Abstract
NADPH oxidase-4 (Nox4) is an important modulator of redox signaling that is inducible at the level of transcriptional expression in multiple cell types. By contrast to other Nox enzymes, Nox4 is continuously active without requiring stimulation. We reported recently that expression of Nox4 is induced in the adult heart as an adaptive stress response to pathophysiological insult. To elucidate the potential downstream target(s) regulated by Nox4, we performed a microarray screen to assess the transcriptomes of transgenic (tg) mouse hearts in which Nox4 was overexpressed. The screen revealed a significant increase in the expression of many antioxidant and detoxifying genes regulated by Nrf2 in tg compared to wild-type (wt) mouse hearts, and this finding was subsequently confirmed by Q-PCR. Expression of glutathione biosynthetic and recycling enzymes was increased in tg hearts and associated with higher levels of both GSH and the ratio of reduced:oxidised GSH, compared to wt hearts. The increases in expression of the antioxidant genes and the changes in glutathione redox effected by Nox4 were ablated in an Nrf2-null genetic background. These data therefore demonstrate that Nox4 can activate the Nrf2-regulated pathway, and suggest a potential role for Nox4 in the regulation of GSH redox in cardiomyocytes.
Abbreviations: NADPH, nicotinamide adenine dinucleotide phosphate; Nrf2, NF-E2-related factor 2; Q-PCR, quantitative polymerase chain reaction; ER, endoplasmic reticulum; EB, embryoid body; αMHC, α myosin heavy chain; βMHC, β myosin heavy chain; MLC2v, myosin regulatory light chain 2; RT, reverse transcriptase; DTT, dithiothreitol; PAGE, polyacrylamide gel electrophoresis; ECL, enhanced chemiluminescence; PBS, phosphate-buffered saline; PVDF, polyvinylidene difluoride; SEM, standard error of the mean; ELISA, enzyme-linked immunosorbent serologic assay
Keywords: Nox4, Nrf2, Cardiomyocytes, Glutathione, Reactive oxygen species
Introduction
The NADPH oxidase family of enzymes (Noxs) produce reactive oxygen species (ROS) as their primary function, and consequently their roles in tightly regulated redox signaling pathways are areas of intense study. The catalytic core of each NADPH oxidase comprises a Nox subunit, 7 members of which have been identified (Nox1–5, Duox-1 and − 2). These are transmembrane proteins which facilitate the transfer of an electron from NADPH within the cell cytosol, via coordinated heme groups to reduce molecular oxygen (O2) to superoxide (O2-) on the opposite side of the membrane [1,2]. Despite the apparent similarity in function of the different isoforms at the biochemical level, different family members display distinct quantitative and qualitative cellular expression patterns, suggesting distinct functions [1–3]. In addition, in cells in which more than one isoform is expressed, the different isoforms typically display contrasting intracellular localisation patterns, again indicating specific functions [4–7].
The activities of different Nox isoforms are subject to very different modes of regulation. Thus Nox1 and Nox2 are acutely regulated by posttranslational mechanisms such as the phosphorylation of regulatory subunits induced by the activities of agonists. [2,3]. By contrast the activity of Nox4 is constitutive and does not require agonist stimulation, or the association of regulatory proteins other than p22phox[8–10]. Consequently its activity is dependent on its level of protein expression, and the ROS it generates is inducible at the level of gene transcription rather than acute, posttranslational mechanisms [11]. Furthermore, many groups have reported that the ROS generated by the different isoforms may differ. Thus while Nox2 is known to generate O2-, the ROS generated by Nox4 have only been convincingly demonstrated using detection systems specific to H2O2 [9,11–13].
Within cardiomyocytes, Nox2 and Nox4 are coexpressed, yet the two isoforms exhibit contrasting patterns of intracellular localisation. Whereas Nox2 is primarily located at the cell membrane [14], Nox4 displays a perinuclear and ER-associated expression pattern [10,15]. Several studies in mouse models of impaired Nox2 activity have demonstrated that Nox2-generated ROS are involved in the development and progression of cardiac hypertrophy, and contractile dysfunction induced by angiotensin II, pressure overload, or myocardial infarction [16–21]. By marked contrast to the apparent detrimental effects of Nox2 activity, Nox4-generated ROS were shown, in both gain-of-function and loss-of-function mouse models, to be beneficial during cardiac remodelling after load-induced stress by enhancing myocardial capillary density and functional cardiac compensation [10]. Endogenous Nox4 expression increases in cardiomyocyes in vivo as a response to pathological stresses of pressure overload or myocardial infarction, and in vitro following hypoxic insult. This increase acts to trigger adaptive stress response(s), including an increase in angiogenesis, through the activation of hypoxia inducible factor 1 [10]. Nox4 has also been suggested to be important in cardiomyocyte differentiation, as specific depletion of Nox4 expression in embryonic stem cells (ESCs) resulted in a reduction in the efficiency of ESC differentiation into beating EBs [22]. The involvement of p38 mitogen activated protein kinase (MAPK) in driving the cardiogenic pathway, downstream of Nox4, was also demonstrated in this study.
The precise molecular targets of oxidation, which mediate the cellular response(s) to increased ROS production by NADPH oxidases in cardiomyocytes and other cells, remain unclear. The most likely mechanism by which NADPH oxidase-generated ROS regulate cellular function is through the reversible thiol oxidation of reactive cysteine residues within regulatory proteins. Potential targets of ROS modification include protein tyrosine phosphatases (PTPs), protein tyrosine kinases, ion channels, and transcription factors [1,23]. One potential target and effector of ROS signaling is the cap ‘n’ collar basic leucine zipper transcription factor, Nrf2. Nrf2 acts as a positive transcriptional regulator of a battery of genes involved in antioxidant and xenobiotic defense systems [24,25]. Under normal, highly reducing cellular conditions Nrf2 is sequestered in the cytoplasm by association with the Kelch-like ECH-associated protein 1 (KEAP 1), and targeted for ubiquitination and proteasomal degradation [26]. However under conditions of increased ROS production, oxidation of critical cysteine residues within KEAP 1 triggers its dissociation from Nrf2, preventing Nrf2 degradation and allowing its translocation into the nucleus [27]. Within the nucleus Nrf2 can bind to cis-acting elements called antioxidant-responsive elements (AREs) or electrophile-responsive elements (EpREs) within the promoters of its target genes to effect their upregulation [28,29]. Recently it has additionally been demonstrated that Nrf2 itself is a potential direct target of oxidation, and that its nuclear distribution and consequent activity can be regulated through modification of specific cysteine residues by oxidation [30]. A number of studies have implicated ROS generated by NADPH oxidases in the activation of Nrf2-dependent pathways [31–35]. However all of these studies have relied on ablating NADPH oxidase activity by the use of pharmacological inhibitors, and direct genetic evidence for the involvement of any specific Nox isoform has so far been lacking.
In this study we aimed to identify downstream targets of Nox4-derived ROS in the heart in vivo. We have made use of a cardiac-specific Nox4 overexpressing transgenic (tg) mouse in which expression of the Nox4 transgene is driven by the αMHC promoter [10]. We have previously shown that Nox4 expression levels are significantly higher in the late stage fetal heart than in the normal adult heart [10]. Here we compare the effects of this decline in Nox4 levels after birth with the αMHC-driven increase in Nox4 expression that occurs at this time in the tg mice. We demonstrate that this increased expression of Nox4 in tg mice results in a highly significant upregulation in the expression of Nrf2-regulated genes, including those involved in glutathione (GSH) biosynthesis and recycling. In addition the early postnatal hearts of the transgenic mice display higher levels of reduced GSH and a higher ratio of GSH/GS-SG than wild-type littermate controls. The involvement of Nrf2 in this phenotype was confirmed in experiments where it was ablated in an Nrf2 null genetic background. These data represent the first definitive genetic demonstration of the activation of the Nrf2-mediated antioxidant pathway(s) by Nox4-derived ROS [36].
Materials and methods
Transgenic mice
Cardiomyocyte-targeted Nox4-transgenic mice have been described previously [10]. Lines were backcrossed > 10 generations onto a C57BL/6 background. Nrf2 null-mice have been described previously [37].
mRNA extraction and microarray analyses
Total RNA was extracted from homogenised heart tissue using the SV total RNA isolation kit (Promega) according to the manufacturer's instructions. RNA quality was assessed on an Agilent 2100 bioanalyser and cRNA was prepared and hybridised as described previously [38]. Two Mouse Expression Arrays (Affimetrix) each representing approximately 23,000 transcripts were screened. Equivalent samples of pooled RNA was used from triplicate heart samples prepared from 2-week-old Nox4-transgenic mouse hearts, or wild-type littermate controls (males and females). Expression levels of transcripts were assessed using statistical algorithms in the Microarray Analysis Suite 5.0 software (Affimetrix). Data were scaled to correct for variations in hybridisation efficiency and laser power, etc. Only genes with a change in expression (signal log ratio ≤ − 1.0 or ≥ + 1.0) between arrays were considered. Genes known to be sex linked in their expression, such as genes involved in X chromosome inactivation, were not included in the analyses.
cDNA synthesis and Q-PCR
Total RNA was reversed-transcribed, using random decamers, with M-MLV RT (Promega) according to the manufacturer's protocol. Control reactions, omitting the RT, were also performed in all cases. Relative gene expression was quantified on an Applied Biosystems 7000 sequence detection system (Applied Biosystems, UK) using SYBR Green and the comparative Ct method, with cytoskeletal β-actin levels used for normalization, as described previously [38]. Forward (F) and reverse (R) primers used to detect mouse transcripts were as follows (all 5′-3′).
βactin F, CTGTCGAGTCGCGTCCACCC; R, ATGCCGGAGCCGTTGTCGAC;
Nox4 F, CCGGACAGTCCTGGCTTATC; R, TGCTTTTATCCAACAATCTTCTTTT;
GSTα2 F, GCTTGATGCCAGCCTTCTG; R, GGCTGCTGATTCTGCTCTTGA;
GCLC F, GTTATGGCTTTGAGTGCTGCAT; R, ATCACTCCCCAGCGACAATC;
Txnrd 1 F, GATGCACCAGGCAGCTTTG; R, TCTTCGACTTTCCAGCCATAGT;
HO 1 F, CAGCCCCACCAAGTTCAAA; R, TCAGGTGTCATCTCCAGAGTG;
NQO 1 F, GCCCGCATGCAGATCCT; R, GGTCTCCTCCCAGACGGTTT;
GSR F, TGGTAGGAAGCCCACCACAA; R, ATTTGGGTCCCGTCCAATG;
Nrf2 F, CTACTCCCAGGTTGCCCACA; R, CGACTCATGGTCATCTACAAATGG.
Western blotting
Snap-frozen tissue was simultaneously homogenised and lysed in hypotonic lysis buffer [50 mM Hepes, pH 7.4, 10 mM KCl, 5 mM EDTA, 5 mM EGTA, 2 mM MgCl2, 2 mM DTT, 0.1% CHAPS, and protease inhibitor cocktail (Sigma)]. Protein concentrations were determined by Bradford assay to ensure equal protein loading and samples (20–50 μg) were then denatured in Laemmli loading buffer. Proteins were separated on either an 8 or 10% SDS-PAGE gel and transferred to PVDF membrane. Membranes were probed with antibodies against peroxiredoxin-SO3 (Abfrontier), Nox2 (BD Transduction), and Nox4 [4]. Blots were probed for β-actin (Sigma) as a loading control. Protein bands were visualised using Amersham Biosciences ECL Western blotting detection reagent (GE Healthcare) according to the manufacturer's instructions.
Nrf2 binding activity
Nuclear extracts were prepared from freshly isolated wt and Nox4-overexpressing tg mice hearts using the NE-PER Nuclear and Cytoplasmic Extraction kit (Pierce Biotechnology) as per the manufacturer's recommendations. Protein concentrations were measured by the Bradford assay and Western blot analysis of nuclear extracts showed a purity of ~ 85%. Nrf2 DNA binding activity was assessed using a TransAM Transcription Factor ELISA kit (Active Motif). The assay consists of oligonucleotides encoding a consensus antioxidant response element (ARE) immobilised to a 96-well plate to which Nrf2 binds. Ten micrograms of nuclear extract was incubated in each well in either the presence or the absence of competing consensus ARE-oligonucleotide for 1 h, as per the manufacturer's instruction. A primary antibody against Nrf2 followed by a horseradish peroxidase (HRP)-conjugated secondary antibody allowed a colorimetric readout of Nrf2 binding activity, which is expressed as OD450 nm ± SD.
Immunocytochemistry
Immunocytochemistry was performed on 6-μm sections, cut from paraffin-embedded wt and tg hearts from 9-day-old mouse pups. Primary antibodies against KEAP 1 (Cell Signaling Technology, Cat. No. 4617) and Nrf2 (Santa Cruz Biotechnology, Cat. No. sc-722) were both used at a dilution of 1:50. Biotinylated-secondary antibodies were detected using a peroxidase-based immunostaining kit (Vectastain Elite ABC kit, Vector Laboratories) and diaminobenzidine (ImmPACT DAB; Vector Laboratories). Sections were counterstained with hematoxylin.
Glutathione assays
Freshly isolated heart tissue was snap-frozen in liquid N2, and subsequently stored at − 80 °C. Samples were homogenised in phosphate-buffered saline containing 2 mM EDTA, at a concentration of 20 mg tissue/ml PBS. GSH assays were performed using the GSH-Glo assay kit (Promega), exactly as described by the manufacturer. To measure total glutathione levels (GSH + GS-SG), extracts (50 μl) were preincubated in a final concentration of 1 mM tris(2-carboxyethyl)phosphine hydrochloride (TCEP; Sigma), for 30 min at room temperature, before proceeding with the assay. From measurements of the total and reduced glutathione levels, the levels of oxidised GS-SG (total-reduced) and the glutathione redox couple (GSH/GS-SG) were calculated.
Statistical analyses
Data are expressed as mean ± SEM. Comparisons between wt and tg cohorts or measurements at different time points were made by unpaired Student's t test.
Results
Expression of Nox4 in the postnatal heart
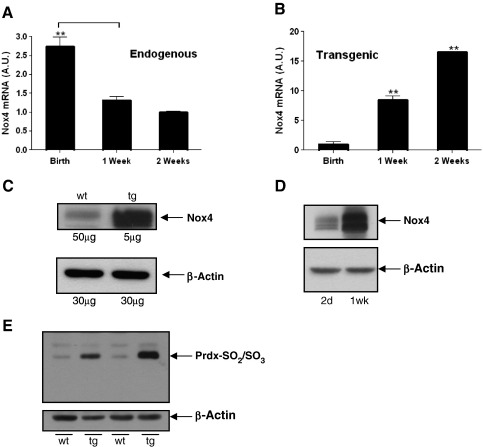
We have demonstrated previously that Nox4 mRNA expression is much higher in the late-staged fetal heart than in the adult heart [10]. We here further investigated the pattern of endogenous Nox4 mRNA expression in mouse hearts immediately after birth, and at 1 week and 2 weeks of age. We found a sharp decline in expression immediately after birth, and in particular during the first week of life (Fig. 1A). Recently we reported the generation of a tg mouse line, which expresses Nox4 specifically in cardiomyocytes, under the control of the mouse αMHC promoter [10]. In rodents, expression of βMHC within the ventricles predominates during embryonic and fetal development while αMHC is strongly upregulated from birth [39]. Accordingly Nox4 mRNA levels increase sharply during the immediate postnatal period in tg mice (Fig. 1B). At the protein level, endogenous Nox4 can be detected in the late stage (15 day) wt embryo heart, and transgenic Nox4 expression at this stage (allowing for the differences in amounts of protein loaded) was estimated to be approximately 70-fold higher (Fig. 1C). This is in broad agreement with measurements of relative levels of Nox4 mRNA in wt and tg hearts at this stage (data not shown). After birth, the decline in the levels of normal Nox4 expression (Fig. 1A and [10]) precludes its detection by Western blotting in our postnatal wt animals. By contrast, transgenic Nox4 protein expression increases sharply after birth, in line with the mRNA expression pattern (Fig. 1D). Thus Nox4 in these tg mice is increasing at a time when the normal endogenous levels decrease.
Fig. 1.
Expression of Nox4 in wt and tg hearts. (A) Q-PCR analyses of relative expression of endogenous Nox4 mRNA, in postnatal hearts at birth (day 0), 1 week, and 2 weeks. (B) Q-PCR analyses of Nox4 transgene expression in Day 0, 1 week, and 2 week postnatal hearts. Hearts samples from 3 littermate animals were analysed in all cases. Values are shown normalised to β-actin, and are plotted as arbitary units (AU). (C) Nox4 protein levels in 15 day wt and tg embryonic hearts (amounts of protein loaded as inducated); (D) Nox4 protein levels in tg hearts, at 2 days and 2 weeks after birth, as indicated (15 μg protein loaded in each case). (E) Analyses of hyperoxidised Prdx in wt and tg littermate controls at 2 weeks after birth, as indicated (15 μg protein loaded in each case). ** P < 0.01.
We reported previously that extracellular H2O2 in tg mouse myocardium was increased by approximately 30% [10]. We have further analysed the intracellular production of H2O2in situ, using an antibody specific to the hyperoxidised active site of four peroxiredoxin isoforms (Prdx 1–4) in which the thiol group within the active site is oxidized to either a sulphinic (SO2) or (irreversibly) to a sulphonic (SO3) group. Previous reports have demonstrated that high levels of hyperoxidised Prdx are observed after ex vivo treatment of heart tissue with H2O2[40]. As shown in Fig. 1E, a significant increase in levels of hyperoxidised Prdx was observed in 2-week-old tg mouse hearts, compared to wt controls, consistent with the production of intracellular H2O2.
Microarray analyses of Nox4-overexpressing hearts
To elucidate the molecular targets of Nox4-generated ROS, we performed a microarray screen, in which we compared the transcriptomes of 2-week-old transgenic and wild-type hearts. At this point, the significant changes in Nox4 expression in tg compared to wt mice, and consequent ROS generation, would be likely to effect dynamic switches in signaling pathways and downstream transcriptional programs. We screened two Mouse Expression Set 430A chips (Affymetrix), using pooled cRNA prepared from equivalent amounts of either 3 tg or 3 wt hearts, and identified 174 characterised, unique genes whose expression was changed at least 2-fold as a result of increased Nox4 expression. Of these, 70 genes were upregulated and 104 genes were downregulated (see Supplementary Data for a list of upregulated genes). It should be noted, however, that the use of only one chip per sample precludes any statistical analyses to evaluate the significance of changes in gene expression of less than 2-fold. Thus the lists of genes presented here may underestimate the downstream transcriptional targets of Nox4 signaling. These microarray data have been submitted to the EBI ArrayExpress database, and can be accessed at http://www.ebi.ac.uk/arrayexpress, Accession Number E-MEXP-3161.
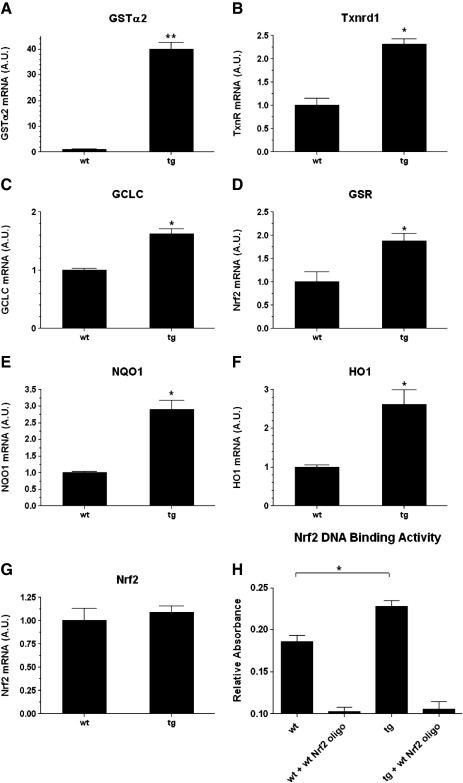
The differentially regulated genes identified were analysed using the Ingenuity Systems Pathway Analysis Software (http://www.ingenuity.com/), which identified cell proliferation and growth as the cellular function which had been most affected by Nox4 overexpression (P = 8.10 × 10-9). In addition the Nrf2-regulated antioxidant response pathway was identified by this application as a canonical pathway that was highly significantly changed (P = 2.27 × 10-6). Thus a significant upregulation was observed in the expression of several genes known to be positively regulated by Nrf2, including glutathione S-transferase α2 (GSTα2), thioredoxin reductase 1 (Txnrd1), and NAD(P)H dehydrogenase, quinone 1 (NQO1). In addition, the expression levels of many other known target genes of Nrf2 activation, such as heme oxygenase (HO 1), glutamate-cysteine ligase, catalytic subunit (GCLC), and glutathione reductase (GSR), were also increased, although less than 2-fold (Table 1). We verified the results of the microarray by individual Q-PCR analyses of multiple RNAs isolated from 2-week-old wt and tg mouse hearts (Fig. 2). In agreement with our microarray data, the most significant change in expression was observed in the case of GSTα2, which was upregulated approximately 40-fold (Fig. 2A), while more modest increases in mRNA levels of other genes were apparent (HO 1, NQO1, GCLC, Txnrd1, GSR; Figs. 2B–F).
Table 1.
Nrf2-regulated genes, found to be upregulated by Nox4 in a microarray screen.
| Gene name | GenBank Accession | Log2 ratio |
|---|---|---|
| aldo keto reductase | AK009462 | 0.3 |
| aldehyde oxidase 1 | NM_009676 | 1.0 |
| activating transcription factor 4 | AV314773 | 1.2 |
| chaperone subunit 7 | NM_007638 | 1.0 |
| dnaJ (Hsp40) homolog, subfamily A, member 2 | C77509 | 0.4 |
| dnaJ (Hsp40) homolog, subfamily A, member 3 | NM_023646 | 0.4 |
| dnaJ (Hsp40) homolog, subfamily B, member 9 | NM_013760 | 0.7 |
| dnaJ (Hsp40) homolog, subfamily B, member 11 | AK010861 | 0.4 |
| dnaJ (Hsp40) homolog, subfamily C, member 3 | BC013766 | 0.2 |
| epoxide hydrolase 1 | NM_010145 | 1.1 |
| ferritin light chain 2 | NM_008049 | 0.6 |
| glutamate-cysteine ligase, catalytic subunit, | BC019374 | 0.9 |
| glutamate-cysteine ligase , modifier subunit | NM_008129 | 0.8 |
| glutathione-S-transferase, alpha 2 | NM_008182 | 5.5 |
| glutathione-S-transferase, alpha 4 | NM_010357 | 0.9 |
| glutathione-S-transferase, mu 2 | NM_008183 | 0.8 |
| glutathione S-transferase, mu 1 | NM_010358 | 0.8 |
| glutathione S-transferase omega 1 | NM_010362 | 0.7 |
| glutathione reductase 1 | NM_010344 | 0.6 |
| heme oxygenase 1 | NM_010442 | 0.4 |
| herpud 1 | AI835088 | 0.5 |
| microsomal glutathione S-transferase 1 | BC009155 | 1.1 |
| NAD(P)H dehydrogenase, quinone 1 | AV158882 | 1.4 |
| peroxiredoxin 1 | NM_011034 | 0.3 |
| related RAS viral (r-ras) oncogene homolog 2 | BE200500 | 0.5 |
| sequestosome 1 | BM232298 | 0.8 |
| small stress protein-like protein (HSP22) | AF250139 | 0.6 |
| superoxide dismutase 1 | BC002066 | 0.3 |
| thioredoxin | NM_011660 | 0.4 |
| thioredoxin reductase 1 | NM_015762 | 1.0 |
| v-maf musculoaponeurotic fibrosarcoma oncogenefamily, protein G | BC002092 | 0.9 |
| valosin containing protein | AI195225 | 0.2 |
Fig. 2.
Expression of Nrf2-regulated genes in Nox4 tg mouse hearts. (A–G) Q-PCR analyses of Nrf2-regulated genes, and Nrf2 itself in 2 week postnatal tg and wt mouse hearts as indicated. Triplicate tg and wt littermate controls were analysed, and relative expression is shown normalised to β-actin in all cases, plotted as arbitary units (AU). (H) Analyses of binding activity of Nrf2 in wt and tg littermate mouse hearts. In both cases binding to the immobilised Nrf2 oligonucleotide is ablated by self-competition (+ wt Nrf2 oligo). ** P < 0.01, * P < 0.05.
We also assayed Nrf2 mRNA levels and DNA binding activity in these hearts. As shown in Fig. 2G, the levels of Nrf2 mRNA were equivalent in the wt and tg groups. However an ELISA-based Nrf2 binding assay demonstrated significantly higher levels of binding to an immobilised consensus ARE in nuclear extracts prepared from tg compared to wt hearts (Fig. 2H).
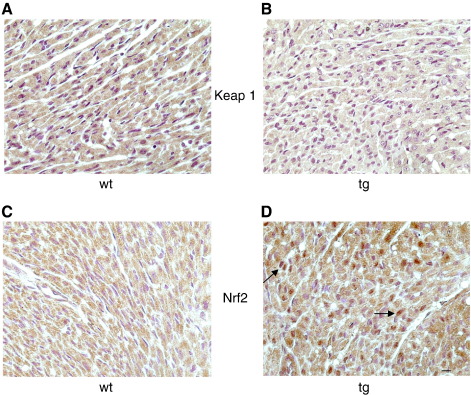
To further validate these findings we performed immunostaining on heart sections of wt and tg hearts using antibodies to KEAP 1 or Nrf2. KEAP 1 staining was cytoplasmic and was markedly higher in the wt, rather than tg hearts (Figs. 3A and B). By contrast Nrf2 staining was clearly localized strongly to the nucleus, specifically in the tg heart sections (Figs. 3C and D).
Fig. 3.
Expression of KEAP 1 and Nrf2 in Nox4 tg mouse hearts. Immunocytochemistry showing stronger cytoplasmic expression of KEAP 1 in wt (A) compared to Nox4 tg (B) hearts (brown stain). Increased nuclear localisation of Nrf2 expression is seen in tg (D) compared to wt (C) hearts (brown stain). Arrows depict strong nuclear expression of Nrf2 that is not apparent in wt sections. Nucleii are stained with hematoxylin (violet) in all panels. Scale bar: 10 μm.
These findings are consistent with the hypothesis that Nox4-generated ROS may upregulate Nrf2 activity by the posttranslational oxidative modification of Keap 1, which results in its degradation and the consequent nuclear translocation of Nrf2 [27].
Glutathione levels are increased in the transgenic hearts
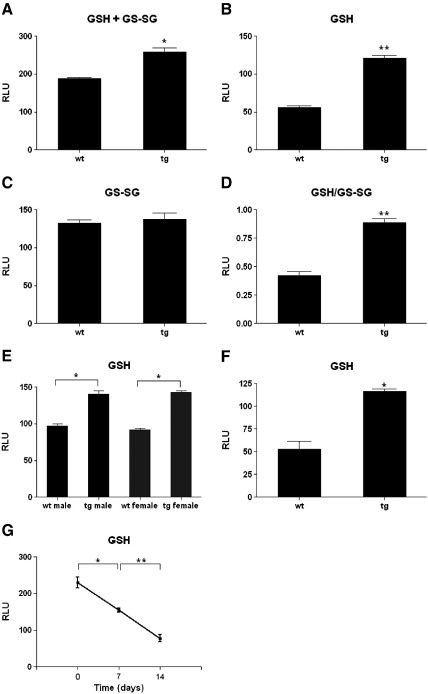
Some of the target genes of Nrf2 activation are involved in both the biosynthesis of GSH, such as GCLC [41], and its redox recyling, such as GSR [42]. Thus Nrf2 has been shown to be a critical regulator of both the level of GSH within the cell and the ratio of oxidised:reduced GSH, i.e., the GSH/GS-SG redox couple. We therefore measured both the total level of GSH (i.e., GSH + GS-SG) and the level of reduced GSH alone, in the hearts of 12-day-old transgenic mouse and their littermate controls, and used these measurements to further calculate the GSH:GS-SG redox couple. As in the microarray analyses, a time point close to 2 weeks was chosen in order to maximise the effects of Nox4 signaling apparent in the hearts. As shown in Fig. 4A, the total levels of glutathione (GSH + GS-SG) were significantly elevated in the tg hearts, and this increase was predominantly due to the increase in the level of reduced (GSH), rather than oxidised (GS-SG), as reflected in the increased GSH:GS-SG ratio (Figs. 4B–D). Because it is desirable, where possible, to use littermate animals for these studies, both males and females were used. Sex differences in the levels of GSH have been demonstrated for aging mice, although in most tissues GSH levels in young mice were reported to be equivalent [43]. We compared GSH levels in male and female wt and tg mice aged 12 days, but found levels to be equivalent between males and females in both the wt and the tg groups (Fig. 4E). To further confirm that the difference in GSH levels observed was due to Nox4 overexpression, we studied a second, independent αMHC-Nox4 tg mouse line and found similar results (Fig. 4F), thereby excluding an integration site effect.
Fig. 4.
Glutathione levels and GSH/GS-SG redox couple in Nox4 tg hearts. Levels of (A) total (GSH + GS-SG), (B) reduced (GSH), and (C) oxidised (GS-SG) glutathione in 12 day postnatal Nox4 tg and littermate wt hearts (males and females). (D) Ratio of GSH/GS-SG in these hearts. Cohorts of 3 tg and 3 wt littermate controls were used for this study. (E) GSH levels in hearts of tg and wt, males and females, from 2 pooled 12 day litters. Cohorts of 3 animals were assessed in each case. (F) GSH levels in 3 tg and 3 wt littermate control hearts of 2 week postnatal mice of a second Nox4 tg line. (G) Time course of endogenous GSH levels in hearts of wt mice. Three littermate samples were assessed at birth (Day 0), 1 week, and 2 weeks. Results are plotted on an arbitrary scale of relative light units (RLU) ** P < 0.01, * P < 0.05.
We also measured the levels of GSH in the hearts of normal wt mice in a postnatal time course. As seen in Fig. 4G, GSH levels were found to decrease markedly during the first 2 weeks of life, in parallel with the observed decrease in expression of endogenous Nox4 (Fig. 1A).
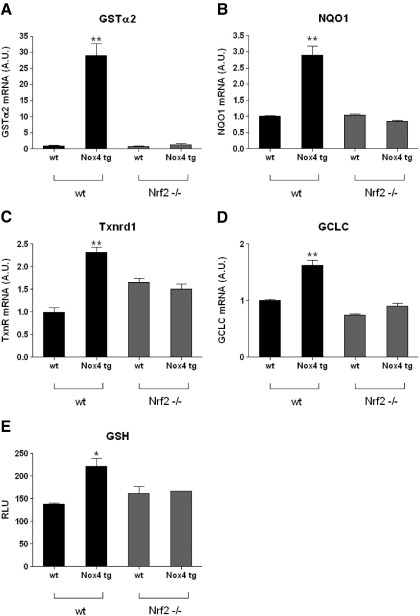
Ablation of effects of Nox4 overexpression in an Nrf2-null genetic background
To confirm that the changes observed in the expression of known Nrf2-regulated genes on overexpression of Nox4 were, indeed, a consequence of the activation of Nrf2, we crossed the Nox4 tg mice onto an Nrf2-null genetic background [37]. The Nrf2-null homozygotic mice were found not to be fertile; thus, we set up crosses between cohorts of heterozygous Nrf2−/+ male mice, and heterozygous Nrf2−/+ females which also carried the Nox4 transgene. The offspring of these crosses were genotyped, and age-matched animals from several different litters (males and females) were pooled from the four groups of interest: Nrf2−/− and Nrf2+/+ mice that did, or did not, carry the Nox4 transgene in each case. We first assessed by Q-PCR the mRNA levels of a subset of the putative Nrf2-regulated genes, which were shown previously to be upregulated by ectopic Nox4 expression. As in the original microarray analyses, RNA was isolated from hearts from 2-week-old mice for this study. As shown in Fig. 5, the increase in transcription on ectopic Nox4 expression of all the genes tested (GSTα2, NQO1, TrxR1, and GCLC) was ablated in the Nrf2-null background. We additionally measured the levels of GSH in samples of the same hearts used to prepare RNA, and again the increase in GSH, apparent in the Nox4tg hearts, was not seen in mice lacking Nrf2 (Fig. 5E). These data are therefore consistent with Nox4-generated ROS both effecting changes in the transcription of antioxidant and phase II detoxifying enzymes and mediating regulation of the GSH cellular redox state within the heart by redox control of Nrf2.
Fig. 5.
Upregulation of Nrf2-regulated genes and GSH levels is ablated in an nrf2-null genetic background. (A–D) Q-PCR analyses of Nrf2-regulated genes in 2 week postnatal Nox4 tg and wt hearts in a normal (black bars) or Nrf2-null genetic background (gray bars), as indicated. Cohorts of 3 age-matched mice (males and females) were used for this study, from pooled litters, and relative expression is shown normalised to β-actin in all cases, plotted as arbitary units (AU). (E) GSH levels in the same cohorts of mice assessed in A–D, plotted on an arbitrary scale of relative light units (RLU). ** P < 0.01, * P < 0.05 in all cases.
Activation of Nrf2 is not apparent in Nox2-overexpressing mice
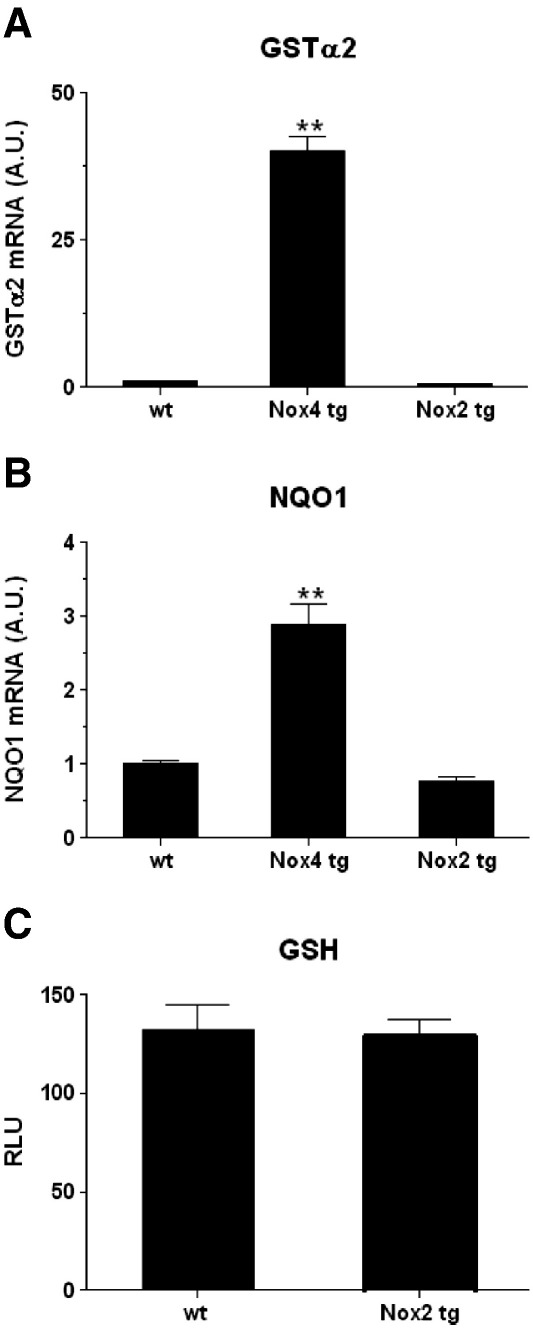
Previous studies using the flavoprotein inhibitor, diphenylene iodonium chloride (DPI), have implicated NADPH oxidase(s) in the activation of Nrf2 in various in vitro settings, but do not inform which specific Nox isoform might be involved [31–35]. To determine whether the ability to activate Nrf2 in cardiomyocytes in vivo is specific to Nox4, we analysed a transgenic mouse line in which Nox2 (the main other Nox isoform expressed in cardiomyocytes) is overexpressed specifically in cardiomyocyes under the regulation of the mouse MLC-2v promoter. These Nox2 tg mice were shown to express high levels of Nox2 mRNA and protein in the postnatal heart (manuscript in preparation). We assessed the mRNA levels of the two Nrf2-regulated genes which were shown to be most significantly increased in the Nox4 tg mouse hearts, GSTα2 and NQO1, but no increase in expression was observed in the Nox2 tgs compared to wt controls (Figs. 6A and B). GSH levels were also found to be unchanged in 2-week-old Nox2 tg mice compared to wt littermate controls (Fig. 6C).
Fig. 6.
Nox2 overexpression does not activate Nrf2. (A, B) Q-PCR analyses of Nrf2-regulated genes in Nox2 tg, Nox4 tg, and wt mouse hearts as indicated. Triplicate heart samples were analysed from adult Nox2 tg and age-matched wt control animals, and 2 week Nox4 tg mice. Relative expression is shown normalised to β-actin in all cases, plotted as arbitary units (AU). ** P < 0.01. C; GSH levels in cohorts of 4 approx 6-week-old Nox2 tg mouse hearts and wt littermate controls (males and females) plotted on an arbitrary scale of relative light units (RLU).
Discussion
Activation of Nrf2-dependent transcription by Nox4 in cardiomyocytes
There is compelling evidence to suggest that ROS generated by Nox4 functions in many different cell types to determine cell fate and function [22,44–48]. Nox4 produces ROS constitutively and consequently its activity reflects its level of expression [11]. Significant changes in intracellular Nox4 mRNA and protein levels are therefore likely to signal regulatory cellular physiological processes. However, the effects of Nox4-dependent signaling appear highly cell-type and/or developmental-stage specific. Thus in cultured vascular smooth muscle cells (VSMCs) in vitro, increased Nox4 expression can act to promote either cellular differentiation or proliferation, dependent on cell culture conditions [44,47]. In preadipocytes and cardiac fibroblasts, increased Nox4 expression acted to promote differentiation into adipocytes and myofibroblasts, respectively [45,46], while increased Nox4 expression in endothelial cells was shown to promote a more proliferative phenotype [49]. It is therefore of significance that in the microarray that we performed here, the cellular function that was identified to be most significantly altered as a result of Nox4 overexpression in cardiomyocytes was cell growth and proliferation. The phenotypic consequences of this altered gene expression are, however, beyond the scope of this paper. It should be noted, however, that Nox4-null animals have been generated and do not display any basal phenotype [10]. This suggests that Nox4 does not play a critical, nonredundant role in cellular differentiation. Future experiments involving inducible and/or stage-specific cardiac ablation of Nox4 during embryogenesis are therefore needed to inform its normal function(s) in cardiomyocyte cell differentiation and growth.
The precise molecular mechanisms which underlie these Nox4-dependent cellular functions, however, are largely unknown. In particular the targets of oxidation of the ROS, generated by Nox4, need to be elucidated. Recently, thiol modification and consequent inhibition of two protein phosphatises, PTP1B and MAPK phosphatase MKP-1, dependent on Nox4-generated ROS, have been demonstrated in endothelial cells and fibroblasts respectively [50,51]. We demonstrate here the ability of Nox4-generated ROS to activate the Nrf2-dependent pathway in cardiomyocytes in vivo. Nrf2 is a positive regulator of a plethora of genes involved in antioxidant and xenobiotic defense. These include the phase II detoxifying enzymes, such as glutathione S-transferases (GSTs) and NAD(P)H:oxioreductase 1 (NQO 1), enzymes involved in the biosynthesis and recycling of glutathione, such as glutamate-cysteine ligase catalytic subunit (GCLC) and glutathione reductase (GSR), and antioxidants such as thioredoxin reductase 1(Txnrd1) and heme oxygenase (HO 1) [24,42,52]. Our microarray screen demonstrated the expression of Nrf2-dependent genes to be highly significantly upregulated by cardiomyocyte-specific overexpression of Nox4. This Nox4-directed upregulation was completely ablated in an Nrf2-null genetic background, demonstrating definitively the involvement of Nrf2 in the molecular pathway.
There have been several reports describing distinct mechanisms of activation of Nrf2, most (but not all) of which appear to be redox-mediated [27,53–57]. In vitro and in vivo, the expression of antioxidant and detoxification enzymes regulated by Nrf2 has been shown to be induced by a variety of chemical agents which act as electrophiles, including diphenols, quinones, isothiocyanates, arsenicals, and heavy metals [58]. Both Nrf2 itself and KEAP1, which acts as a negative regulator of Nrf2 activity, are susceptible to oxidative thiol modifications of reactive cysteines which regulate their functions [27,30], and are therefore potential targets of NADPH oxidase-generated ROS. In addition phoshorylation of Nrf2 by protein kinase C has been shown to facilitate the dissociation of Nrf2 from KEAP1; a process which may also be redox regulated [59]. Our data are consistent with activation of Nrf2 by mechanism(s) involving oxidative posttranslational modification. Thus mRNA levels of Nrf2 were not altered in the Nox4 tg mice, whereas the nuclear binding activity of Nrf2 was significantly increased by Nox4 overexpression. Several previous reports have suggested that ROS generated by NADPH oxidases can activate Nrf2 in vitro[31–35]. However, all of these studies have been based on the inhibition of the oxidase activity by nonspecific agents such as DPI and apocynin, and the specificity of both these reagents has been questioned [60]. The data presented here represent the first direct genetic evidence that ROS generated in situ by an NADPH oxidase, i.e., Nox4, can activate Nrf2.
It is now clear that different Nox isoforms can effect distinct signaling responses in cells in which they are coexpressed [4,6,7]. This likely results from differences in the intracellular localisation of the Nox isoforms, the levels and species of ROS generated, and the duration of production of the ROS. By contrast to Nox4, cardiac-specific overexpression of Nox2 did not result in activation of Nrf2 in our Nox2 tg mice. One potential explanation for this difference is that constitutive, continuous production of ROS is required to modulate Nrf2.
In addition to Nrf2-regulated genes, we also found many other genes that were upregulated in the Nox4 tg hearts, which remained altered in the Nrf2-null genetic background (data not shown). These results suggest that Nox4-derived ROS can regulate other pathways distinct from those controlled by Nrf2.
Regulation of GSH levels by Nox4 during cellular differentiation
Nrf2 has been suggested previously to be a key regulator of GSH redox in a variety of cellular settings [61,62]. We show here that in cardiomyocytes Nox4-generated ROS regulates both the level and the redox state of GSH through modulation of Nrf2 activity. In addition to its role as part of an integrated antioxidant system, the GSH redox state per se is known to be a critical regulator of skeletal muscle differentiation [63]. In addition, endogenous H2O2 was shown to regulate the GSH redox state during skeletal muscle cell differentiation in vitro, although the source of the ROS was not identified [61]. Nonetheless, the H2O2 produced during the differentiation process was shown to act in a strikingly similar fashion to the ROS generated in the Nox4 tg mouse hearts, described here. Namely the H2O2 acted via Nrf2 to mediate the cellular GSH redox state by activation of the GSH biosynthetic and recycling enzmes, GCLC and GSR. The authors further demonstrated the involvement of phosphatidylinositol 3-kinase in the activation of Nrf2, downstream of the ROS production. Nox4 activity has been demonstrated to be critical for efficient cardiomyocyte differentiation, in vitro. Our data may therefore suggest that modulation of GSH levels through regulation of Nrf2 may be an important mechanism which underlies the emerging role of Nox4 in cellular differentiation [22,44–48].
Role of Nox4 in cardiomyocytes
Diverse functions, including gene expression, cell-cycle progression, apoptosis, and metabolism, have also been suggested to be regulated by the intracellular GSH redox state [64–66]. Few studies have so far focused on the specific role of Nox4 with cardiomyocytes in vivo. We found that GSH levels drop sharply in the postnatal heart, concomitant with a decrease in Nox4 expression. During this time cardiomyocytes undergo many complex changes in their physiology as they adapt to the demands of their altered environment [67]. Therefore the regulation of GSH levels via Nrf2 to influence the profound changes in cardiomyocyte proliferation, cell growth, and metabolism that occur in the postnatal heart may be an important role of Nox4.
A function for Nox4 in the adult heart has also been demonstrated in that, as a response to pathophysiological stress, its expression is increased and it effects an adaptive cardioprotective response [10]. Therefore higher levels of expression of Nox4 in cardiomyocytes associate with a more fetal phenotype, that can be reactivated in the adult heart as a response to stress [10,68]. Recently it has also been suggested that Nrf2 is a mediator of this cardioprotective adaptive response to hemodynamic stress [69], and again suggests that activation of Nrf2 is a molecular mechanism underlying Nox4 function in the cardiomyocyte.
Taken together, the results of this study suggest that Nox4 may be an important regulator of GSH redox state in cardiomyocytes, through the modulation of Nrf2 activity.
The following are the supplementary materials related to this article.
Genes significantly upregulated by Nox4 overexpression
Acknowledgments
We thank Estibaliz Aldecoa-Otalora Astarloa for technical assistance with microarray analyses and Mr. Yurdakal Mustapha for histological assistance. This work was supported by the British Heart Foundation (BHF) Grants PG/08/110/26228 and RG/08/011/25922, a BHF Centre of Excellence award, (2E/08/003), and a Leducq Foundation Transatlantic Network of Excellence award (09CV DO1).
References
- 1.Bedard K., Krause K.H. The NOX family of ROS-generating NADPH oxidases: physiology and pathophysiology. Physiol. Rev. 2007;87:245–313. doi: 10.1152/physrev.00044.2005. [DOI] [PubMed] [Google Scholar]
- 2.Lambeth J.D. NOX enzymes and the biology of reactive oxygen. Nat. Rev. Immunol. 2004;4:181–189. doi: 10.1038/nri1312. [DOI] [PubMed] [Google Scholar]
- 3.Brown D.I., Griendling K.K. Nox proteins in signal transduction. Free Radic. Biol. Med. 2009;47:1239–1253. doi: 10.1016/j.freeradbiomed.2009.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anilkumar N., Weber R., Zhang M., Brewer A., Shah A.M. Nox4 and nox2 NADPH oxidases mediate distinct cellular redox signaling responses to agonist stimulation. Arterioscler. Thromb. Vasc. Biol. 2008;28:1347–1354. doi: 10.1161/ATVBAHA.108.164277. [DOI] [PubMed] [Google Scholar]
- 5.Helmcke I., Heumuller S., Tikkanen R., Schroder K., Brandes R.P. Identification of structural elements in Nox1 and Nox4 controlling localization and activity. Antioxid. Redox Signal. 2009;11:1279–1287. doi: 10.1089/ars.2008.2383. [DOI] [PubMed] [Google Scholar]
- 6.Hilenski L.L., Clempus R.E., Quinn M.T., Lambeth J.D., Griendling K.K. Distinct subcellular localizations of Nox1 and Nox4 in vascular smooth muscle cells. Arterioscler. Thromb. Vasc. Biol. 2004;24:677–683. doi: 10.1161/01.ATV.0000112024.13727.2c. [DOI] [PubMed] [Google Scholar]
- 7.Van Buul J.D., Fernandez-Borja M., Anthony E.C., Hordijk P.L. Expression and localization of NOX2 and NOX4 in primary human endothelial cells. Antioxid. Redox Signal. 2005;7:308–317. doi: 10.1089/ars.2005.7.308. [DOI] [PubMed] [Google Scholar]
- 8.Ambasta R.K., Kumar P., Griendling K.K., Schmidt H.H., Busse R., Brandes R.P. Direct interaction of the novel Nox proteins with p22phox is required for the formation of a functionally active NADPH oxidase. J. Biol. Chem. 2004;279:45935–45941. doi: 10.1074/jbc.M406486200. [DOI] [PubMed] [Google Scholar]
- 9.Martyn K.D., Frederick L.M., von Loehneysen K., Dinauer M.C., Knaus U.G. Functional analysis of Nox4 reveals unique characteristics compared to other NADPH oxidases. Cell. Signal. 2005;18:69–82. doi: 10.1016/j.cellsig.2005.03.023. [DOI] [PubMed] [Google Scholar]
- 10.Zhang M., Brewer A.C., Schroder K., Santos C.X., Grieve D.J., Wang M., Anilkumar N., Yu B., Dong X., Walker S.J., Brandes R.P., Shah A.M. NADPH oxidase-4 mediates protection against chronic load-induced stress in mouse hearts by enhancing angiogenesis. Proc. Natl Acad. Sci. U. S. A. 2010;107:18121–18126. doi: 10.1073/pnas.1009700107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Serrander L., Cartier L., Bedard K., Banfi B., Lardy B., Plastre O., Sienkiewicz A., Forro L., Schlegel W., Krause K.H. NOX4 activity is determined by mRNA levels and reveals a unique pattern of ROS generation. Biochem. J. 2007;406:105–114. doi: 10.1042/BJ20061903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dikalov S.I., Dikalova A.E., Bikineyeva A.T., Schmidt H.H., Harrison D.G., Griendling K.K. Distinct roles of Nox1 and Nox4 in basal and angiotensin II-stimulated superoxide and hydrogen peroxide production. Free Radic. Biol. Med. 2008;45:1340–1351. doi: 10.1016/j.freeradbiomed.2008.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nisimoto Y., Jackson H.M., Ogawa H., Kawahara T., Lambeth J.D. Constitutive NADPH-dependent electron transferase activity of the Nox4 dehydrogenase domain. Biochemistry. 2010;49:2433–2442. doi: 10.1021/bi9022285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heymes C., Bendall J.K., Ratajczak P., Cave A.C., Samuel J.L., Hasenfuss G., Shah A.M. Increased myocardial NADPH oxidase activity in human heart failure. J. Am. Coll. Cardiol. 2003;41:2164–2171. doi: 10.1016/s0735-1097(03)00471-6. [DOI] [PubMed] [Google Scholar]
- 15.Ago T., Kuroda J., Pain J., Fu C., Li H., Sadoshima J. Upregulation of Nox4 by hypertrophic stimuli promotes apoptosis and mitochondrial dysfunction in cardiac myocytes. Circ. Res. 2010;106:1253–1264. doi: 10.1161/CIRCRESAHA.109.213116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bendall J.K., Cave A.C., Heymes C., Gall N., Shah A.M. Pivotal role of a gp91phox-containing NADPH oxidase in angiotensin II-induced cardiac hypertrophy in mice. Circulation. 2002;105:293–296. doi: 10.1161/hc0302.103712. [DOI] [PubMed] [Google Scholar]
- 17.Byrne J.A., Grieve D.J., Bendall J.K., Li J.M., Gove C., Lambeth J.D., Cave A.C., Shah A.M. Contrasting roles of NADPH oxidase isoforms in pressure-overload versus angiotensin II-induced cardiac hypertrophy. Circ. Res. 2003;93:802–805. doi: 10.1161/01.RES.0000099504.30207.F5. [DOI] [PubMed] [Google Scholar]
- 18.Doerries C., Grote K., Hilfiker-Kleiner D., Luchtefeld M., Schaefer A., Holland S.M., Sorrentino S., Manes C., Schieffer B., Drexler H., Landmesser U. Critical role of the NAD(P)H oxidase subunit p47phox for left ventricular remodeling/dysfunction and survival after myocardial infarction. Circ. Res. 2007;100:894–903. doi: 10.1161/01.RES.0000261657.76299.ff. [DOI] [PubMed] [Google Scholar]
- 19.Looi Y.H., Grieve D.J., Siva A., Walker S.J., Anilkumar N., Cave A.C., Marber M., Monaghan M.J., Shah A.M. Involvement of Nox2 NADPH oxidase in adverse cardiac remodeling after myocardial infarction. Hypertension. 2008;51:319–325. doi: 10.1161/HYPERTENSIONAHA.107.101980. [DOI] [PubMed] [Google Scholar]
- 20.Nakagami H., Takemoto M., Liao J.K. NADPH oxidase-derived superoxide anion mediates angiotensin II-induced cardiac hypertrophy. J. Mol. Cell. Cardiol. 2003;35:851–859. doi: 10.1016/s0022-2828(03)00145-7. [DOI] [PubMed] [Google Scholar]
- 21.Satoh M., Ogita H., Takeshita K., Mukai Y., Kwiatkowski D.J., Liao J.K. Requirement of Rac1 in the development of cardiac hypertrophy. Proc. Natl Acad. Sci. U. S. A. 2006;103:7432–7437. doi: 10.1073/pnas.0510444103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li J., Stouffs M., Serrander L., Banfi B., Bettiol E., Charnay Y., Steger K., Krause K.H., Jaconi M.E. The NADPH oxidase NOX4 drives cardiac differentiation: role in regulating cardiac transcription factors and MAP kinase activation. Mol. Biol. Cell. 2006;17:3978–3988. doi: 10.1091/mbc.E05-06-0532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen K., Craige S.E., Keaney J.F., Jr. Downstream targets and intracellular compartmentalization in Nox signaling. Antioxid. Redox Signal. 2009;11:2467–2480. doi: 10.1089/ars.2009.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Itoh K., Chiba T., Takahashi S., Ishii T., Igarashi K., Katoh Y., Oyake T., Hayashi N., Satoh K., Hatayama I., Yamamoto M., Nabeshima Y. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem. Biophys. Res. Commun. 1997;236:313–322. doi: 10.1006/bbrc.1997.6943. [DOI] [PubMed] [Google Scholar]
- 25.Jaiswal A.K. Nrf2 signaling in coordinated activation of antioxidant gene expression. Free Radic. Biol. Med. 2004;36:1199–1207. doi: 10.1016/j.freeradbiomed.2004.02.074. [DOI] [PubMed] [Google Scholar]
- 26.Kobayashi M., Yamamoto M. Molecular mechanisms activating the Nrf2-Keap1 pathway of antioxidant gene regulation. Antioxid. Redox Signal. 2005;7:385–394. doi: 10.1089/ars.2005.7.385. [DOI] [PubMed] [Google Scholar]
- 27.Yamamoto T., Suzuki T., Kobayashi A., Wakabayashi J., Maher J., Motohashi H., Yamamoto M. Physiological significance of reactive cysteine residues of Keap1 in determining Nrf2 activity. Mol. Cell. Biol. 2008;28:2758–2770. doi: 10.1128/MCB.01704-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Friling R.S., Bensimon A., Tichauer Y., Daniel V. Xenobiotic-inducible expression of murine glutathione S-transferase Ya subunit gene is controlled by an electrophile-responsive element. Proc. Natl Acad. Sci. U. S. A. 1990;87:6258–6262. doi: 10.1073/pnas.87.16.6258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rushmore T.H., Morton M.R., Pickett C.B. The antioxidant responsive element. Activation by oxidative stress and identification of the DNA consensus sequence required for functional activity. J. Biol. Chem. 1991;266:11632–11639. [PubMed] [Google Scholar]
- 30.Li W., Yu S.W., Kong A.N. Nrf2 possesses a redox-sensitive nuclear exporting signal in the Neh5 transactivation domain. J. Biol. Chem. 2006;281:27251–27263. doi: 10.1074/jbc.M602746200. [DOI] [PubMed] [Google Scholar]
- 31.Churchman A.T., Anwar A.A., Li F.Y., Sato H., Ishii T., Mann G.E., Siow R.C. Transforming growth factor-beta1 elicits Nrf2-mediated antioxidant responses in aortic smooth muscle cells. J. Cell. Mol. Med. 2009;13:2282–2292. doi: 10.1111/j.1582-4934.2009.00874.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee I.T., Wang S.W., Lee C.W., Chang C.C., Lin C.C., Luo S.F., Yang C.M. Lipoteichoic acid induces HO-1 expression via the TLR2/MyD88/c-Src/NADPH oxidase pathway and Nrf2 in human tracheal smooth muscle cells. J. Immunol. 2008;181:5098–5110. doi: 10.4049/jimmunol.181.7.5098. [DOI] [PubMed] [Google Scholar]
- 33.Papaiahgari S., Kleeberger S.R., Cho H.Y., Kalvakolanu D.V., Reddy S.P. NADPH oxidase and ERK signaling regulates hyperoxia-induced Nrf2-ARE transcriptional response in pulmonary epithelial cells. J. Biol. Chem. 2004;279:42302–42312. doi: 10.1074/jbc.M408275200. [DOI] [PubMed] [Google Scholar]
- 34.Papaiahgari S., Zhang Q., Kleeberger S.R., Cho H.Y., Reddy S.P. Hyperoxia stimulates an Nrf2-ARE transcriptional response via ROS-EGFR-PI3K-Akt/ERK MAP kinase signaling in pulmonary epithelial cells. Antioxid. Redox Signal. 2006;8:43–52. doi: 10.1089/ars.2006.8.43. [DOI] [PubMed] [Google Scholar]
- 35.Sekhar K.R., Crooks P.A., Sonar V.N., Friedman D.B., Chan J.Y., Meredith M.J., Starnes J.H., Kelton K.R., Summar S.R., Sasi S., Freeman M.L. NADPH oxidase activity is essential for Keap1/Nrf2-mediated induction of GCLC in response to 2-indol-3-yl-methylenequinuclidin-3-ols. Cancer Res. 2003;63:5636–5645. [PubMed] [Google Scholar]
- 36.Brewer A.C., Murray V.A., Shah A.M. Regulation of the antioxidant pathway by NOX4 in the postnatal heart (abstract) Free Radic. Biol. Med. 2010;49:S126. [Google Scholar]
- 37.McMahon M., Itoh K., Yamamoto M., Chanas S.A., Henderson C.J., McLellan L.I., Wolf C.R., Cavin C., Hayes J.D. The cap'n'collar basic leucine zipper transcription factor Nrf2 (NF-E2 p45-related factor 2) controls both constitutive and inducible expression of intestinal detoxification and glutathione biosynthetic enzymes. Cancer Res. 2001;61:3299–3307. [PubMed] [Google Scholar]
- 38.Alexandrovich A., Arno M., Patient R.K., Shah A.M., Pizzey J.A., Brewer A.C. Wnt2 is a direct downstream target of GATA6 during early cardiogenesis. Mech. Dev. 2006;123:297–311. doi: 10.1016/j.mod.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 39.Weiss A., Leinwand L.A. The mammalian myosin heavy chain gene family. Annu. Rev. Cell Dev. Biol. 1996;12:417–439. doi: 10.1146/annurev.cellbio.12.1.417. [DOI] [PubMed] [Google Scholar]
- 40.Schroder E., Brennan J.P., Eaton P. Cardiac peroxiredoxins undergo complex modifications during cardiac oxidant stress. Am. J. Physiol. Heart Circ. Physiol. 2008;295:H425–H433. doi: 10.1152/ajpheart.00017.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moinova H.R., Mulcahy R.T. An electrophile responsive element (EpRE) regulates beta-naphthoflavone induction of the human gamma-glutamylcysteine synthetase regulatory subunit gene. Constitutive expression is mediated by an adjacent AP-1 site. J. Biol. Chem. 1998;273:14683–14689. doi: 10.1074/jbc.273.24.14683. [DOI] [PubMed] [Google Scholar]
- 42.Harvey C.J., Thimmulappa R.K., Singh A., Blake D.J., Ling G., Wakabayashi N., Fujii J., Myers A., Biswal S. Nrf2-regulated glutathione recycling independent of biosynthesis is critical for cell survival during oxidative stress. Free Radic. Biol. Med. 2009;46:443–453. doi: 10.1016/j.freeradbiomed.2008.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang H., Liu H., Liu R.M. Gender difference in glutathione metabolism during aging in mice. Exp. Gerontol. 2003;38:507–517. doi: 10.1016/s0531-5565(03)00036-6. [DOI] [PubMed] [Google Scholar]
- 44.Clempus R.E., Sorescu D., Dikalova A.E., Pounkova L., Jo P., Sorescu G.P., Schmidt H.H., Lassegue B., Griendling K.K. Nox4 is required for maintenance of the differentiated vascular smooth muscle cell phenotype. Arterioscler. Thromb. Vasc. Biol. 2007;27:42–48. doi: 10.1161/01.ATV.0000251500.94478.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cucoranu I., Clempus R., Dikalova A., Phelan P.J., Ariyan S., Dikalov S., Sorescu D. NAD(P)H oxidase 4 mediates transforming growth factor-beta1-induced differentiation of cardiac fibroblasts into myofibroblasts. Circ. Res. 2005;97:900–907. doi: 10.1161/01.RES.0000187457.24338.3D. [DOI] [PubMed] [Google Scholar]
- 46.Schroder K., Wandzioch K., Helmcke I., Brandes R.P. Nox4 acts as a switch between differentiation and proliferation in preadipocytes. Arterioscler. Thromb. Vasc. Biol. 2009;29:239–245. doi: 10.1161/ATVBAHA.108.174219. [DOI] [PubMed] [Google Scholar]
- 47.Sturrock A., Cahill B., Norman K., Huecksteadt T.P., Hill K., Sanders K., Karwande S.V., Stringham J.C., Bull D.A., Gleich M., Kennedy T.P., Hoidal J.R. Transforming growth factor-beta1 induces Nox4 NAD(P)H oxidase and reactive oxygen species-dependent proliferation in human pulmonary artery smooth muscle cells. Am. J. Physiol. Lung Cell. Mol. Physiol. 2006;290:L661–L673. doi: 10.1152/ajplung.00269.2005. [DOI] [PubMed] [Google Scholar]
- 48.Sturrock A., Huecksteadt T.P., Norman K., Sanders K., Murphy T.M., Chitano P., Wilson K., Hoidal J.R., Kennedy T.P. Nox4 mediates TGF-beta1-induced retinoblastoma protein phosphorylation, proliferation, and hypertrophy in human airway smooth muscle cells. Am. J. Physiol. Lung Cell. Mol. Physiol. 2007;292:L1543–L1555. doi: 10.1152/ajplung.00430.2006. [DOI] [PubMed] [Google Scholar]
- 49.Petry A., Djordjevic T., Weitnauer M., Kietzmann T., Hess J., Gorlach A. NOX2 and NOX4 mediate proliferative response in endothelial cells. Antioxid. Redox Signal. 2006;8:1473–1484. doi: 10.1089/ars.2006.8.1473. [DOI] [PubMed] [Google Scholar]
- 50.Chen K., Kirber M.T., Xiao H., Yang Y., Keaney J.F., Jr. Regulation of ROS signal transduction by NADPH oxidase 4 localization. J. Cell Biol. 2008;181:1129–1139. doi: 10.1083/jcb.200709049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu R.M., Choi J., Wu J.H., Gaston Pravia K.A., Lewis K.M., Brand J.D., Mochel N.S., Krzywanski D.M., Lambeth J.D., Hagood J.S., Forman H.J., Thannickal V.J., Postlethwait E.M. Oxidative modification of nuclear mitogen-activated protein kinase phosphatase 1 is involved in transforming growth factor beta1-induced expression of plasminogen activator inhibitor 1 in fibroblasts. J. Biol. Chem. 2010;285:16239–16247. doi: 10.1074/jbc.M110.111732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kensler T.W., Wakabayashi N., Biswal S. Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu. Rev. Pharmacol. Toxicol. 2007;47:89–116. doi: 10.1146/annurev.pharmtox.46.120604.141046. [DOI] [PubMed] [Google Scholar]
- 53.Calay D., Rousseau A., Mattart L., Nuyens V., Delporte C., Van A.P., Moguilevsky N., Arnould T., Boudjeltia K.Z., Raes M. Copper and myeloperoxidase-modified LDLs activate Nrf2 through different pathways of ROS production in macrophages. Antioxid. Redox Signal. 2010;13:1491–1502. doi: 10.1089/ars.2009.2971. [DOI] [PubMed] [Google Scholar]
- 54.Cho M.K., Kim W.D., Ki S.H., Hwang J.I., Choi S., Lee C.H., Kim S.G. Role of Galpha12 and Galpha13 as novel switches for the activity of Nrf2, a key antioxidative transcription factor. Mol. Cell. Biol. 2007;27:6195–6208. doi: 10.1128/MCB.02065-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jyrkkanen H.K., Kansanen E., Inkala M., Kivela A.M., Hurttila H., Heinonen S.E., Goldsteins G., Jauhiainen S., Tiainen S., Makkonen H., Oskolkova O., Afonyushkin T., Koistinaho J., Yamamoto M., Bochkov V.N., Yla-Herttuala S., Levonen A.L. Nrf2 regulates antioxidant gene expression evoked by oxidized phospholipids in endothelial cells and murine arteries in vivo. Circ. Res. 2008;103:e1–e9. doi: 10.1161/CIRCRESAHA.108.176883. [DOI] [PubMed] [Google Scholar]
- 56.Kang H.J., Hong Y.B., Kim H.J., Bae I. CR6-interacting factor 1 (CRIF1) regulates NF-E2-related factor 2 (NRF2) protein stability by proteasome-mediated degradation. J. Biol. Chem. 2010;285:21258–21268. doi: 10.1074/jbc.M109.084590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang L., Chen Y., Sternberg P., Cai J. Essential roles of the PI3 kinase/Akt pathway in regulating Nrf2-dependent antioxidant functions in the RPE. Invest. Ophthalmol. Vis. Sci. 2008;49:1671–1678. doi: 10.1167/iovs.07-1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Prestera T., Holtzclaw W.D., Zhang Y., Talalay P. Chemical and molecular regulation of enzymes that detoxify carcinogens. Proc. Natl Acad. Sci. U. S. A. 1993;90:2965–2969. doi: 10.1073/pnas.90.7.2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huang H.C., Nguyen T., Pickett C.B. Phosphorylation of Nrf2 at Ser-40 by protein kinase C regulates antioxidant response element-mediated transcription. J. Biol. Chem. 2002;277:42769–42774. doi: 10.1074/jbc.M206911200. [DOI] [PubMed] [Google Scholar]
- 60.Jaquet V., Scapozza L., Clark R.A., Krause K.H., Lambeth J.D. Small-molecule NOX inhibitors: ROS-generating NADPH oxidases as therapeutic targets. Antioxid. Redox Signal. 2009;11:2535–2552. doi: 10.1089/ars.2009.2585. [DOI] [PubMed] [Google Scholar]
- 61.Ding Y., Choi K.J., Kim J.H., Han X., Piao Y., Jeong J.H., Choe W., Kang I., Ha J., Forman H.J., Lee J., Yoon K.S., Kim S.S. Endogenous hydrogen peroxide regulates glutathione redox via nuclear factor erythroid 2-related factor 2 downstream of phosphatidylinositol 3-kinase during muscle differentiation. Am. J. Pathol. 2008;172:1529–1541. doi: 10.2353/ajpath.2008.070429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Morito N., Yoh K., Itoh K., Hirayama A., Koyama A., Yamamoto M., Takahashi S. Nrf2 regulates the sensitivity of death receptor signals by affecting intracellular glutathione levels. Oncogene. 2003;22:9275–9281. doi: 10.1038/sj.onc.1207024. [DOI] [PubMed] [Google Scholar]
- 63.Ardite E., Barbera J.A., Roca J., Fernandez-Checa J.C. Glutathione depletion impairs myogenic differentiation of murine skeletal muscle C2C12 cells through sustained NF-kappaB activation. Am. J. Pathol. 2004;165:719–728. doi: 10.1016/s0002-9440(10)63335-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dalton T.P., Chen Y., Schneider S.N., Nebert D.W., Shertzer H.G. Genetically altered mice to evaluate glutathione homeostasis in health and disease. Free Radic. Biol. Med. 2004;37:1511–1526. doi: 10.1016/j.freeradbiomed.2004.06.040. [DOI] [PubMed] [Google Scholar]
- 65.Reddy N.M., Kleeberger S.R., Bream J.H., Fallon P.G., Kensler T.W., Yamamoto M., Reddy S.P. Genetic disruption of the Nrf2 compromises cell-cycle progression by impairing GSH-induced redox signaling. Oncogene. 2008;27:5821–5832. doi: 10.1038/onc.2008.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wu G., Fang Y.Z., Yang S., Lupton J.R., Turner N.D. Glutathione metabolism and its implications for health. J. Nutr. 2004;134:489–492. doi: 10.1093/jn/134.3.489. [DOI] [PubMed] [Google Scholar]
- 67.Hew K.W., Keller K.A. Postnatal anatomical and functional development of the heart: a species comparison. Birth Defects Res. B Dev. Reprod. Toxicol. 2003;68:309–320. doi: 10.1002/bdrb.10034. [DOI] [PubMed] [Google Scholar]
- 68.Heineke J., Molkentin J.D. Regulation of cardiac hypertrophy by intracellular signalling pathways. Nat. Rev. Mol. Cell Biol. 2006;7:589–600. doi: 10.1038/nrm1983. [DOI] [PubMed] [Google Scholar]
- 69.Li J., Ichikawa T., Villacorta L., Janicki J.S., Brower G.L., Yamamoto M., Cui T. Nrf2 protects against maladaptive cardiac responses to hemodynamic stress. Arterioscler. Thromb. Vasc. Biol. 2009;29:1843–1850. doi: 10.1161/ATVBAHA.109.189480. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Genes significantly upregulated by Nox4 overexpression