Abstract
Background
Amodiaquine (AQ) along with sulphadoxine-pyrimethamine (SP) offers effective and cheaper treatment against chloroquine-resistant falciparum malaria in many parts of sub-Saharan Africa. Considering the previous history of hepatitis, agranulocytosis and neutrocytopenia associated with AQ monotherapy, it becomes imperative to study the toxicity of co-administration of AQ and SP. In this study, toxicity and resulting global differential gene expression was analyzed following exposure to these drugs in experimental Swiss mice.
Methods
The conventional markers of toxicity in serum, oxidative stress parameters in tissue homogenates, histology of liver and alterations in global transcriptomic expression were evaluated to study the toxic effects of AQ and SP in isolation and in combination.
Results
The combination therapy of AQ and SP results in more pronounced hepatotoxicity as revealed by elevated level of serum ALT, AST with respect to their individual drug exposure regimen. Furthermore, alterations in the activity of major antioxidant enzymes (glutathione peroxidase, superoxide dismutase, catalase, glutathione reductase), indicating the development of oxidative stress, was more significant in AQ+SP combination therapy. cDNA microarray results too showed considerably more perturbed gene expression following combination therapy of AQ and SP as compared to their individual drug treatment. Moreover, a set of genes were identified whose expression pattern can be further investigated for identifying a good biomarker for potential anti-malarial hepatotoxicity.
Conclusion
These observations clearly indicate AQ+SP combination therapy is hepatotoxic in experimental Swiss mice. Microarray results provide a considerable number of potential biomarkers of anti-malarial drug toxicity. These findings hence will be useful for future drug toxicity studies, albeit implications of this study in clinical conditions need to be monitored with cautions.
Background
Malaria remains to be the major killer disease in the developing countries that affects lives of more than 500 million people and kills about two million of them annually [1]. Most of the drugs that are used to treat malaria can be broadly grouped into 4-aminoquinolines, 8-aminoquinolines, anti-folates, artemisinin derivatives, hydroxyl naphthoquinones and certain class of antibiotics, such as doxycycline and clindamycin. 4-aminoquiniline derivatives, such as chloroquine and amodiaquine (AQ), have been the first-line drugs against malaria for past several decades. Development of resistance against these drugs in several parts of world necessitated the use of other drugs along with it for efficient treatment. Malaria treatment guidelines issued by WHO also recommends the use of AQ and SP combinations for the treatment of chloroquine-resistant malaria [2-4]. Many clinical trials and field studies, carried primarily in African countries, showed that AQ in combination with SP was very effective in controlling cases of malaria [5-7]. Although resistance against sulphadoxine-pyrimethamine has also been reported in parts of East Africa [8-10], it remains a good choice for rest of the world, including West Africa [11]. Notwithstanding their utility in controlling malaria, most of these anti-malarials are also associated with risk of drug-induced toxicity [12-14]. In spite of wide use of AQ and SP as anti-malarials, there is dearth of scientific literature describing their potential toxicity [15].
Liver is a vital organ of body and mainly involved in drug metabolism and its biotransformation. Its unique position and crucial link with gastro-intestinal tract renders it highly vulnerable to drug induced toxicity [16,17]. Previously, Noel et al. had described toxicity and gene expression alterations in murine liver following exposure to the anti-relapse anti-malarial drugs primaquine [18] and bulaquine [19]. High throughput gene expression profiling facilitates prediction of toxicity and interpretation of mechanism of toxicity based on distinct gene expression changes. The simplest approach to identify genes of potential interest through several related experiments is to search for those that are consistently either up- or down-regulated [18-20]. Therefore, an attempt was made to delineate the mechanism of anti-malarial drug toxicity in liver tissue following exposure to AQ and SP combination in murine models.
Methods
Animal groups, drug administration and tissue collection
10-12 weeks old, male Swiss albino mice (Mus musculus), weighing 25-30 g (Central Drug Research Institute, Lucknow, India) were randomly assigned to control and treatment groups. All animal procedures were performed following IAEC approval (115/07/Toxicol./IAEC dated 11.9.2007) and in compliance to institutional animal ethics guidelines. The animals were acclimated to optimal conditions of temperature (25 ± 2°C) and light/dark cycle (12 h each) before initiation of drug administration. The doses for AQ and SP were calculated from human therapeutic doses [21] based on equivalent body surface area index [22]. The duration of dosing in mice was also similar to the human therapeutic regime. Animals were divided into four groups each consisting of six animals and were given following dosages orally.
Group 1: 1% DMSO-treated controls, for three consecutive days
Group 2: AQ, 120 mg/kg for three consecutive days
Group 3: 300 mg/kg sulphadoxine and 15 mg/kg pyrimethamine on day one
Group 4: 120 mg/kg AQ and SP, 300 mg/kg and 15 mg/kg respectively, on day one followed by 120 mg/kg AQ, on day two and three
All animals were sacrificed by cervical dislocation on day four of study and liver was taken out after perfusion with normal saline and a part of it is kept at -70°C until further analysis. Prior to sacrifice blood was taken out from cardiac puncture from each animal and left undisturbed for 30 minutes for serum separation. A part of liver tissue was immediately fixed in 10% formal saline for histological investigations.
Serum biochemistry and liver histology
Alanine aminotransferase (ALT), aspartate aminotransferase (AST) [markers of hepatotoxicity] levels were estimated in the serum with automated biochemical analyzer using the kits (Beckmann). Fixed liver tissues were washed overnight, dehydrated through graded alcohols and embedded in paraffin wax. Serial sections of 5 μm thickness were stained with haematoxylin and eosin (H&E) for histological examination.
Biochemical estimation of antioxidant enzymes in liver tissue fraction
Markers of oxidative stress {tissue levels of lipid peroxidation; LPO [a measure of malondialdehyde (MDA) concentration] and reduced glutathione level; GSH} and enzyme activities of major antioxidant enzymes (glutathione peroxidase, superoxide dismutase, catalase, glutathione reductase) were estimated in liver tissue homogenates using standard tests [23-27].
RNA isolation, cDNA labeling and hybridization
50 mg frozen liver tissue was crushed in liquid nitrogen and immediately homogenized (Heidolph, Germany) in 1 ml of TRI reagent (Sigma, St. Louis, MO, USA) to isolate total RNA. RNA samples with approximately 2:1 ratio of 28S:18S rRNA and 260/280 values ≥ 1.8 were used for gene expression analysis. Equal amount of RNA from individuals of the same group was pooled to eliminate inter-individual variations. 25 μg of pooled RNA was converted into labeled cDNA using CyScribe First Strand cDNA-labeling kit (Amersham, Buckinghamshire, UK) following manufacturer's protocol. Labeled cDNA was purified with GFX columns as per manufacturer's guidelines and subsequently concentrated by evaporation under vacuum after estimating the percent incorporation of the dyes with a spectrophotometer (Thermo, Waltham, MA, USA). Dye swap technical replicate experiments were performed with aliquots of same RNA preparation to address inconsistencies regarding dye incorporation and other technical means of variance. The Cy5- and Cy3-labeled cDNA samples were mixed in CyScribe Hyb buffer (Amersham, Buckinghamshire, UK) containing 10 μg/ml sheared salmon sperm DNA and 10 μg/ml yeast tRNA (Ambion, Austin, Texas, USA) as blocking agents. The labeled sample was hybridized to mouse 22.4k arrays [28] for 18 h at 42°C.
Scanning and microarray data analysis
The arrays were washed and subsequently scanned to collect raw data with Array Scanner III supported with Image-Quant version 5 (Molecular Dynamics). Intensity values were extracted from the scanned images with ArrayVision version 8 (Imaging research, GE healthcare Biosciences Corp., Piscataway, NJ, USA). Raw intensity data was analyzed with Avadis Express version 4.3 (Strand life Sciences, Bangalore, India) and the background corrected intensities were LOWESS normalized (Cy5 against Cy3) to obtain log (base 2) ratios. Furthermore, log2 values of duplicate spots were averaged in order to get a single mean value to perform k-means clustering with MeV version 3.1 [TM4, The Institute of Genomic Research [29]]. Each expression cluster was further clustered hierarchically with Euclidean distance matrix and average linkage to identify gene with similar expression patterns. Raw and log transformed data (series accession no. GSE 17392) has been submitted to Gene Expression Omnibus database [30] and conforms to MIAME guidelines developed by microarray gene expression data (MGED) society.
Real time-PCR
mRNA was reverse transcribed according to the manufacturer's instruction (First Strand cDNA Synthesis Kit for RT-PCR, Invitrogen, California, USA). PCRs were performed on a Light Cycler 480 System (Roche Diagnostics) in 96-well plates. Each reaction was carried out in 20 μl reaction volume comprising of SYBR Green qPCR Master Mix (Invitrogen, California, USA), cDNA template, 200 nM of forward and reverse primers and nuclease-free water. Serial dilutions of genomic DNA (250-0.08 ng) were used to generate a quantitative PCR standard curve. The LightCycler protocol was: 2 min. of UDG incubation (Invitrogen, California USA) at 50°C followed by 10 min. of 95°C hot-start enzyme activation; 40 cycles of 95°C denaturation for 15 s, 60°C annealing and elongation. Melting curve analysis temperatures were 95°C for 5 s, 70°C for 60 s, and then heating to 95°C. Water was used as the template for negative control amplifications included with each PCR. Data were analyzed using the Roche LightCycler 480 software and Cp was calculated by the Second Derivate Maximum Method [31]. The amount of the target mRNA was examined and normalized to the GAPDH gene mRNA. The relative expression ratio of a target gene was calculated as described by Pfaffl [32], based on real-time PCR efficiencies. Results reported were obtained from at least three biological replicates and PCR runs were repeated at least twice.
Statistical analysis
Data were expressed as the mean ± standard error of the means (S.E.M.). Group means were compared by one-way analysis of variance (ANOVA) with Newman-Keuls post analysis test. The differences in the data obtained were considered statistically significant when the P-value was less than 0.05. All statistical analysis was done through using Prism ver.5 (GraphPad Software Inc., USA).
Results
Effect of amodiaquine and sulphadoxine-pyrimethamine treatment on the biomarkers of hepatotoxicity and oxidative stress
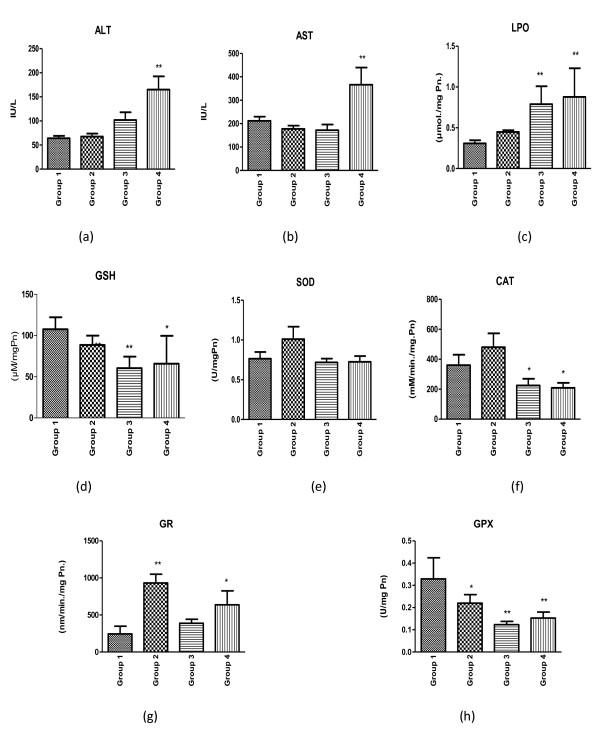
Treatment of AQ at 120 mg/kg does not impart hepatotoxicity or oxidative stress, as levels of ALT, AST, LPO and GSH were comparable to that of untreated control. Although administration of SP does not cause any elevation in level of ALT or AST, it causes appreciable oxidative stress, as a significant elevation in LPO and a decrease in GSH were observed in mice dosed with SP. Interestingly, co-administration of AQ and SP (i.e. AQ+SP) causes both hepatotoxicity as well as oxidative stress as evident from marked increase in ALT, AST, LPO and decrease in GSH (Figure 1a, b, c and 1d).
Figure 1.
(a - h) Assessment of markers of hepatotoxicity and oxidative stress following exposure of AQ, SP and AQ+SP. Group 1: untreated control; Group 2: treated with AQ, 120 mg/kg body wt[AQ]; Group 3: treated with sulphadoxine (300 mg/kg) and pyrimethamine (15 mg/kg) [SP]; Group 4: Co-treatment of AQ and SP[AQ+SP]; [*(P < 0.05), ** (P < 0.01), *** (P < 0.001)].
Effect of amodiaquine and sulphadoxine-pyrimethamine treatment on antioxidant enzymes in liver tissue fraction
Effects of AQ and SP treatment on enzymatic activities of SOD, catalase, GR and GPx, which are the major antioxidant enzymes in liver tissue fraction, were investigated. SOD activity was not altered after AQ and SP treatment, while catalase and GPx activities were drastically reduced by the treatment of SP and AQ+SP. However, AQ administration did cause a moderate, statistically non-significant, increase in the activity of SOD and catalase. However, activity of GR was increased by administration of AQ and AQ+SP combination (Figure 1e, f, g and 1h).
cDNA Microarray analysis of differential gene expression in murine liver and kidney exposed to anti-malarials amodiaquine and sulphadoxine-pyrimethamine
Following AQ administration in murine liver, a total of 133 probes were differentially regulated, of which 60 were up-regulated and 73 down-regulated. Some of these are listed in Table 1. Major important up-regulated probes following AQ dosing included the TAP binding gene involved in antigen processing, the neogenin gene involved in ATP binding, the dihydropyrimidinase like 5 gene involved in axon guidance, the ankyrin repeat domain 6 gene involved in DNA binding and genes for GATA binding protein 2 involved in DNA binding and transcription. Some of the important down-regulated probes following AQ administration included the DEAD box polypeptide 6 gene involved in ATP-dependent helicase activity, the voltage dependent calcium channel L type alpha 1 C subunit gene involved in calcium channel activity, the lipoma HMGIC fusion partner-like 2 gene involved in general metabolism, and the GCN5 gene involved in N-acetyl transferase activity.
Table 1.
List of important differentially expressed probes after administration of AQ in murine Liver.
| Spot labels | Fold Change | Gene Name/Description | GO : Biological function |
|---|---|---|---|
| 11544 | 2.27 | TAP binding protein | antigen processing |
| 20523 | 2.73 | RAB39B, member RAS oncogene family | GTP binding |
| 18680 | 2.49 | Neogenin | ATP binding |
| 11260 | 2.18 | ATPase, Ca++ transporting, plasma membrane 1 | ATP binding |
| 12910 | 2.20 | Dihydropyrimidinase-like 5 | axon guidance |
| 18325 | 3.18 | Calcium channel, voltage-dependent, L type, alpha 1D subunit | calcium channel activity |
| 9921 | 3.51 | Ankyrin repeat domain 6 | DNA binding |
| 18064 | 6.11 | GATA binding protein 2 | DNA binding |
| 20032 | 2.51 | Parathyroid hormone receptor 1 | G-protein coupled receptor activity |
| 12886 | 3.62 | Transmembrane protein with EGF-like and two follistatin-like domains 1 | growth factor activity |
| 21365 | 3.33 | Solute carrier family 38, member 1 | L-glutamine transport |
| 492 | 6.38 | DNA segment, Chr 5, Wayne State University 178, expressed | phospholipid biosynthesis |
| 22462 | -4.53 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 6 | ATP-dependent helicase activity |
| 10941 | -2.32 | Calcium channel, voltage-dependent, L type, alpha 1C subunit | calcium channel activity |
| 2989 | -2.69 | Nucleoporin 153 | DNA binding |
| 22225 | -4.12 | Suppressor of variegation 4-20 homolog 1 (Drosophila) | histone lysine N-methyltransferase activity |
| 15707 | -2.09 | Lipoma HMGIC fusion partner-like 2 | metabolism |
| 9653 | -2.55 | GCN5 general control of amino acid synthesis-like 2 (yeast) | N-acetyltransferase activity |
| 7818 | -2.15 | Nuclear receptor subfamily 3, group C, member 2 | transcription factor activity |
Fold change (FC) >2(Up-regulated) and FC<-2 (Down-regulated) and P < 0.01.
Administration of SP in murine liver leads to differential regulation of 156 probes of which 90 were up-regulated and 66 down-regulated, some of which are listed in Table 2. Some of the important up-regulated probes following SP treatment included the DEAH box polypeptide 15 gene involved in ATP-dependent helicase activity, the transketolase gene involved in calcium ion binding, the procollagen type VI alpha 2 genes mainly involved in cell adhesion, the procollagen lysine 2-oxoglutarate 5-dioxygenase 2 genes involved in endopeptidase inhibitor activity and a gene coding for RNA binding motif protein X. Major down-regulated probes following SP administration included the CDC42 effector protein 1 (Rho GTPase binding) involved in signal transduction, the serine (or cysteine) peptidase inhibitor clade B member 6a gene involved in endopeptidase inhibitor activity and the cytochrome c oxidase subunit VIb polypeptide 2 gene involved in electron transfer.
Table 2.
List of Important differentially expressed probes after administration of SP in murine Liver.
| Spot labels | Fold Change | Gene Name/Description | GO : Biological function |
|---|---|---|---|
| 5638 | 2.01 | Histone deacetylase 9 | Histone deacetylase activity |
| 728 | 2.04 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 42 | ATP binding |
| 15419 | 2.33 | DEAH (Asp-Glu-Ala-His) box polypeptide 15 | ATP-dependent helicase activity |
| 1745 | 2.66 | Transketolase | Calcium ion binding |
| 14698 | 2.76 | Procollagen, type VI, alpha 2 | Cell adhesion |
| 17252 | 2.02 | Procollagen lysine, 2-oxoglutarate 5-dioxygenase 2 | Endopeptidase inhibitor activity |
| 2107 | 2.62 | Lymphocyte antigen 6 complex, locus G6C | Extracellular space |
| 14620 | 2.11 | RNA binding motif protein, X chromosome retrogene | RNA binding |
| 21645 | 2.11 | RNA binding motif protein 28 | RNA binding |
| 12990 | 2.11 | Eukaryotic translation initiation factor 1 | Translation factor activity |
| 21844 | -3.44 | CDC42 effector protein (Rho GTPase binding) 1 | Signal transduction |
| 8922 | -2.20 | Serine (or cysteine) peptidase inhibitor, clade B, member 6a | Endopeptidase inhibitor activity |
| 5874 | -7.13 | Cytochrome c oxidase subunit VIb polypeptide 2 | Unknown |
| 6344 | -5.56 | Tumor protein D52 | ------------- |
| 11499 | -2.21 | Unknown | ------------- |
| 11550 | -2.54 | RIKEN cDNA 4930471M23 gene | ------------- |
| 11891 | -2.26 | TBC1 domain family, member 19 | ------------- |
Fold change (FC) >2(Up regulated) and FC<-2 (Down regulated) and P < 0.01.
Co-administration of AQ and SP for three consecutive days resulted in differential regulation of 231 probes, including 118 up-regulated and 113 down-regulated probes (Table 3). Major up-regulated probes following co-exposure of AQ and SP included genes having a cysteine and histidine-rich domain (CHORD) containing zinc-binding protein 1, mainly involved in calcium ion binding, the solute carrier family 28 (sodium-coupled nucleoside transporter) member 3 gene involved in ion transport, the Kelch-like 2 Mayven (Drosophila) gene involved in actin binding, the integrin beta 8 gene involved in cell adhesion, the gene for suppression of tumorigenicity (colon carcinoma) involved in cell migration and mortality factor 4 like 1 gene involved in cell proliferation. Some of the major down-regulated probes following AQ+SP treatment were the lysophospholipase 3 gene involved in acyltransferase activity, the mitogen-activated protein kinase 14 gene involved in ATP binding, the transforming growth factor beta receptor I gene involved in ATP binding, the procollagen type VI alpha3 gene involved in cell adhesion, the gene for microfibrillar-associated protein 4 involved mainly in cell adhesion, the BTB and CNC homology 2 genes involved in DNA binding and the CXXC finger 1 (PHD domain) gene also involved in DNA binding.
Table 3.
List of Important differentially expressed probes after administration of AQ+SP in murine Liver.
| Spot labels | Fold Change |
Gene Name/Description | GO: Biological Function |
|---|---|---|---|
| 3557 | 2.03 | Cysteine and histidine-rich domain (CHORD)-containing, zinc-binding protein 1 | Calcium ion binding |
| 1410 | 3.02 | Solute carrier family 28 (sodium-coupled nucleoside transporter), member 3 | Integral to plasma membrane |
| 16564 | 2.50 | Kelch-like 2, Mayven (Drosophila) | Actin binding |
| 5639 | 2.51 | Integrin beta 8 | Cell adhesion |
| 10049 | 2.49 | Suppression of tumorigenicity 14 (colon carcinoma) | Cell migration |
| 21956 | 2.25 | Mortality factor 4 like 1 | Cell proliferation |
| 14809 | 2.61 | Zinc finger, SWIM domain containing 4 | Cellular component |
| 21155 | 2.42 | Metal response element binding transcription factor 1 | DNA binding |
| 21758 | 3.61 | Regulatory factor X, 3 (influences HLA class II expression) | DNA binding |
| 1649 | 5.71 | Polymerase (RNA) III (DNA directed) polypeptide F | DNA binding |
| 20006 | 3.53 | Cytochrome b5 type B | Electron transport |
| 12949 | 2.47 | GTP binding protein (gene overexpressed in skeletal muscle) | GTP binding |
| 492 | 2.47 | DNA segment, Chr 5, Wayne State University 178, expressed | Integral to membrane |
| 11468 | 3.82 | DNA segment, Chr 18, ERATO Doi 653, expressed | Integral to membrane |
| 9206 | 2.18 | Dystrobrevin binding protein 1 | Muscle development |
| 12725 | 3.03 | Dolichyl-di-phosphooligosaccharide-protein glycotransferase | N-linked glycosylation via asparagine |
| 14957 | 3.64 | Adenosine deaminase, RNA-specific, B2 | RNA binding |
| 15208 | 2.70 | Solute carrier family 25 (mitochondrial carrier, glutamate), member 22 | Transporter activity |
| 22601 | 3.18 | Ubiquitin-conjugating enzyme E2D 2 | Ubiquitin-dependent protein catabolism |
| 10183 | -2.83 | Lysophospholipase 3 | Acyltransferase activity |
| 6999 | -3.22 | Mitogen activated protein kinase 14 | ATP binding |
| 22383 | -2.63 | Transforming growth factor, beta receptor I | ATP binding |
| 2877 | -2.03 | Procollagen, type VI, alpha 3 | Cell adhesion |
| 18522 | -3.11 | Microfibrillar-associated protein 4 | Cell adhesion |
| 874 | -3.24 | BTB and CNC homology 2 | DNA binding |
| 11690 | -8.17 | CXXC finger 1 (PHD domain) | DNA binding |
| 13862 | -2.05 | GLI-Kruppel family member GLI3 | DNA binding |
| 19141 | -2.44 | Protein disulfide isomerase associated 6 | DNA binding |
| 17585 | -2.07 | AT rich interactive domain 5B (Mrf1 like) | DNA binding |
| 18686 | -3.25 | Cytochrome b5 type B | Electron transport |
| 16362 | -3.08 | Proteasome (prosome, macropain) subunit, beta type 2 | Endopeptidase activity |
| 20020 | -4.67 | Phosphatidylinositol 3-kinase, C2 domain containing, alpha polypeptide | Glycerophospholipid metabolism |
| 13958 | -2.83 | RAS related protein 1b | GTP binding |
| 11531 | -3.27 | Zinc metallopeptidase, STE24 homolog (S. cerevisiae) | Hydrolase activity |
| 13203 | -2.64 | SH2-B PH domain containing signaling mediator 1 | Intracellular signaling cascade |
| 19040 | -2.26 | Malate dehydrogenase 2, NAD (mitochondrial) | Malate dehydrogenase activity |
| 5575 | -2.06 | Ring finger protein (C3HC4 type) 19 | Protein ubiquitination |
| 6605 | -3.30 | Cleavage and polyadenylation specific factor 4 | RNA binding |
Fold change (FC) >2(Up regulated) and FC<-2 (Down regulated) and P < 0.01.
Real time quantitative PCR analysis showed that most of genes that are differentially expressed in microarray produced similar results in PCR too, i.e. the genes which are up-regulated in microarray are up-regulated in real time PCR too and vice versa (Table 4).
Table 4.
List of genes with their description and expression results by Q-PCR and microarray following treatment with AQ and SP in murine liver.
| Gene Symbol | Gene Name/Description | Q-PCR fold change | Microarray Result Up regulated(▲)/Down regulated (▼) |
|---|---|---|---|
| ADRA1B | Adrenergic receptor, alpha 1b | -6 | ▲ |
| CYP1A2 | Cytochrome P450, family 1, subfamily a, polypeptide 2 | -6 | ▼ |
| CYP2E1 | Cytochrome P450, family 2, subfamily e, polypeptide 1 | -5 | ▼ |
| SC4MOL | Sterol-C4-methyl oxidase-like | -4 | ▼ |
| H2DM | Histocompatibility 2, class II, | 1.2 | ▲ |
| RAC | RAS-related C3 botulinum | -1.8 | ▼ |
| MCM4 | Minichromosome maintenance deficient 4 homolog | 5 | ▲ |
| VKORC | Vitamin K epoxide reductase complex, | -1.5 | ▼ |
| SC5D | Sterol-C5-desaturase (fungal ERG3, delta-5-desaturase) homolog | 7 | ▲ |
| ADH1 | Alcohol dehydrogenase 1 (class I) | 4.8 | ▲ |
| GADD45 | Growth arrest and DNA-damage-inducible 45 gamma | 3.2 | ▲ |
| UGT2B1 | UDP glucuronosyltransferase 2 family, polypeptide B1 | 5 | ▲ |
| MCM5 | Minichromosome maintenance deficient 5 homolog | 2.5 | ▲ |
| GCGR | Glucagon receptor | 8 | ▲ |
Note the direction similarity among Q-PCR and microarray findings for gene expression results.
Discussion
Amodiaquine and sulphadoxine-pyrimethamine offer a great potential as effective anti-malarial against chloroquine-resistant malaria and has been used in many parts of Africa as first-line anti-malarial treatment. However, considering the previous history of drug-induced hepatitis, oxidative stress associated with these drugs particularly AQ, it becomes imperative to study the toxicity associated with these drugs and their combination in liver tissue.
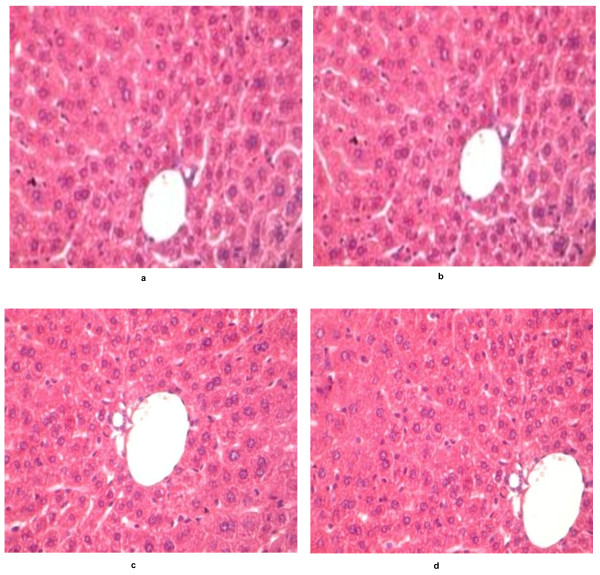
Dosages and duration of AQ and SP treatment in Swiss mice was according to the human therapeutic equivalent dose and malaria treatment regimen suggested by WHO guidelines [4]. This observation was that only co-treatment of AQ and SP (AQ+SP) as recommended combination therapy regimen produces toxicity and not their individual exposure. However, treatment with SP alone does produces appreciable oxidative stress leading to a conclusion that observed hepatotoxicity and oxidative stress in AQ+SP group might be a result of either SP toxicity alone or an additive effect of both these drugs. Interestingly, none of these drugs or drug combinations results in alterations in normal liver histology as no histopathological damage was observed in any sections of liver tissues (Figure 2).
Figure 2.
Murine liver cross-sections treated with amodiaquine and sulphadoxine: (a) untreated control, (b) treated with 120 mg/kg of AQ, (c) treated with 300 mg/kg of sulphadoxine and 15 mg/kg of pyrimethamine (SP), (d) Co-exposure of 120 mg/kg AQ and 300 mg/kg sulphadoxine along with 15 mg/kg of pyrimethamine (AQ+SP).
Previous reports showed that anti-malarials, particularly chloroquine, produce oxidative stress in liver tissue [14], and it was also interesting to study the alteration in antioxidant profile of major antioxidant enzyme present in liver tissue fraction. Results showed that the activity of SOD was not affected either by the treatment of AQ or SP or their combination (AQ+SP). However, the level of GPx was significantly reduced in all three treatment groups and catalase activity was reduced in SP and AQ+SP group in murine liver fraction. The decrease in the activity of GPx observed in this study might be the result of a decrease in GSH content; a measure substrate in GPX catalyzed reaction. Interestingly, GR activity was observed to increase in AQ and AQ+SP. The alterations in activities of antioxidant enzymes of liver observed in the present study were an indication of oxidative injury brought by the AQ and SP dosing.
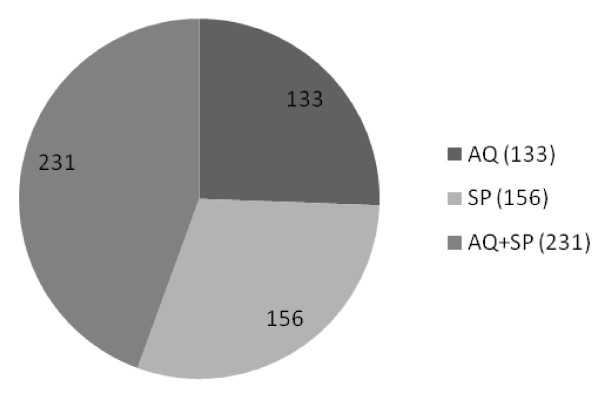
High throughput expression profiling facilitates the prediction and mechanism of toxicity based on distinct gene expression changes. Therefore, the study of differential gene expression in murine liver at high statistical stringency (i.e. P < 0.01 and expression fold change >2) clearly indicated that the molecular mechanism of AQ and SP induced oxidative stress. Furthermore, validation of microarray findings using qRT-PCR further substantiates these results, which is the most sensitive and accurate method for validating microarray-based differential expression of genes [33]. The pattern of differential expression of genes in combination therapy, i.e. the AQ+SP treated groups, were on an expected line with biochemical observations, showing more robust expression pattern than either of the drug given alone. Here the number of differentially expressed probes was 231, far more differentially expressed genes than AQ (133) or SP (156) alone. Of the 231 differentially expressed genes in murine liver after AQ+SP treatment, the number of up-regulated (118) and of down-regulated (113) probes was almost similar (Figure 3).
Figure 3.
Total number of differentially expressed genes following exposure to anti-malarial drugs in murine liver.
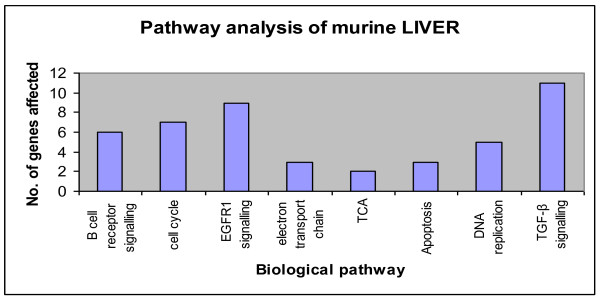
GenMAPP and MAPPFinder tools [34] were utilized to enlist the various biological pathways that are perturbed following exposure to AQ, SP or their combination (AQ+SP). The pathways that are most affected are signaling pathways, carbohydrate metabolism, oxidative stress and drug metabolism (Figure 4). These observations suggest that anti-malarial drug exposure imparts stress in liver tissue causing changes in mRNA expression level of antioxidant pathway and major drug metabolism pathway.
Figure 4.
Important biological pathways regulated by administration of all the three dose categories (AQ, SP and AQ+SP) in murine liver.
One of the many genes that are up-regulated in murine liver following exposure to AQ, SP and their co-treatment i.e. AQ+SP includes EPRS (glutamyl-prolyl tRNA synthetase). EPRS is a multifunctional aminoacyl-tRNA synthetase that catalyzes the aminoacylation of glutamic acid and proline tRNA species [35]. Sampath et al. [36] showed that EPRS has a regulated, noncanonical activity that blocks synthesis of ceruloplasmin. Fall in the level of ceruloplasmin which is the major copper carrier protein, is an indication of hepatic stress [37], so the elevation in the level of EPRS following anti-malarial drug treatment can explain the observed hepatic stress. Supervilin (SVIL) is another gene that is consistently up-regulated in murine liver following exposure to AQ, SP and their combination. This gene codes for a protein, which is tightly associated with both actin filaments and plasma membranes, suggesting that it forms a link between the actin cytoskeleton and the membrane. An up-regulated SVIL (which is required for membrane integrity) following drug treatment may be an explanation for the rise in lipid peroxidation level observed in the present study. It appears that membrane damage following anti-malarial drug treatment is an inducing factor for up regulation of supervillin. Some of the many genes that were up-regulated in the present study include HSP90ab1, PAWR, and IKbRb among others. An up-regulated HSP90ab1 indicates that anti-malarial drug exposure has resulted in the development of hepatic stress. The PAWR genes are found to be transcriptionally induced by apoptotic signals in the rat ventral prostate [38]. Woronicz et al. [39] observed that IKbRb activates NF-kappa-B when overexpressed and phosphorylate serine residues 32 and 36 of I-kappa-B-alpha and 19 and 23 of I-kappa-B-beta. Therefore, upregulated PAWR and IKbRb in murine liver is an indication of cellular toxicity and inflammatory responses within liver hepatocytes following anti-malarial exposure.
One of the several genes that were down-regulated following anti-malarial exposure in murine liver is the myotubularin related protein 2 (MTMR2) gene. The MTMR2 gene encodes a protein that belongs to the myotubularin family, which is characterized by the presence of a phosphatase domain. Berger et al. [40] determined that mouse MTMR2 gene dephosphorylates phosphatidylinositol 3-phosphate (PI3P) and phosphatidylinositol 3, 5-bisphosphate (PI3, 5P2) with high efficiency and peak activity at neutral pH. A perturbation in phosphatidylinositol pathway resulting from down regulated MTMR2 expression is an indication of disturbances of signaling pathways following anti-malarial treatment.
Conclusion
Both biochemical and microarray results suggest that combination therapy of AQ and SP are more damaging than their individual monotherapies. Microarray results further suggests that present anti-malarial combination therapies lead to inflammatory responses and perturbed signaling cascade leading to general hepatic stress as observed in biochemical evaluation of liver tissue. Furthermore, expression level of EPRS, SVIL, PAWR, and MTMR2 can be good markers for anti-malarial drug induced hepatotoxicity. Hence, the present study can help in understanding anti-malarial drug induced toxicity. However, the clinical implication of the study needs to be evaluated further with caution as this study in experimental mice may not hold equally good in case of malaria prophylaxis and treatment for human population.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
SKM Conceptual design of work, experimental work, data analysis and manuscript writing. PS Experimental work and data analysis. SKR Conceptual design of work, overall supervision of work and manuscript writing. All authors read and approved the final manuscript
Contributor Information
Shrawan Kumar Mishra, Email: shrawancdri@gmail.com.
Prabhat Singh, Email: prabhat_singh@cdri.res.in.
Srikanta Kumar Rath, Email: skrath@cdri.res.in.
Acknowledgements
This work was supported by grant to SKR from the Council of Scientific and Industrial Research (CSIR) network project (NWP0034). Authors SKM and PS are recipients of Senior Research Fellowship from Indian Council of Medical Research, and Department of Biotechnology, India respectively. The paper bears CDRI communication number 8051.
References
- Snow RW, Guerra CA, Noor AM, Myint HY, Hay SI. The global distribution of clinical episodes of Plasmodium falciparum malaria. Nature. 2005;434:214–217. doi: 10.1038/nature03342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowunmi A. A randomized comparison of chloroquine, amodiaquine and their combination with pyrimethamine-sulfadoxine in the treatment of acute, uncomplicated, Plasmodium falciparum malaria in children. Ann Trop Med Parasitol. 2002;96:227–238. doi: 10.1179/000349802125000763. [DOI] [PubMed] [Google Scholar]
- Lederman ER, Maguire JD, Sumawinata IW, Chand K, Elyazar I, Estiana L, Sismadi P, Bangs MJ, Baird JK. Combined chloroquine, sulfadoxine/pyrimethamine and primaquine against Plasmodium falciparum in Central Java, Indonesia. Malar J. 2006;5:108. doi: 10.1186/1475-2875-5-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malaria treatment guidelines. World Health Organisation (WHO), Geneva. 2010.
- Bell DJ, Nyirongo SK, Mukaka M, Zijlstra EE, Plowe CV, Molyneux ME, Ward SA, Winstanley PA. Sulfadoxine-pyrimethamine-based combinations for malaria: a randomised blinded trial to compare efficacy, safety and selection of resistance in Malawi. PLoS One. 2008;3:e1578. doi: 10.1371/journal.pone.0001578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez MA, Cortes LJ, Guerra AP, Knudson A, Usta C, Nicholls RS. Efficacy of the amodiaquine+sulfadoxine-pyrimethamine combination and of chloroquine for the treatment of malaria in Cordoba, Colombia, 2006. Biomedica. 2008;28:148–159. [PubMed] [Google Scholar]
- Sowunmi A, Balogun T, Gbotosho GO, Happi CT, Adedeji AA, Bolaji OM, Fehintola FA, Folarin OA. Activities of artemether-lumefantrine and amodiaquine-sulfalene-pyrimethamine against sexual-stage parasites in falciparum malaria in children. Chemotherapy. 2008;54:201–208. doi: 10.1159/000140463. [DOI] [PubMed] [Google Scholar]
- The East African Network for Monitoring Antimalarial Treatment (EANMAT) The efficacy of antimalarial monotherapies, sulphadoxine-pyrimethamine and amodiaquine in East Africa: implications for sub-regional policy. Trop Med Int Health. 2003;10:860–867. doi: 10.1046/j.1360-2276.2003.01114.x. [DOI] [PubMed] [Google Scholar]
- Talisuna AO, Nalunkuma-Kazibwe A, Bakyaita N, Langi P, Mutabingwa TK, Watkins WW, Van Marck E, D'Alessandro U, Egwang TG. Efficacy of sulphadoxine-pyrimethamine alone or combined with amodiaquine or chloroquine for the treatment of uncomplicated falciparum malaria in Ugandan children. Trop Med Int Health. 2004;9:222–229. doi: 10.1046/j.1365-3156.2003.01187.x. [DOI] [PubMed] [Google Scholar]
- Salle B, editor. Discussion Paper for the RBM PARTNERS' MEETING. WHO, Geneva; 2002. pp. 26–28.http://www.emro.who.int/rbm/Publications/DiscussionPaper02.pdf [Google Scholar]
- White NJ. Antimalarial drug resistance. J Clin Invest. 2004;113:1084–1092. doi: 10.1172/JCI21682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzur JR. Letter: Quinidine hepatotoxicity. JAMA. 1976;235:908. doi: 10.1001/jama.235.9.908d. [DOI] [PubMed] [Google Scholar]
- Farver DK, Lavin MN. Quinine-induced hepatotoxicity. Ann Pharmacother. 1999;33:32–34. doi: 10.1345/aph.18172. [DOI] [PubMed] [Google Scholar]
- Pari L, Murugavel P. Protective effect of alpha-lipoic acid against chloroquine-induced hepatotoxicity in rats. J Appl Toxicol. 2004;24:21–26. doi: 10.1002/jat.940. [DOI] [PubMed] [Google Scholar]
- Taylor RJ, White NJ. Antimalarial drug toxicity. Drug Safety. 2004;27:25–61. doi: 10.2165/00002018-200427010-00003. [DOI] [PubMed] [Google Scholar]
- Lee WM. Drug-induced hepatotoxicity. N Engl J Med. 2003;349:474–485. doi: 10.1056/NEJMra021844. [DOI] [PubMed] [Google Scholar]
- Minami K, Saito T, Narahara M, Tomita H, Kato H, Sugiyama H, Katoh M, Nakajima M, Yokoi T. Relationship between hepatic gene expression profiles and hepatotoxicity in five typical hepatotoxicant-administered rats. Toxicol Sci. 2005;87:296–305. doi: 10.1093/toxsci/kfi235. [DOI] [PubMed] [Google Scholar]
- Noel S, Sharma S, Shanker R, Rath SK. Primaquine-induced differential gene expression analysis in mice liver using DNA microarrays. Toxicology. 2007;239:96–107. doi: 10.1016/j.tox.2007.06.098. [DOI] [PubMed] [Google Scholar]
- Noel S, Sharma S, Shankar R, Rath SK. Identification of differentially expressed genes after acute exposure to bulaquine (CDRI 80/53) in mice liver. Basic Clin Pharmacol Toxicol. 2008;103:522–529. doi: 10.1111/j.1742-7843.2008.00279.x. [DOI] [PubMed] [Google Scholar]
- Blomme EA, Yang Y, Waring JF. Use of toxicogenomics to understand mechanisms of drug-induced hepatotoxicity during drug discovery and development. Toxicol Lett. 2009;186:22–31. doi: 10.1016/j.toxlet.2008.09.017. [DOI] [PubMed] [Google Scholar]
- WHO Malaria Factsheet. http://www.who.int/mediacentre/factsheets/fs094/en
- Freireich EJ, Gehan EA, Rall DP, Schmidt LH, Skipper HE. Quantitative comparison of toxicity of anticancer agents in mouse, rat, hamster, dog, monkey, and man. Cancer Chemother Rep. 1966;50:219–244. [PubMed] [Google Scholar]
- Ohkawa H, Ohishi N, Yagi K. Assay for lipid peroxides in animal tissues by thiobarbituric acid reaction. Anal Biochem. 1979;95:351–358. doi: 10.1016/0003-2697(79)90738-3. [DOI] [PubMed] [Google Scholar]
- ELLMAN GL. Tissue sulfhydryl groups. Arch Biochem Biophys. 1959;82:70–77. doi: 10.1016/0003-9861(59)90090-6. [DOI] [PubMed] [Google Scholar]
- Kakkar P, Das B, Viswanathan PN. A modified spectrophotometric assay of superoxide dismutase. Indian J Biochem Biophys. 1984;21:130–132. [PubMed] [Google Scholar]
- Sinha AK. Colorimetric assay of catalase. Anal Biochem. 1972;47:389–394. doi: 10.1016/0003-2697(72)90132-7. [DOI] [PubMed] [Google Scholar]
- Wendel A. In: Enzymatic Basis of Detoxication. Jakoby WB, editor. Academic Press, New York; 1980. Glutathione peroxidase; pp. 333–348. [Google Scholar]
- Website describing the distribution of probes in microarray slides used in the experiments. http://www.microarrays.ca
- Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- Gene Expression Omnibus Database. http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=vjopbqokouiiqny&acc=GSE17392
- Luu-The V, Paquet N, Calvo E, Cumps J. Improved real-time RT-PCR method for high-throughput measurements using second derivative calculation and double correction. Biotechniques. 2005;38:287–293. doi: 10.2144/05382RR05. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuaqui RF, Bonner RF, Best CJ, Gillespie JW, Flaig MJ, Hewitt SM, Phillips JL, Krizman DB, Tangrea MA, Ahram M, Linehan WM, Knezevic V, Emmert-Buck MR. Post-analysis follow-up and validation of microarray experiments. Nat Genet. 2002;32(Suppl):509–514. doi: 10.1038/ng1034. [DOI] [PubMed] [Google Scholar]
- GenMAPP-Pathway analysis tool. http://www.genmapp.org
- Cerini C, Kerjan P, Astier M, Gratecos D, Mirande M, Semeriva M. A component of the multisynthetase complex is a multifunctional aminoacyl-tRNA synthetase. EMBO J. 1991;10:4267–4277. doi: 10.1002/j.1460-2075.1991.tb05005.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampath P, Mazumder B, Seshadri V, Gerber CA, Chavatte L, Kinter M, Ting SM, Dignam JD, Kim S, Driscoll DM, Fox PL. Noncanonical function of glutamyl-prolyl-tRNA synthetase: gene-specific silencing of translation. Cell. 2004;119:195–208. doi: 10.1016/j.cell.2004.09.030. [DOI] [PubMed] [Google Scholar]
- Scheinberg IH, Gitlin D. Deficiency of ceruloplasmin in patients with hepatolenticular degeneration (Wilson's disease) Science. 1952;116:484–485. doi: 10.1126/science.116.3018.484. [DOI] [PubMed] [Google Scholar]
- Johnstone RW, See RH, Sells SF, Wang J, Muthukkumar S, Englert C, Haber DA, Licht JD, Sugrue SP, Roberts T, Rangnekar VM, Shi Y. A novel repressor, par-4, modulates transcription and growth suppression functions of the Wilms' tumor suppressor WT1. Mol Cell Biol. 1996;16:6945–6956. doi: 10.1128/mcb.16.12.6945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woronicz JD, Gao X, Cao Z, Rothe M, Goeddel DV. IkappaB kinase-beta: NF-kappaB activation and complex formation with IkappaB kinase-alpha and NIK. Science. 1997;278:866–869. doi: 10.1126/science.278.5339.866. [DOI] [PubMed] [Google Scholar]
- Berger P, Bonneick S, Willi S, Wymann M, Suter U. Loss of phosphatase activity in myotubularin-related protein 2 is associated with Charcot-Marie-Tooth disease type 4B1. Hum Mol Genet. 2002;11:1569–1579. doi: 10.1093/hmg/11.13.1569. [DOI] [PubMed] [Google Scholar]