Abstract
Background
TGF-β acts as a suppressor of primary tumour initiation but has been implicated as a promoter of the later malignant stages. Here associations with risk of invasive breast cancer are assessed for SNPs tagging seventeen genes in the canonical TGF-β ALK5/SMADs 2&3 and ALK1/SMADs 1&5 signalling pathways: LTBP1, LTBP2, LTBP4, TGFB1, TGFB2, TGFB3, TGFBR1(ALK5), ALK1, TGFBR2, Endoglin, SMAD1, SMAD2, SMAD3, SMAD4, SMAD5, SMAD6 and SMAD72.
Methods
354 tag SNPs (minor allele frequency>0.05) were selected for genotyping in a staged study design using 6,703 cases and 6,840 controls from the SEARCH study. Significant associations were meta-analysed with data from the NCI Polish Breast Cancer Study (PBCS) (1,966 cases and 2,347 controls) and published data from the Breast Cancer Association Consortium (BCAC).
Results
Associations of three SNPs, tagging TGFB1 (rs1982073), TGFBR1 (rs10512263) and TGFBR2 (rs4522809) were detected in SEARCH; however associations became weaker in meta-analyses including data from PBCS and BCAC. Tumour sub-type analyses indicated that the TGFB1 rs1982073 association may be confined to increased risk of developing progesterone receptor negative (PR−) tumours (1.18 (95% CI 1.09-1.28), 4.1×10−5 (P value for heterogeneity of ORs by PR status = 2.3 × 10−4)). There was no evidence for breast cancer risk associations with SNPs in the endothelial-specific pathway utilising ALK1/SMADs 1&5 that promotes angiogenesis.
Conclusion
Common variation in the TGF-β ALK5/SMADs 2&3 signalling pathway, which initiates signalling at the cell surface to inhibit cell proliferation, might be related to risk of specific tumour sub-types.
Impact
The subtype specific associations require very large studies to be confirmed.
Keywords: TGF-beta, Breast cancer, Susceptibility, Tagging, Genotyping
Introduction
Variants that increase risk of breast cancer fall into three categories: rare, high-penetrance alleles; moderate-risk alleles; and common, low-risk alleles. Rare high/moderate risk inherited mutations (BRCA1, BRCA2, TP53, CHEK2, PALB2 and BRIP1) account for approximately 27% of excess familial risk of breast cancer (1). These risk alleles were identified by linkage analysis/candidate gene resequencing approaches and extensive efforts to find more susceptibility alleles have been unsuccessful. The remaining risk is likely to be caused by the cumulative effect of multiple moderate/lower-risk inherited variants (2). Confirming this, genome-wide association studies (GWAS) have already identified 17 common, low penetrance loci; for example FGFR2, TNRC9/LOC643714 and NEK10/SLC4A7 (3-7). Few common SNP associations, CASP8 D302H and TGFB1 L10P (rs1982073) (8), have been identified via the candidate gene approach, which has been hampered by both a lack of understanding of the biology of breast cancer and small, underpowered studies. The TGFB1 Pro allele, which has been shown to increase TGFB1 secretion in vitro (9), has been reported to be associated with an increased risk of breast cancer relative to the Leu allele: OR 1.08, 95% CI 1.04-1.11, P = 2.8×10−5 (8). The functionally relevant repeat length polymorphism in exon 1 of the TGFBR1 (TGFBR1*6A) has also been examined as a candidate; a recent meta-analysis of 15 breast cancer studies including 10,826 cases and 12,964 controls reported a significant association for allelic effect (6A vs. 9A): OR 1.16, 95% CI 1.01-1.34, P = 0.04 (10).
Other variants in TGF-β signalling pathway genes have also been studied in relation to association with breast cancer risk, with the main focus previously being a SNP in the promoter region of TGFB1 (c-509t) (3987 cases, 3867 controls: OR 1.25 95% CI 1.06-1.48, P = 0.009) (9). The c-509t variant is in linkage disequilibrium (LD) with the L10P polymorphism previously discussed, (r2=0.69 in Stage 1 data presented here) and probably marks the same causal variant.
SNP associations in TGF-β signalling pathway genes have not been detected by recent breast cancer GWAS (3-7).
There is accumulating evidence that the TGF-β signalling pathway (Figure 1) has a dual role, acting both in initial tumour development and in later tumour progression. A broad range of evidence for TGF-β acting as a tumour suppressor gene comes from humans, in particular from studies of epithelial colorectal tumours, and from transgenic mouse studies. However, when a primary tumour has been established, TGF-β may act to enhance subsequent tumour progression, based on studies of human cells in vitro and of transgenic mouse models in which TGF-β signalling is modulated (11, 12). Studies in endothelial cells in vitro have indicated that TGF-β may regulate angiogenesis via the balance of signalling through two pathways, activated by the anti-angiogenic receptor ALK5 (TGFBR1) and the pro-angiogenic receptor ALK1 (Figure 1) (13-15). It has been proposed that shifting the balance to a pro-angiogenic role for TGF-β may be a significant mechanism by which it enhances tumour progression. The expression of TGF-β signalling factor phospho-SMAD2 has recently been reported to be greater in human breast cancers associated with lymph node metastasis, consistent with a pro-progression role for TGF-β (16). TGF-β signalling patterns were found to vary with age and pathological features of prognostic significance, further supporting the proposal that the pathway may be associated with disease progression in humans.
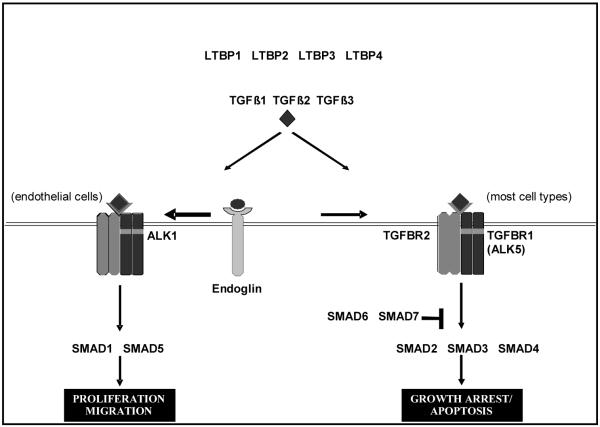
Figure 1. TGF-β signalling pathway.
Once cleaved from latent TGF-β binding protein (four isoforms, LTBP1-4) the mature and active dimeric TGF-β (three isoforms, TGFB1-3) can bind to heteromeric receptor complexes formed by type I (TGFBR1; ALK5 and ALK1 (activin A receptor type II-like 1)) receptors and type II (TGFBR2) receptors, initiating signal transduction. The binding of TGF-β to the receptor complex is facilitated by type III co-receptors, such as endoglin. Endoglin is expressed at particularly high levels in endothelial cells (32). Activation of receptor kinase domains initiates downstream signalling by phosphorylation of transcription factors e.g. SMAD1-5 (SMAD family members). The SMAD complex migrates to the nucleus and acts as a transcription factor to regulate cell and phenotype specific patterns of gene expression. SMAD6 and SMAD7 block phosphorylation of SMAD2 or SMAD3 and act as inhibitors of the TGF-β signalling pathway (15, 32). SMADs are widely expressed in most adult tissue and cell types indicating that this TGF-β signalling mechanism is ubiquitous.
Functionally relevant gene variants have been previously identified in two TGF-β signalling pathway genes, supporting the established rationale for additional studies of candidate genes that belong to cancer-related pathways (17). TGF-β signalling pathway genes are attractive candidates.
In this association study using invasive breast cancer cases and controls we comprehensively tagged the common variants in 17 genes comprising both arms of the TGF-β signalling pathways (Figure 1).
Materials and Methods
Study populations
As patients were recruited into the Studies of Epidemiology and Risk Factors in Cancer Heredity (SEARCH) study, samples were collected into three stages, each totalling approximately 2,300 cases and 2,300 controls. Different controls were used for each stage. The geographical and ethnic background of both cases and controls is similar, with 99.7% being Caucasian. Given that only 0.31% of cases and controls are non-Caucasian (Eurasians) the chance of false positives or negatives as a result of these differences is minimal.
Epithelial breast cancer cases were drawn from SEARCH (18), an ongoing population-based collection of breast cancer cases ascertained through the Eastern Cancer Registration and Information Centre (19). Ethical approval was obtained from the Anglia and Oxford Multicentre Research Committee and informed consent was obtained from each patient.
All women diagnosed with invasive, epithelial breast cancer under the age of 55 years between 1 January 1991 and 30 June 1996 and who were alive at the start of the study (prevalent cases) as well as women under the age of 70 years who were diagnosed from 1996 onwards (incident cases) were eligible for inclusion. Breast cancer cases were ascertained by both medical records and pathological reports. Cases were randomly selected for Stage 1 from the first 3,500 recruited with sufficient quantity of DNA extracted from the blood collected. Stage 2 comprises of the remainder of these plus the next 900 incident cases recruited, with Stage 3 comprising the next incident cases recruited. 64% eligible cases and 41% invited controls provided blood samples (Supplementary Table 1). The proportion of prevalent cases was higher in the first two stages; Stage 1 (33%), Stage 2 (20%), Stage 3 (0.5%). Median age at diagnosis was similar in Stage 1 and Stage 2 (51 and 52 years old respectively) but higher in Stage 3 (56 years old for the incident cases and 68 years old for the prevalent cases) as the selection criteria for Stage 3 differed in age from previous stages. For Stages 1 and 2 combined 27% were prevalent cases with a median age of 48 years and 73% were incident cases with a median age of 54 years. For Stages 1, 2 and 3 combined, the median age for incident cases was 54 years old and for the prevalent cases was 49 years old. There were no substantive differences in the morphology, histopathological grade or clinical stage of the cases by stage or by prevalent/incident status. Despite the different proportion of prevalent cases by Stage, survival bias has not been observed for TGFB1 SNPs in the samples comprising Stage 1 of this study (20) and we have no evidence that allele frequencies of the SNPs studied here will be affected by the inclusion of prevalent and incident cases in SEARCH.
For Stages 1 and 2, volunteer female controls were selected in approximate order of recruitment from the Norfolk component of the European Prospective Investigation of Cancer (EPIC) (21). EPIC is a prospective study of diet and cancer being carried out in nine European countries. The EPIC-Norfolk cohort comprises 25,000 individuals resident in Norfolk, East Anglia. Controls were aged between 45-74 years and were recruited between 1992 and 1994 from the same geographic region as cases. The female controls for Stage 3 were recruited from General Practitioners as part of SEARCH (Supplementary Table 1). Subjects diagnosed with breast cancer were excluded.
Hidden population structure was minimised by selecting cases and controls from the same region of the UK (3, 6-8, 22). Other sources of false positive findings were limited by the use of the same DNA extraction method for all samples, uniform handling and storage of DNA, interspersed arraying of cases and controls on the same plate and manual inspection of all genotype clusters.
SNPs were genotyped in a three-staged SEARCH study design, each stage comprising of 2,200 (Stages 1 and 2) or 2,303 (Stage 3) breast cancer cases and 2,280 controls (different for each stage). If a SNP showed marginally significant associations it was taken through to the next stage (see selection criteria). Data from each stage were combined for analysis as the power was greater than using each stage as an independent replication. A maximum of 6,703 cases and 6,840 controls were available for genotyping.
Additional genotyping data were obtained from the NCI Polish Breast Cancer Study (PBCS) to increase the power to confirm or negate any putative associations. This is a population-based study that included 1,966 incident cases diagnosed from 2000-2003 in Warsaw and Łódź, and 2,347 randomly selected controls obtained from population lists of all residents of Poland, and matched to the cases on age and city (23).
Selection of tagging SNPs (tSNPs)
The aim of selecting tSNPs was to tag efficiently all of the common variations in a gene by genotyping a relatively small subset of the common variants to gain information on the entire region. Primary genotype data for each gene were obtained predominantly from the International HapMap project (24) on 30 Caucasian parent-offspring trios from Utah, US (Centre d’Etude du Polymorphisme Humain data). SNP genotype data on 90 individuals representative of the US population from the NIEHS project (25) were used if the data from HapMap were not sufficient. Any unidentified SNPs are likely to be tagged by a tSNP selected for this region. The programme Tagger, accessed via Haploview, was used to select tSNPs using the pairwise function (26) with the aim of selecting a minimal set of markers (tSNPs) such that all common alleles (defined as a MAF >0.05) captured are correlated at an rp2 >0.8 with a selected tSNP. If a tSNP failed to manufacture or genotype, an alternative tSNP was selected; with 354 tSNPs genotyped in total.
Genotyping
The selected tSNPs were genotyped in Stage 1 using either the Taqman® ABI PRISM 7900 sequence detection system (Applied Biosystems) (using 10 ng DNA) or the GenomeLab™ SNPstream® Genotyping system (Beckman Coulter) (using 8 ng genomic DNA) according to the manufacturer’s instructions. If a tSNP was selected for Stage 2, a Taqman assay was designed and used for genotyping on Stage 2 and Stage 3. For accuracy and consistency, Stage 1 genotyping performed by SNPstream was regenerated using a Taqman assay. Taqman genotyping resulted in a slightly higher call rate with no discordant replicate samples and it was preferable to combine data that had been generated by the same technology. Sequences for SNPs were retrieved and annotated using Seq4SNP software (27) or manually using databases available from NCBI (28) or ENSEMBL (29).
Genotyping accuracy for Taqman and SNPstream
Each 384-well plate included two controls with no DNA. Genotyping 12 replicate samples from each plate on a separate 384 well plate ensured genotyping accuracy. For genotypes generated using the Taqman platform, there was 100% concordance between the replicates (excluding failed replicate samples). For genotypes generated using the SNPstream platform there was some discordance (maximum of 3.6% discordance for any SNP assay, excluding failed replicate samples). Failed genotypes were not repeated. The rate for failed genotypes did not exceed 3% for any of the SNPs under study by Taqman. There is no evidence for greater deviation from Hardy-Weinberg equilibrium (HWE) than expected for the number of assays, indicating that genotyping quality was high. Of the 354 SNPs genotyped in Stage 1, 25 (7%) exhibited significant (P <0.05) deviation from HWE – see Supplementary Table 2.
Statistical methods
For each polymorphism, deviation of the genotype frequencies from those expected under HWE was assessed in the controls by a Chi2 test. Genotype frequencies in cases and controls were compared using a Chi2 test with 2 degrees of freedom (df) (Pheterogeneity), and the Armitage trend test (Chi2 with 1df) for the trend in breast cancer risk with number of rare alleles (Ptrend). The relative risks of breast cancer for heterozygotes and for rare homozygotes, relative to common homozygotes, were estimated as odds ratios (ORs) with associated 95% confidence intervals (CI). Meta-analysis was performed by STATA version 9.0 (Stata Corporation, College Station, TX), weighting the studies by size.
Selection criteria
Selection criteria for genotyping in further stages were Chi2 test Ptrend <0.1 or Chi2 test Pheterogeneity <0.1 revised to Chi2 test Ptrend <0.05 for TGFB1, TGFB2, TGFBR1 (ALK5), ALK1, TGFBR2 and Endoglin SNPs. The selection criteria were revised as it became clear that potential associations at the less stringent cut-off were not replicated in further stages. The number of tSNPs selected for Stage 2 from genes studied was further reduced by analysing correlations between the significant tSNPs, thus avoiding selecting several tSNPs that detected the same association (data not shown). A tSNP in TGFB1, rs1982073, was also included for genotyping in Stage 2 as it has previously been shown to be associated with breast cancer risk.
Analysis by receptor status
Relative risk estimates for specific tumour subtypes defined by ER and PR status were estimated using polychromous logistic regression models adjusted by study comparing cases within a tumour subgroup category to all controls. Data on hormone receptor status were not available for all cases; the number of cases with data available are indicated in Table 1.
Table 1a.
Association between SNPs in the TGFB pathway and breast cancer risk by ER status of the tumors
| SNP (Gene) | Genotype | Controls | ER+ | OR | 95% CI | P | ER− | OR | 95% CI | P | P-Het* |
|---|---|---|---|---|---|---|---|---|---|---|---|
| rs10512263 (TGFBR1 ) |
AA | 7782 | 3994 | 1.00 | 1282 | 1.00 | |||||
| AG | 1314 | 573 | 0.85 | 0.76 - 0.94 | 0.002 | 175 | 0.85 | 0.72 - 1.01 | 0.060 | ||
| GG | 58 | 20 | 0.67 | 0.40 - 1.12 | 0.126 | 9 | 0.99 | 0.49 - 2.01 | 0.982 | ||
| Per allele | 9154 | 4587 | 0.85 | 0.77 - 0.93 | 0.001 | 1466 | 0.83 | 0.71 - 0.97 | 0.021 | 0.851 | |
| rs4522809 (TGFBR2 ) |
AA | 2650 | 1369 | 1.00 | 441 | 1.00 | |||||
| AG | 4486 | 2275 | 0.98 | 0.90 - 1.07 | 0.659 | 690 | 0.94 | 0.82 - 1.07 | 0.339 | ||
| GG | 2024 | 931 | 0.89 | 0.80 - 0.99 | 0.025 | 325 | 1.00 | 0.86 - 1.17 | 0.991 | ||
| Per allele | 9160 | 4575 | 0.95 | 0.90 - 1.00 | 0.033 | 1456 | 0.98 | 0.90 - 1.06 | 0.572 | 0.450 | |
| rs1982073 (TGFB1 ) |
AA | 3382 | 1625 | 1.00 | 476 | 1.00 | |||||
| AG | 4256 | 2087 | 1.02 | 0.94 - 1.11 | 0.583 | 663 | 1.09 | 0.96 - 1.23 | 0.192 | ||
| GG | 1295 | 688 | 1.11 | 0.99 - 1.24 | 0.063 | 231 | 1.22 | 1.03 - 1.45 | 0.020 | ||
| Per allele | 8933 | 4400 | 1.04 | 0.99 - 1.10 | 0.103 | 1370 | 1.12 | 1.03 - 1.22 | 0.006 | 0.112 |
P value for heterogeneity of ORs by ER status of the tumors
Analyses are adjusted by study
Results and Discussion
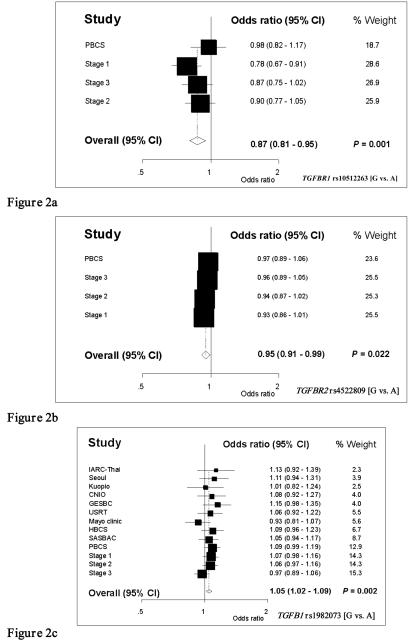
Assays for a total of 354 SNPs were successfully generated, together tagging 92% of the 1254 common variants with r2 >0.8 identified in the 17 genes (Supplementary Table 3). These assays were used to genotype the 2200 cases and 2280 controls from the SEARCH study in Stage 1. Twenty-one of the 354 SNPs were selected, on the basis of Armitage-Cochrane test significance levels, for progression into Stage 2 (a further 2200 cases and 2280 controls from the SEARCH study) and six of these subsequently progressed to Stage 3 (a further 2303 cases and 2280 controls from the SEARCH study). Of these, five SNPs were genotyped in incident cases and controls from PBCS. Data are shown for all three stages in Supplementary Table 2 and weighted meta-analyses of all three stages and the PBCS data are shown in Figure 2.
Figure 2.
Relative risks of breast cancer estimated as OR and 95% CI by study/stage and meta-analysis for overall OR and 95% CI. (a) TGFBR1 rs10512263 [G vs. A], and (b) TGFBR2 rs4522809 [G vs. A] include data from SEARCH stages 1, 2 and 3 and PBCS. (c) TGFB1 rs1982073 [G vs. A (Pro vs. Leu) includes data from SEARCH Stages 1, 2 and 3 and Breast Cancer Association Consortium (BCAC) (including PBCS). Studies have been weighted by size
In addition to the previously reported association with TGFB1 Leu10Pro (rs1982073), two SNPs showed marginally significant associations with risk of invasive breast cancer (meta-analysis of the SEARCH and PBCS studies). In TGFBR1, the minor G allele of SNP rs10512263 showed a protective effect (OR [G vs. A] 0.87, 95% CI 0.81-0.95, P = 0.001). In TGFBR2, the G allele of rs4522809 also had a marginally protective effect (OR [G vs. A] 0.95, 95% CI 0.91-0.99, P = 0.02). The magnitudes of these effects are not markedly altered by removal of the hypothesis generating data (SEARCH Stage 1) (OR 0.91, 95% CI 0.83-1.00, P = 0.050) (OR 0.96, 95% CI 0.91-1.01, P = 0.094) for TGFBR1 and TGFBR2 respectively.
For rs1982073 a meta-analysis of all previously published data from the BCAC consortium (8) together with new data generated by this study is presented in Figure 2c. There is a dose-dependent association of the proline-encoding allele with increased risk of invasive breast cancer (OR [Pro vs. Leu] 1.05, 95% CI 1.02-1.09, P = 0.002).
It has been suggested that this Pro allele association may be confined to an increased risk of developing PR− tumours (8). The SEARCH and PBCS data presented here, (including SEARCH Stage 3 and the entire cohort of PBCS in addition to the data previously published) (Table 1b) support that suggestion (PR− OR [Pro vs. Leu] 1.18, 95% CI 1.09-1.28, P = 4.1×10−5; PR+ OR [Pro vs. Leu] 1.01, 95% CI 0.94 – 1.08, P = 0.85). In this context it is of interest that TGFB1 inhibits the PR expression in human endometrial stromal cells in vitro via SMAD signalling (30). If, as is likely, the same effect occurs in breast epithelial cells in vivo, the observation may account, at least in part, for the association of the Pro allele (rs1982073) in TGFB1 with PR− tumours, as the Pro allele causes substantial increases in TGFB1 secretion in vitro compared with the Leu allele.
Table 1b.
Association between SNPs in the TGFB pathway and breast cancer risk by PR status of the tumors
| SNP | Genotype | Controls | PR+ | OR | 95% CI | P | PR− | OR | 95% CI | P | P-Het** |
|---|---|---|---|---|---|---|---|---|---|---|---|
| rs10512263 (TGFBR1 ) |
AA | 7782 | 2118 | 1.00 | 1360 | 1.00 | |||||
| AG | 1314 | 306 | 0.90 | 0.78 - 1.02 | 0.107 | 193 | 0.93 | 0.79 - 1.09 | 0.374 | ||
| GG | 58 | 9 | 0.60 | 0.30 - 1.22 | 0.162 | 11 | 1.23 | 0.64 - 2.38 | 0.537 | ||
| Per allele | 9154 | 2433 | 0.88 | 0.78 - 1.00 | 0.045 | 1564 | 0.96 | 0.82 1.11 | 0.571 | 0.709 | |
| rs4522809 (TGFBR2 ) |
AA | 2650 | 734 | 1.00 | 461 | 1.00 | |||||
| AG | 4486 | 1207 | 0.99 | 0.89 - 1.09 | 0.788 | 756 | 1.00 | 0.88 1.14 | 0.999 | ||
| GG | 2024 | 486 | 0.90 | 0.79 - 1.02 | 0.094 | 340 | 1.04 | 0.89 1.21 | 0.652 | ||
| Per allele | 9160 | 2427 | 0.95 | 0.89 - 1.01 | 0.114 | 1557 | 1.02 | 0.94 1.10 | 0.677 | 0.286 | |
| rs1982073 (TGFB1 ) |
AA | 3382 | 868 | 1.00 | 476 | 1.00 | |||||
| AG | 4256 | 1072 | 0.97 | 0.87 - 1.07 | 0.510 | 692 | 1.12 | 0.98 1.27 | 0.092 | ||
| GG | 1295 | 353 | 1.04 | 0.90 - 1.19 | 0.613 | 277 | 1.44 | 1.22 1.69 | 1.5E-05 | ||
| Per allele | 8933 | 2293 | 1.01 | 0.94 - 1.08 | 0.853 | 1445 | 1.18 | 1.09 1.28 | 4.1E-05 | 2.3E-04 |
P value for heterogeneity of ORs by PR status of the tumors
Analyses are adjusted by study
In light of this finding, tumour sub-type associations of the TGFBR1 and TGFBR2 SNPs have also been investigated (Tables 1a and b). For rs10512263 the protective effect is more pronounced in PR+ (OR [G vs. A] 0.88, 95% CI 0.78-1.00, P = 0.05) than in PR− tumours (OR [G vs. A] 0.96, 95% CI 0.82-1.11, P = 0.57). For rs4522809 the marginal protective association of the G allele is confined to ER+ tumours (OR [G vs. A] 0.95, 95% CI 0.90-1.00, P = 0.03). Confirmation of these early indications of possible tumour sub-type differences would require much larger studies.
The effects of these SNPs on the strength of TGFB1 signalling mediated by TGFBR1 (ALK5) and TGFBR2 on PR and ER expression in breast epithelial cells may provide a biological consistency test for the SNP associations with tumour sub-type. It would be predicted, for example, that the protective G allele for the TGFBR1 SNP would be associated with reduced TGF-β signalling and higher PR expression. However the associations for TGFBR1 (ALK5) and TGFBR2 need to be interpreted with caution as they would not remain significant after Bonferroni adjustment for multiple testing (354 tSNPs, P = 1.4×10−4). Potentially contradicting our findings, TGFBR2 expression has recently been associated with prognostically favourable small tumours among ER− tumours (16).
If these SNP associations are true, the questions of how they exert their effects remain. SNP rs1982073 may be a causal variant. It is exonic and affects the signal peptide, responsible for secretion of TGF-β from the cell. In previous studies we have shown that rs1982073 causes a 2.8 fold increase in the amount of protein secreted in vitro (9). SNP rs10512263 is located in intron 1 of TGFBR1, in a region that is conserved between species. It exhibits no strong LD with any other known variants. It is possible that this SNP is a causal variant with an unknown function or it may be marking the causal SNP. The functionally relevant TGFBR1*6A SNP that has previously been associated with breast cancer risk (10) has not been genotyped as part of the HapMap project or within our own cohorts and it is currently beyond the means of this study to determine the LD between this SNP and SNP rs10512263. SNP rs4522809 is situated in intron 2 of the TGFBR2 gene and is most likely marking a putative causal variant. Identifying disease-associated functionally-relevant alleles in candidate genes can be more readily interpreted (31).
No tSNPs in the genes of the endothelial-specific ALK1/SMADs 1&5 pathway, which promotes angiogenesis (14, 15), have been identified as associated with breast cancer risk. The balance of signalling through the two pathways may regulate TGF-β angiogenic activity but there is no detectable effect of SNPs on genes that regulate the pro-angiogenic pathway specifically. Any pro-angiogenic effects of SNPs are therefore likely to be restricted to effects on genes in the TGFBR1 (ALK5) pathway that weaken the anti-proliferative effect of TGF-β and indirectly rebalance signalling towards the pro-angiogenic pathway.
From this tag SNP study it has been possible to exclude large associations with invasive breast cancer of the common variants in the LTBP1, LTBP2, LTBP4, TGFB2, TGFB3, ALK1, Endoglin, SMAD1, SMAD2, SMAD3, SMAD4, SMAD5, SMAD6 and SMAD7 genes. It remains possible that associations of moderate effect size could have been missed (as false negatives). This is illustrated by the example of rs1982073, which would not have been selected for Stage 2 and 3 without prior data. However, in the case of TGFB1, the association of a correlated (r2=0.61 in Stage 1) SNP rs4803455 was identified (see Supplementary Table 2), demonstrating the effectiveness of the study to capture SNP associations.
The study provides some evidence that SNPs in the TGFB1, TGFBR1 (ALK5) and TGFBR2 genes may play a role in breast cancer development, although further sub-group analyses are needed to confirm these associations and any associations with hormone receptor type-specific disease. However this study and numerous breast cancer GWAS indicate that it is highly unlikely that other common variants in the TGF-β signalling pathway contribute significantly to breast cancer risk.
Supplementary Material
Acknowledgements
Don Conroy for providing technical support, Helen Field for providing bioinformatic support and the members of the SEARCH study team, particularly Melanie Maranian.
Footnotes
Approved Human Gene Nomenclature Committee gene names: ACVRL1 (for ALK1) and ENG (for Endoglin)
References
- 1.Ghoussaini M, Pharoah PD. Polygenic susceptibility to breast cancer: current state-of-the-art. Future Oncol. 2009;5(5):689–701. doi: 10.2217/fon.09.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pharoah PDP, Antoniou A, Bobrow M, Zimmern RL, Easton DF, Ponder BAJ. Polygenic susceptibility to breast cancer and implications for prevention. Nat Genet. 2002;31(1):33–36. doi: 10.1038/ng853. [DOI] [PubMed] [Google Scholar]
- 3.Easton DF, Pooley KA, Dunning AM, Pharoah PD, Thompson D, Ballinger DG, et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447(7148):1087–1093. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hunter DJ, Kraft P, Jacobs KB, Cox DG, Yeager M, Hankinson SE, et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat Genet. 2007;39(7):870–874. doi: 10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stacey SN, Manolescu A, Sulem P, Rafnar T, Gudmundsson J, Gudjonsson SA, et al. Common variants on chromosomes 2q35 and 16q12 confer susceptibility to estrogen receptor-positive breast cancer. Nat Genet. 2007;39(7):865–869. doi: 10.1038/ng2064. [DOI] [PubMed] [Google Scholar]
- 6.Ahmed S, Thomas G, Ghoussaini M, Healey CS, Humphreys MK, Platte R, et al. Newly discovered breast cancer susceptibility loci on 3p24 and 17q23.2. Nat Genet. 2009;41(5):585–590. doi: 10.1038/ng.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Turnbull C, Ahmed S, Morrison J, Pernet D, Renwick A, Maranian M, et al. Genome-wide association study identifies five new breast cancer susceptibility loci. Nat Genet. 2010;42(6):504–507. doi: 10.1038/ng.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cox A, Dunning AM, Garcia-Closas M, Balasubramanian S, Reed MW, Pooley KA, et al. A common coding variant in CASP8 is associated with breast cancer risk. Nat Genet. 2007;39(3):352–358. doi: 10.1038/ng1981. [DOI] [PubMed] [Google Scholar]
- 9.Dunning AM, Ellis PD, McBride S, Kirschenlohr HL, Healey CS, Kemp PR, et al. A Transforming Growth Factor{beta}1 Signal Peptide Variant Increases Secretion in Vitro and Is Associated with Increased Incidence of Invasive Breast Cancer. Cancer Research. 2003;63(10):2610–2615. [PubMed] [Google Scholar]
- 10.Liao RY, Mao C, Qiu LX, Ding H, Chen Q, Pan HF. TGFBR1*6A/9A polymorphism and cancer risk: a meta-analysis of 13,662 cases and 14,147 controls. Mol Biol Rep. 2010;33:3227–3232. doi: 10.1007/s11033-009-9906-7. [DOI] [PubMed] [Google Scholar]
- 11.Akhurst RJ, Balmain A. Genetic events and the role of TGF beta in epithelial tumour progression. J Pathol. 1999;187(1):82–90. doi: 10.1002/(SICI)1096-9896(199901)187:1<82::AID-PATH248>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 12.Xu Y, Pasche B. TGF-beta signalling alterations and susceptibility to colorectal cancer. Hum Mol Genet. 2007;15(16):R14–20. doi: 10.1093/hmg/ddl486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goumans MJ, Valdimarsdottir G, Itoh S, Rosendahl A, Sideras P, ten Dijke P. Balancing the activation state of the endothelium via two distinct TGF-beta type I receptors. EMBO J. 2002;21(7):1743–1753. doi: 10.1093/emboj/21.7.1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goumans MJ, Valdimarsdottir G, Itoh S, Lebrin F, Larsson J, Mummery C, et al. Activin receptor-like kinase (ALK)1 is an antagonistic mediator of lateral TGFbeta/ALK5 signaling. Mol Cell. 2003;12(4):817–828. doi: 10.1016/s1097-2765(03)00386-1. [DOI] [PubMed] [Google Scholar]
- 15.Goumans MJ, Lebrin F, Valdimarsdottir G. Controlling the angiogenic switch: a balance between two distinct TGF-b receptor signaling pathways. Trends Cardiovasc Med. 2003;13(7):301–307. doi: 10.1016/s1050-1738(03)00142-7. [DOI] [PubMed] [Google Scholar]
- 16.Figueroa JD, Flanders KC, Garcia-Closas M, Anderson WF, Yang XR, Matsuno RK, et al. Expression of TGF-beta signaling factors in invasive breast cancers: relationships with age at diagnosis and tumor characteristics. Breast Cancer Res Treat. 2010;121(3):727–35. doi: 10.1007/s10549-009-0590-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pasche B, Yi N. Candidate gene association studies: successes and failures. Current Opinion in Genetics & Development. 2010;20(3):257–61. doi: 10.1016/j.gde.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. http://www.srl.cam.ac.uk/search/Homepage.htm.
- 19. http://www.ecric.org.uk.
- 20.Goode EL, Dunning AM, Kuschel B, Healey CS, Day NE, Ponder BA, et al. Effect of Germ-Line Genetic Variation on Breast Cancer Survival in a Population-based Study. Cancer Res. 2002;62:3052–3057. [PubMed] [Google Scholar]
- 21.Day N, Oakes S, Luben R, Khaw KT, Bingham S, Welch A, et al. EPIC-Norfolk: study design and characteristics of the cohort: European Prospective Investigation of Cancer. Br J Cancer. 1999;80(1):95–103. [PubMed] [Google Scholar]
- 22.The Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garcia-Closas M, Egan KM, Newcomb PA, Brinton LA, Titus-Ernstoff L, Chanock S, et al. Polymorphisms in DNA double-strand break repair genes and risk of breast cancer: two population-based studies in USA and Poland, and meta-analyses. Hum Genet. 2006;119:376–388. doi: 10.1007/s00439-006-0135-z. [DOI] [PubMed] [Google Scholar]
- 24. http://www.hapmap.org.
- 25. http://egp.gs.washington.edu/T.html.
- 26.de Bakker Paul. http://www.broad.mit.edu/mpg/tagger/
- 27.Field HI, Scollen SA, Luccarini C, Baynes C, Morrison J, Dunning AM, et al. Seq4SNPs: new software for retrieval of multiple, accurately annotated DNA sequences, ready formatted for SNP assay design. BMC Bioinformatics. 2009;12(10):180. doi: 10.1186/1471-2105-10-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. http://www.ncbi.nlm.nih.gov/projects/SNP/
- 29. http://www.ensembl.org/index.html.
- 30.Kane N, Jones M, Brosens JJ, Saunders PT, Kelly RW, Critchley HO. Transforming Growth Factor-β1 Attenuates Expression of Both the Progesterone Receptor and Dickkopf in Differentiated Human Endometrial Stromal Cells. Molecular Endocrinology. 2008;22(3):716–728. doi: 10.1210/me.2007-0316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dong DM, Potter JD, White E, Ulrich CM, Cardon LR, Peters U. Genetic Susceptibility to Cancer: the Role of Polymorphisms in Candidate Genes. JAMA. 2008;299(20):2423–2436. doi: 10.1001/jama.299.20.2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Massague J, Chen YG. Controlling TGF-beta signaling. Genes and Development. 2000;14(6):627–644. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.