Most strikingly, ALDH1B1 was abundantly expressed in adenocarcinomas originating from these tissues and particularly in colonic adenocarcinoma, where extensive staining of ALDH1B1 was observed in 98% tumor samples. Given that only a minor proportion of cells stained positively for ALDH1B1 in the normal colon, this increase in ALDH1B1 expression in colonic cancer tissues is quite dramatic.
Using the same set of samples, we also examined the expression of ALDH1A1. In normal tissues, ALDH1A1-positive cells were rare (in the breast) or represent a small number of cells (in the colon, ovary and lung). In adenocarcinomas of colon, lung and ovary, approximately 30% tumors showed immunopositivity for ALDH1A1 and among them 20–30% of cancer cells were positively stained. Interestingly, in the 34 breast adenocarcinoma specimens examined in this study, only one case revealed positive staining for ALDH1A1. Taken together, our data are in agreement with other reports (7;9;10). More importantly, in the present study, we have observed that the expression of ALDH1B1 is predominant relative to that of ALDH1A1 in human adenocarcinoma. It should be noted that the anti-ALDH1B1 antibody does have some degree of cross-reactivity towards recombinant ALDH1A1 protein (See Supplementary Material). However, the IHC signal of ALDH1B1 in both normal and cancerous tissues exhibits a punctate pattern which is indicative of organelle-associated expression, rather than diffuse cytoplasmic staining. Therefore, we believe that the detected IHC signal using this antibody represents the true staining for ALDH1B1 protein.
It has been shown that a subset of cells from normal or cancer tissues that are selected by high ALDH activity using the ALDEFLUOR assay possess the features of stem cells (6–12;18). To date, only ALDH1A1 has been tested for the ALDEFLUOR assay (3), whereas the catalytic characteristics of other ALDH isozymes in the ALDEFLUOR assay are not yet defined. Among the nineteen human ALDH isozymes, a few of them have been characterized biochemically (2;14;19). While these ALDHs exhibit distinct substrate specificity, they also show an overlapping spectrum for aldehyde substrates. This current study as well as others (11;20) have shown that upregulation of ALDH isozymes, other than ALDH1A1, occur in certain types of human cancer. It is therefore worthy of speculation that the selective expression of ALDH isozyme(s) in malignant tissues may be organ- or tumor-specific. Identifying the tumor-specific ALDH isozyme(s) and understanding their mechanistic roles in tumor formation warrants future studies.
The observation that ALDH1B1 is dramatically induced in colonic adenocarcinoma is of particular interest. First, the distribution of ALDH1B1 IHC-signal in the normal colon suggests that ALDH1B1 protein may be strictly expressed in the stem-like cells. It is proposed that the presence of cells having high ALDH activity in stem cell compartments of various tissues is associated with ALDH-mediated RA signaling in regulating the cellular differentiation (5). To date, the catalytic capacity of ALDH1B1 in converting retinol to RA is unclear. Interestingly, a correlation between the dynamic expression of ALDH1B1 and the granulocytic development of hematopoietic stem cells has been reported (21). Future studies aiming at understanding the metabolic roles of ALDH1B1 in RA metabolism are warranted. Second, in colonic adenocarcinoma, almost the entire tumor parenchyma is populated by ALDH1B1-positive cells. This phenomenon could be explained by ALDH1B1-positive cells of the normal colon serving as progenitors for malignant cells during tumorigenesis. Studies to test this hypothesis will provide important information regarding a potential role of ALDH1B1 in colon CSCs. Alternatively, during the process of tumor formation in the colon, ALDH1B1 expression is turned on in malignant cells and is favorable for cellular survival. It is known that chronic alcohol consumption is a strong risk factor for colon cancer, because the ethanol metabolite acetaldehyde is highly carcinogenic (22). Given that ALDH1B1 plays an active role in acetaldehyde metabolism (14), it is not unlikely that the aberrant expression of ALDH1B1 in colon cancer cells is associated with one of the major etiological factors for colon cancer. Regardless of the underlying mechanisms, the dramatic induction of ALDH1B1 protein specifically in colon adenocarcinoma may have great clinical implication, making this ALDH isozyme a promising biomarker for human colon cancer.
Research Highlights.
ALDH1B1 and ALDH1A1 are differentially expressed in normal human tissues.
ALDH1B1 is expressed at higher levels than ALDH1A1 in human epithelial cancers.
In normal human colon ALDH1B1 is expressed strictly in the stem cell compartments.
In human colonic adenocarcinoma ALDH1B1 is intensively expressed in all cancer cells.
Supplementary Material
Acknowledgement
This work was supported by National Institutes of Health (NIH) grants AA017754.
Abbreviations
- ALDH
Aldehyde dehydrogenase
- LPO
lipid peroxidation
- BAAA
BODIPY aminoacetaldehyde
- RA
retinoic acid
- HSC
hematopoietic stem cell
- CSC
cancer stem cell
- TARP
Tissue Array Research Program
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- 1.Vasiliou V, Pappa A, Estey T. Role of human aldehyde dehydrogenases in endobiotic and xenobiotic metabolism. Drug Metab Rev. 2004;36:279–299. doi: 10.1081/dmr-120034001. [DOI] [PubMed] [Google Scholar]
- 2.Marchitti SA, Brocker C, Stagos D, et al. Non-P450 aldehyde oxidizing enzymes: the aldehyde dehydrogenase superfamily. Expert.Opin.Drug Metab Toxicol. 2008;4:697–720. doi: 10.1517/17425250802102627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Storms RW, Trujillo AP, Springer JB, et al. Isolation of primitive human hematopoietic progenitors on the basis of aldehyde dehydrogenase activity. Proc.Natl.Acad.Sci.U.S.A. 1999;96:9118–9123. doi: 10.1073/pnas.96.16.9118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Purton E. Roles of retinoids and retinoic acid receptors in the regulation of hematopoietic stem cell self-renewal and differentiation. PPAR.Res. 2007;2007:87934. doi: 10.1155/2007/87934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chute JP, Muramoto GG, Whitesides J, et al. Inhibition of aldehyde dehydrogenase and retinoid signaling induces the expansion of human hematopoietic stem cells. Proc.Natl.Acad.Sci.U.S.A. 2006;103:11707–11712. doi: 10.1073/pnas.0603806103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ran D, Schubert M, Pietsch L, et al. Aldehyde dehydrogenase activity among primary leukemia cells is associated with stem cell features and correlates with adverse clinical outcomes. Exp.Hematol. 2009;37:1423–1434. doi: 10.1016/j.exphem.2009.10.001. [DOI] [PubMed] [Google Scholar]
- 7.Ginestier C, Hur MH, Charafe-Jauffret E, et al. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell. 2007;1:555–567. doi: 10.1016/j.stem.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ucar D, Cogle CR, Zucali JR, et al. Aldehyde dehydrogenase activity as a functional marker for lung cancer. Chem.Biol.Interact. 2009;178:48–55. doi: 10.1016/j.cbi.2008.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deng S, Yang X, Lassus H, et al. Distinct expression levels and patterns of stem cell marker, aldehyde dehydrogenase isoform 1 (ALDH1), in human epithelial cancers. PLoS.One. 2010;5:e10277. doi: 10.1371/journal.pone.0010277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang EH, Hynes MJ, Zhang T, et al. Aldehyde dehydrogenase 1 is a marker for normal and malignant human colonic stem cells (SC) and tracks SC overpopulation during colon tumorigenesis. Cancer Res. 2009;69:3382–3389. doi: 10.1158/0008-5472.CAN-08-4418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van den HC, van der HG, Cheung H, et al. High aldehyde dehydrogenase activity identifies tumor-initiating and metastasis-initiating cells in human prostate cancer. Cancer Res. 2010;70:5163–5173. doi: 10.1158/0008-5472.CAN-09-3806. [DOI] [PubMed] [Google Scholar]
- 12.Dembinski JL, Krauss S. Characterization and functional analysis of a slow cycling stem cell-like subpopulation in pancreas adenocarcinoma. Clin.Exp.Metastasis. 2009;26:611–623. doi: 10.1007/s10585-009-9260-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tanei T, Morimoto K, Shimazu K, et al. Association of breast cancer stem cells identified by aldehyde dehydrogenase 1 expression with resistance to sequential Paclitaxel and epirubicin-based chemotherapy for breast cancers. Clin.Cancer Res. 2009;15:4234–4241. doi: 10.1158/1078-0432.CCR-08-1479. [DOI] [PubMed] [Google Scholar]
- 14.Stagos D, Chen Y, Brocker C, et al. Aldehyde dehydrogenase 1B1: molecular cloning and characterization of a novel mitochondrial acetaldehyde-metabolizing enzyme. Drug Metab Dispos. 2010;38:1679–1687. doi: 10.1124/dmd.110.034678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coskran TM, Morton D, Menniti FS, et al. Immunohistochemical localization of phosphodiesterase 10A in multiple mammalian species. J.Histochem.Cytochem. 2006;54:1205–1213. doi: 10.1369/jhc.6A6930.2006. [DOI] [PubMed] [Google Scholar]
- 16.Patel M, Lu L, Zander DS, et al. ALDH1A1 and ALDH3A1 expression in lung cancers: correlation with histologic type and potential precursors. Lung Cancer. 2008;59:340–349. doi: 10.1016/j.lungcan.2007.08.033. [DOI] [PubMed] [Google Scholar]
- 17.Li T, Su Y, Mei Y, et al. ALDH1A1 is a marker for malignant prostate stem cells and predictor of prostate cancer patients' outcome. Lab Invest. 2010;90:234–244. doi: 10.1038/labinvest.2009.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen YC, Chen YW, Hsu HS, et al. Aldehyde dehydrogenase 1 is a putative marker for cancer stem cells in head and neck squamous cancer. Biochem.Biophys.Res.Commun. 2009;385:307–313. doi: 10.1016/j.bbrc.2009.05.048. [DOI] [PubMed] [Google Scholar]
- 19.Brocker C, Lassen N, Estey T, et al. Aldehyde dehydrogenase 7A1 (ALDH7A1) is a novel enzyme involved in cellular defense against hyperosmotic stress. J.Biol.Chem. 2010;285:18452–18463. doi: 10.1074/jbc.M109.077925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marchitti SA, Orlicky DJ, Brocker C, et al. Aldehyde dehydrogenase 3B1 (ALDH3B1): immunohistochemical tissue distribution and cellular-specific localization in normal and cancerous human tissues. J.Histochem.Cytochem. 2010;58:765–783. doi: 10.1369/jhc.2010.955773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luo P, Wang A, Payne KJ, et al. Intrinsic retinoic acid receptor alpha-cyclin-dependent kinase-activating kinase signaling involves coordination of the restricted proliferation and granulocytic differentiation of human hematopoietic stem cells. Stem Cells. 2007;25:2628–2637. doi: 10.1634/stemcells.2007-0264. [DOI] [PubMed] [Google Scholar]
- 22.Seitz HK, Becker P. Alcohol metabolism and cancer risk. Alcohol Res.Health. 2007;30 38-7. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.