Abstract
Recent data indicate that sustained elevations in plasma insulin suppress the mRNA for IRS-2, a component of the insulin signaling pathway in liver, and that this deficiency contributes to hepatic insulin resistance and inappropriate gluconeogenesis. Here, we use nuclear run-on assays to show that insulin inhibits transcription of the IRS-2 gene in the livers of intact rats. Insulin also inhibited transcription of a reporter gene driven by the human IRS-2 promoter that was transfected into freshly isolated rat hepatocytes. The human promoter contains a heptanucleotide sequence, TGTTTTG, that is identical to the insulin response element (IRE) identified previously in the promoters of insulin-repressed genes. Single base pair substitutions in this IRE decreased transcription of the IRS-2-driven reporter in the absence of insulin and abolished insulin-mediated repression. We conclude that insulin represses transcription of the IRS-2 gene by blocking the action of a positive factor that binds to the IRE. Sustained repression of IRS-2, as occurs in chronic hyperinsulinemia, contributes to hepatic insulin resistance and accelerates the development of the diabetic state.
Keywords: insulin resistance, type 2 diabetes, gluconeogenesis, phosphoenolpyruvate carboxykinase
The ingestion of a glucose-containing meal induces the pancreas to secrete insulin, which suppresses the synthesis of glucose by the liver and its release into the circulation, thereby preventing hyperglycemia. In insulin-resistant states, insulin no longer suppresses hepatic gluconeogenesis, and the consequent glucose overproduction contributes to hyperglycemia and diabetes mellitus (1). Recent insights into the mechanism of hepatic insulin resistance emerged from observations in two mouse models, ob/ob and lipodystrophic mice, both of which manifest hyperglycemia and hyperinsulinemia caused by deficiencies of the adipocyte-derived hormone leptin. In both of these insulin-resistant states, the liver exhibited a marked reduction in IRS-2, an intracellular signaling protein that is phosphorylated by the insulin receptor in response to insulin (2). Phosphorylated IRS-2 binds and activates phosphatidylinositol 3-kinase (PI3K), thereby triggering a signaling cascade that is responsible for many actions of insulin (3). The importance of IRS-2 in the liver was established earlier by the observation that mice with a genetic deficiency of IRS-2 manifest enhanced hepatic gluconeogenesis and a resistance to insulin-mediated suppression of hepatic glucose output (4, 5).
In the ob/ob and lipodystrophic mice, the down-regulation of hepatic IRS-2 protein was attributed to a down-regulation of IRS-2 mRNA in response to chronic hyperinsulinemia (2). This conclusion was supported by experiments with freshly isolated rat hepatocytes. When these cells were isolated initially, they contained relatively high levels of IRS-2 mRNA. However, when the cells were incubated with high concentrations of insulin for 12 h, the amount of IRS-2 mRNA fell markedly. The resultant deficiency of IRS-2 protein was correlated with a failure of insulin to activate PI3K to a normal extent, and this loss led to a marked reduction in the phosphorylation and activation of Akt (also called protein kinase B), a crucial downstream kinase whose activity is stimulated indirectly by PI3K (3, 6). The hypothesis that chronic hyperinsulinemia suppresses IRS-2 mRNA was supported by the observation that hepatic IRS-2 levels rose when plasma insulin levels fell upon treatment of the ob/ob and lipodystrophic mice with leptin. The rise in IRS-2 mRNA and protein was associated with a restoration of the normal ability of insulin to suppress acutely the enzymes of gluconeogenesis (2, 7).
The question arises as to whether insulin suppresses hepatic IRS-2 by the same mechanism by which it suppresses other genes in this organ. Among the insulin-repressed genes in liver, the most well studied is the gene encoding phosphoenolpyruvate carboxykinase (PEPCK), an enzyme of gluconeogenesis (8). Extensive studies by Granner and coworkers (9, 10) have identified a heptanucleotide (TGTTTTG) in the enhancer region of this gene that serves as an insulin response element (IRE). Similar, but not identical, sequences have been identified in other insulin-repressed genes, including insulin-like growth factor-binding protein-1 (IGFBP-1), tyrosine aminotransferase, glucose-6-phosphatase, apolipoprotein C III, and aspartate aminotransferase [see Hall et al. (9) for references]. In the PEPCK gene, the IRE appears to be a binding site for a positive transcription factor that is activated by glucocorticoids. In the presence of insulin, this positive factor no longer acts, and transcription is reduced. Several candidates for this insulin-sensitive factor have been proposed, including members of the forkhead/winged-helix family, such as FKHR and FKHRL1 (9, 11–13). However, to date, none of these proteins has been shown to bind to the heptanucleotide in vitro with the same specificity as that required for insulin-sensitive activation in vivo (9).
In the current studies, we use nuclear run-on assays to confirm that insulin inhibits transcription of the IRS-2 gene in rat liver. We identify a heptanucleotide sequence (TGTTTTG) in the 5′ flanking region of the human IRS-2 gene that is a perfect match with the IRE of the PEPCK gene, and we use reporter gene assays to show that this sequence is required for insulin-mediated repression of IRS-2 gene transcription in hepatocytes. These results provide a potential mechanism for the insulin-mediated repression of the IRS-2 gene and thereby help to explain the hepatic insulin resistance that accompanies the sustained hyperinsulinemia of type 2 diabetes and lipodystrophy.
Materials and Methods
Reagents.
We obtained DMEM (catalog no. 11885-084) from GIBCO/BRL; dexamethasone from Biomol (Plymouth Meeting, PA); and bovine insulin from Sigma. Lipoprotein-deficient serum (density > 1.215 g/ml) was prepared by ultracentrifugation as described (14).
Nuclear Run-On Assay.
This assay was carried out with rat liver nuclei according to a modification of a previously described procedure (15, 16). Briefly, liver tissue (≈3 g) was homogenized with a Dounce homogenizer in 30 ml of buffer A [10 mM Hepes, pH 7.4/60 mM KCl/2 M sucrose/10% (vol/vol) glycerol/1 mM DTT/10 mM sodium EDTA/15 mM spermine/0.5 mM spermidine]. The homogenate was layered over a 10-ml cushion of buffer A in ultra-clear tubes and centrifuged at 24,000 rpm at 4°C for 40 min in a Sorvall AH629 rotor. The resulting pellet was resuspended in 2 ml of buffer B (75 mM Hepes, pH 7.5/40% glycerol/60 mM KCl/5 mM MgCl2/0.1 mM sodium EDTA/0.5 mM spermine/1 mM spermidine/0.5 mM DTT). Aliquots of the isolated nuclei (0.2 ml; 50 A260 units/ml) were frozen in liquid nitrogen and stored at −80°C until use.
For the run-on assay, each 0.2-ml aliquot of nuclei was washed twice by centrifugation in buffer C (20 mM Tris⋅chloride, pH 8.0/20% glycerol/0.14 M KCl/10 mM MgCl2/1 mM MnCl2/14 mM 2-mercaptoethanol). Each reaction mixture contained the following components in a final volume of 0.3 ml: 0.2 ml of washed rat liver nuclei in buffer C, 10 μM creatine phosphate, 16 units/ml creatine kinase, 0.5 mM CTP, 0.5 mM GTP, 1 mM ATP, 0.1 μM UTP, and 100 μCi of [α-32P]UTP (800 Ci/mmol; Amersham Pharmacia). After incubation for 20 min at 26°C, labeled nascent RNA transcripts were extracted with phenol/chloroform, and aliquots of the extracted RNA (≈8 × 106 cpm) were hybridized at 42°C for 72 h to nitrocellulose membranes containing linearized cDNAs for rat fatty acid synthase (7.2 kb), rat PEPCK (2.26 kb), mouse IRS-1 (3.7 kb), mouse IRS-2 (4.0 kb), rat sterol regulatory element-binding protein-1 (SREBP-1) (3.4 kb), and rat cyclophilin (750 bp). The IRS-2 and IRS-1 cDNAs were kindly provided by Takashi Kadowaki (Tokyo University, Tokyo) and Ryutaro Komuro (University of Texas Southwestern Medical Center, Dallas), respectively. All other cDNAs were cloned by PCR using cDNA libraries from rat liver as the template and oligonucleotide primers that were designed from published sequences. All cDNAs were subcloned into pCRII vector (Invitrogen).
IRS-2 Promoter–Reporter Constructs.
The human IRS-2 promoter (base pairs −1051 to −116, relative to the translation start site) (17) was amplified from the genomic clone BACH-222E8, which was obtained from a human bacterial artificial chromosome library (Incyte Genomics, St. Louis). PCR was carried out with the following primers: 5′ primer, 5′-TACGGCATTGACGGTACCCCGCCCATGCTTCCACTGCAAGGCC-3′, and 3′ primer, 5′-TGAGTCAATGCGTACGGTACCATCACGCGTCCCTCGGGCCCAGGCGGT-3′ (17). The resulting PCR product was cloned into pCR2.1 by using the TOPO PCR TA Cloning Kit (Invitrogen) and designated pCR2.1-IRS2. The entire IRS-2 promoter fragment was excised from pCR2.1-IRS2 with HindIII and XhoI and ligated into the pGL3-Basic Luciferase Vector (Promega). The resulting plasmid was designated pIRS2-Luciferase(wt). A mutant pIRS2-Luciferase plasmid containing three base pair substitutions in the IRE sequence was constructed by using pIRS2-Luciferase(wt) as a template and the QuickChange Site-Directed Mutagenesis Kit (Stratagene). In this mutant, designated pIRS-Luciferase(mut-3bp), the first, third, and fifth nucleotides spanning base pairs −574 to −568 in the IRS-2 promoter were changed from 5′-TGTTTTG-3′ to 5′-AGATCTG-3′. Plasmids containing single base pair substitutions in the IRE sequence of pIRS2-Luciferase were also constructed from pIRS2-Luciferase(wt) by using the QuickChange Site-Directed Mutagenesis Kit. The integrity of all plasmids was verified by DNA sequencing.
Primary Rat Hepatocytes.
Male Sprague–Dawley rats (200–250 g; obtained from Harlan Breeders, Indianapolis) were housed in colony cages, maintained on a 12-h light/12-h dark cycle, and fed Teklad 4% Mouse/Rat Diet no. 7001 (Harlan Teklad, Madison, WI). Nonfasted rats were killed at the beginning of the dark cycle, and primary hepatocytes were isolated by the collagenase method as described (18, 19) with the following minor modifications. After perfusion and collagenase digestion, the liver was removed, and the dissociated cells were dispersed by shaking. The cells were filtered at 4°C through gauze into an equal volume of ice-cold medium A [DMEM supplemented with 5% (vol/vol) newborn calf lipoprotein-deficient serum, 100 units/ml sodium penicillin, and 100 μg/ml streptomycin sulfate]. The hepatocytes were pelleted by centrifugation and washed twice at 4°C with the same buffer. Aliquots of 1.2 × 106 cells were plated onto 60-mm mouse collagen IV-coated dishes (Fisher Scientific; catalog no. 08-774-32). The cells were incubated at 37°C in 5% CO2 in medium A for 16 h before transfection.
Plasmid Transfections and Assays of Luciferase and Alkaline Phosphatase.
For all studies, duplicate dishes of hepatocytes were placed in serum-free RPMI medium 1640 supplemented with 100 units/ml penicillin and 100 μg/ml streptomycin sulfate and cotransfected with 3 μg per dish of the wild-type or mutant pIRS2-Luciferase plasmid and 0.1 μg per dish of a control pCMV-SEAP plasmid encoding human placental alkaline phosphatase (Tropix, Bedford, MA) by using Fugene 6 (Roche Molecular Biochemicals).
After transfection, cells and medium were harvested for measurement of luciferase (20) and alkaline phosphatase (21) activities, respectively, as described in the indicated reference. Luciferase activity of cell extracts was measured with the Luciferase Assay Kit (Promega) according to the manufacturer's protocol. Photon production was detected as relative light units by using an Optima II luminometer (MGM Instruments, Hamden, CT). The amount of luciferase activity in transfected cell extracts (relative light units) was normalized to the amount of alkaline phosphatase activity measured in the medium from the same dish of transfected cells (see below).
Alkaline phosphatase activity in the medium was measured by using the Phospha-Light Chemiluminescent Reporter Gene Assay System (Tropix). From each dish of cells, 0.5 ml of medium was centrifuged at 15,000 × g for 15 min at 4°C. Duplicate aliquots of the supernatant (33 μl) were each mixed with 0.1 ml of 1× dilution buffer, incubated at 65°C for 30 min to inactivate nonplacental alkaline phosphatase, and assayed for placental alkaline phosphatase activity according to the manufacturer's instructions. Chemiluminescence was quantified with an Optima II luminometer.
Results
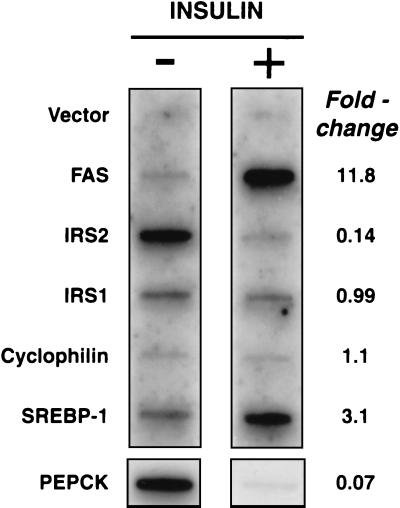
Fig. 1 shows the results of a nuclear run-on assay designed to measure the effects of insulin on the relative rates of transcription of IRS-2 and other genes in rat liver. The animals were rendered insulin-deficient by treatment with streptozotocin, which destroys the insulin-secreting cells of the pancreas (19, 22). After 36 h, one group was treated with insulin s.c., and the other group received only vehicle. After an additional 6 h, during which time food was withheld, the animals were killed, and the liver nuclei were isolated and incubated with [α-32P]UTP. The labeled transcripts were hybridized to various cDNA probes that had been immobilized on nitrocellulose and then visualized by autoradiography. As expected, insulin treatment elevated transcription of the fatty acid synthase gene by nearly 12-fold and reduced transcription of the PEPCK gene by >90%. The transcription of IRS-2 was also reduced by >85%. The transcription of IRS-1, the other prominent IRS isoform in liver, was unchanged by insulin treatment. The transcription rate for SREBP-1, which represents the sum of the two isoforms SREBP-1a and SREBP-1c, was elevated by 3-fold. As an internal control, we measured the transcription of cyclophilin, which was unaffected by insulin.
Figure 1.
Insulin-mediated inhibition of IRS-2 gene transcription in rat liver. Nuclear run-on assays of nascent 32P-labeled RNA transcripts were performed with nuclei isolated from the livers of streptozotocin-treated rats that were injected s.c. with or without insulin as indicated. Male Sprague–Dawley rats were treated with streptozotocin for 36 h and then injected with insulin or vehicle for 6 h, as described (19). For each experimental group, liver tissue from three rats was pooled, homogenized, and subjected to nuclear run-on assay, as described in Materials and Methods. After hybridization with the indicated cDNA probe, the filters for all probes were exposed to Kodak X-Omat film with Bio-Max (Kodak) intensifying screens at −80°C for 48 h, except for the PEPCK probe, which was exposed for 8 h. Relative levels of the transcripts for each gene were quantified with a Fuji Bio-Imaging analyzer. A blank value corresponding to the vector band was subtracted from all values.
Inspection of the 5′ flanking region of the mouse IRS-2 gene revealed the heptanucleotide TGTTTTG at position −1571 to −1565 relative to the translation start site (23) (Fig. 2). The same sequence is present in the human IRS-2 at position −574 to −568 (17). This heptanucleotide sequence is identical to the IRE that was identified previously in the rat PEPCK enhancer by Hall et al. (9). To determine the function of this sequence in the human IRS-2 gene, we prepared an expression plasmid in which the 5′ flanking region of the human IRS-2 gene (base pairs −1051 to −116) drove transcription of a luciferase reporter gene. We also prepared a mutant version containing transversions in three nucleotides within this sequence (5′-TGTTTTG-3′ mutated to 5′-AGATCTG-3′). This mutant plasmid was predicted to be inactive for insulin-regulated transcription based on the prior studies of Hall et al. (9).
Figure 2.
Putative IRE sequence in promoter region of mouse and human IRS-2 genes as compared with rat PEPCK gene. Nucleotide numbering at the right refers to the translation start site, where +1 is the A of the ATG. Arrows below the human IRS-2 sequence denote the three base pair changes contained in the pIRS2-Luciferase(mut-3bp) reporter gene. Mouse IRS-2 sequence is from Sun et al. (23). Human IRS-2 sequence is from Vaβen et al. (17). Rat PEPCK sequence is from Beale et al. (32).
To measure insulin-regulated transcription, we isolated hepatocytes from fed rats. After a 16-h preliminary incubation, the wild-type and mutant plasmids were introduced into the cells by transfection. We also introduced a control plasmid encoding human placental alkaline phosphatase driven by the cytomegalovirus (CMV) promoter. The cells were incubated in serum-free medium containing various concentrations of insulin for 16 h before harvest for measurement of luciferase and alkaline phosphatase activities. The data were corrected for transfection efficiency by dividing the luciferase activity by the alkaline phosphatase activity. As shown in Fig. 3A, in the absence of insulin, the wild-type IRS-2 promoter produced higher luciferase activity than the mutant promoter. In the presence of insulin, the activity of the wild-type promoter was reduced so that it became equal to the mutant promoter, which was not affected by insulin. When the cells were incubated with the glucocorticoid dexamethasone, in the absence of insulin, the activity of the wild-type promoter was increased slightly (Fig. 3, compare A with B). This difference was abolished by insulin. Neither dexamethasone nor insulin had an effect on the mutant promoter. These data are similar to those of Granner and coworkers (9, 10) on the PEPCK promoter. They indicate that the IRE in the IRS-2 promoter is a binding site for a positive transcription factor whose activity is abolished by insulin.
Figure 3.
Insulin-mediated inhibition of IRS-2 promoter activity in transfected rat hepatocytes incubated in the absence (A) and presence (B) of dexamethasone. The wild-type construct, pIRS2-Luciferase(wt), contains a ≈1-kb fragment corresponding to base pairs −1051 to −116 of the human IRS-2 promoter linked to a luciferase reporter gene. The mutant pIRS2-Luciferase construct, pIRS2-Luciferase(mut-3bp), contains the same human IRS-2 promoter fragment except that the IRE sequence (base pairs −574 to −568) was mutated from 5′-TGTTTTG-3′ to 5′-AGATCTG-3′. Primary rat hepatocytes were isolated and plated as described in Materials and Methods. Hepatocytes were cotransfected with 0.1 μg per dish of the reference reporter plasmid pCMV-SEAP and 3 μg per dish of either pIRS2-Luciferase(wt) (▴) or pIRS2-Luciferase(mut-3bp) (●) as indicated. After transfection, the cells were switched from medium A to serum-free RPMI medium 1640 and incubated for 8 h at 37°C, after which the medium was supplemented with the indicated concentration of insulin in the absence (A) or presence (B) of 5 μM dexamethasone. After an additional incubation for 16 h, the cells and medium were harvested for measurement of luciferase and alkaline phosphatase activities, respectively. Alkaline phosphatase activity was used as the internal reference to normalize for transfection efficiency. The value for wild-type pIRS2-luciferase activity in the absence of insulin was arbitrarily set as 1. Each value represents the average of duplicate incubations. This experiment was repeated three times with similar results.
Fig. 4 shows the time course of suppression of IRS-2 promoter activity by insulin as measured with the luciferase reporter assay in isolated rat hepatocytes. Insulin exerted a near-complete inhibitory effect when added to the cells 8 h before harvest. The insulin effect at 16 h was completely blocked by the inclusion of the PI3K inhibitor wortmannin (1 μM) in the incubation medium (J.Z., J.L.G., and M.S.B., unpublished observations).
Figure 4.
Time course of insulin-mediated inhibition of IRS-2 promoter activity in transfected rat hepatocytes. Primary rat hepatocytes were isolated and plated as described in Materials and Methods. Hepatocytes were cotransfected with pCMV-SEAP plasmids and either pIRS2-Luciferase(wt) (▴) or pIRS2-Luciferase(mut-3bp) (●) as described in the legend to Fig. 3. Six hours after transfection (zero time for the experiment), the serum-free RPMI medium 1640 in all dishes was supplemented with 5 μM dexamethasone, and the dishes were incubated an additional 24 h before harvest. Insulin (100 nM) was added to duplicate dishes at the indicated time before harvest. Luciferase and alkaline phosphatase activities were measured as described in the legend to Fig. 3. The value for wild-type pIRS2-Luciferase activity in the absence of insulin was arbitrarily set as 1. Each value represents the average of duplicate incubations. This experiment was repeated three times with similar results.
To further dissect the roles of individual nucleotides of the heptanucleotide consensus sequence, we prepared a series of IRS-2-luciferase plasmids with single transversion mutations in or around the core element. The plasmids were introduced into fresh hepatocytes by transfection, and luciferase was measured after incubation for 16 h with dexamethasone in the absence or presence of 100 nM insulin (Fig. 5A). Fig. 5B shows the calculated value for percent inhibition by insulin for each plasmid. As expected, the activity of the wild-type IRS-2 promoter was reduced by 75% in the presence of insulin. Mutation of base pairs 1–4 of the heptanucleotide sequence markedly reduced the insulin-sensitive component of transcription. As a result, the promoter activity was relatively low, and it was nearly the same in the absence and presence of insulin. Mutation of base pairs 5 and 7 increased transcription slightly, but insulin sensitivity was retained. Mutation of base pair 6 reduced transcription equally in the absence and presence of insulin, and thus the relative suppression by insulin was retained. Mutation of either of the two base pairs that flank the heptanucleotide had no effect.
Figure 5.
Single point mutation analysis of the IRE sequence in human IRS-2 promoter (A and B) and in rat PEPCK promoter (C). (A and B) Primary rat hepatocytes were isolated and plated as described in Materials and Methods. Cells were cotransfected with 0.1 μg per dish pCMV-SEAP and 3 μg per dish of either pIRS2-Luciferase(wt) or the indicated mutant version in which the indicated single base pair substitution was made in the IRE sequence. After incubation for 8 h at 37°C, the serum-free RPMI medium 1640 was supplemented with 5 μM dexamethasone in the absence (open bar) or presence (closed bar) of 100 nM insulin. After an additional 16-h incubation, the cells and medium were harvested for measurement of luciferase and alkaline phosphatase activities, respectively. (A) IRS-2-mediated luciferase activity in the absence and presence of insulin for each mutant construct is plotted above the corresponding mutated base pair. The bases in the heptanucleotide IRE are numbered 1–7. The value for the wild-type construct was arbitrarily set at 1. (B) The percent inhibition of IRS-2-mediated luciferase activity in mutant constructs by insulin is plotted below the corresponding mutated base pair. Results in A and B are plotted as the mean ± SEM of four independent experiments, each of which was done in duplicate. (C) The percent inhibition of PEPCK-mediated chloramphenicol acetyltransferase (CAT) activity in mutant constructs by insulin is plotted below the corresponding mutated base pair. These data are replotted from Hall et al. (9).
For purposes of comparison, Fig. 5C shows previously published data of Hall et al. (9), who used a reporter assay to measure the transcriptional activity of the PEPCK promoter in a transfected rat liver cell line. The PEPCK and IRS-2 promoters both showed a requirement for base pairs 2–4 of the heptanucleotide to achieve a complete response to insulin. Compared with the PEPCK promoter, the IRS-2 promoter showed a stronger requirement for T1 and a lesser requirement for T4. Whether these differences reflect true differences in the factor recognition motif, or whether they represent subtle differences in the assay methodology, remains to be determined.
Discussion
The current data provide a potential mechanism for the recently recognized ability of insulin to down-regulate the IRS-2 mRNA in rodent hepatocytes. The nuclear run-on assays with rat liver indicate that insulin mediates this effect by decreasing the transcription of the IRS-2 gene. The promoter reporter assays, performed in transiently transfected primary rat hepatocytes, indicate that transcriptional inhibition is mediated by the same heptanucleotide sequence that mediates insulin's inhibition of the PEPCK and IGFBP-1 genes. The transfection data are consistent with a model in which a positive factor binds to this heptanucleotide in the absence of insulin, thereby activating transcription. In the presence of insulin, this factor does not act, and transcription falls to the level that is seen with enhancers that lack this heptanucleotide.
Based on studies with other IRE-containing promoters, it is likely that the positive factor that binds to the IRS-2 IRE belongs to the forkhead/winged-helix family of transcription factors. Studies with the PEPCK and IGFBP-1 promoters first cast suspicion on HNF-3, a member of this family whose action is stimulated by glucocorticoids (24, 25). However, there is an imperfect correlation between the ability of mutant IREs to bind to HNF3 in vitro and the ability of these mutants to respond to the insulin-inhibitable transcription factor in intact cells (26).
Recently, intense interest has been expressed in another subgroup of the forkhead/winged-helix family that includes the human proteins FKHR and FKHRL1. This interest was stimulated by genetic studies in the roundworm Caenorhabditis elegans. These studies defined an insulin response pathway that proceeds through the C. elegans homologues of the insulin receptor, the catalytic subunit of PI3K, and Akt (27, 28). Loss-of-function mutations in any of these genes produce a state in which the worms behave as though they are deprived of energy-supplying nutrients. They store increased amounts of fat, and they enter spontaneously into the dauer larval stage, a state of developmental arrest and reduced metabolic activity. If the worms express alleles that retain some function, they emerge from the dauer larval stage, whereupon they show increased longevity. This complex phenotype is abolished by loss-of-function mutations in a gene, daf-16, which encodes a transcription factor of the forkhead/winged-helix family (29, 30). These studies indicate that, in C. elegans, the insulin pathway inactivates DAF-16, and they immediately suggested that insulin might also inactivate a forkhead transcription factor in vertebrates.
Subsequent studies of mammalian forkhead proteins revealed that they bind specifically to the PEPCK and IGFBP-1 IREs and they are phosphorylated by Akt in vitro [see Hall et al. (9) for references]. When overexpressed in mammalian cells by transfection, FKHR and FKHRL1 stimulate transcription of promoters that contain IREs, and this stimulation is abolished by insulin (9, 12). Moreover, C. elegans DAF-16 activates the IGFBP-1 promoter when expressed in human HepG2 cells, and this action is opposed by insulin (31).
The identity of FKHRL1 as the insulin-inhibited transcription factor was called into question recently by studies showing that PEPCK IREs with a specific point mutation could not support insulin-inhibitable transcription, even though FKHRL1 bound to this mutant sequence in vitro with the same affinity that it showed for the wild-type IRE (9). The mutation in question consisted of a thymidine-to-adenine transversion at base pair 4 of the heptanucleotide sequence (see Fig. 5). This mutation is designated “m7” in the paper by Hall et al. (9). We observed that this mutation also caused a reduction in the insulin sensitivity of transcription when it was placed into the IRS-2 promoter. However, the reduction was not as complete as it was for the PEPCK promoter. It is possible that FKHRL1 is the IRE-specific factor and that it binds less well to the T4 to A mutant sequence in the nucleus than it does in the in vitro gel retardation assay. Further studies are necessary to clarify this point.
The finding of a functional IRE in the IRS-2 promoter creates a paradox that must be explained. For purposes of the following discussion, we assume that insulin inactivates IRE-dependent transcription by phosphorylating a forkhead protein and that the same protein controls two target genes, IRS-2 and PEPCK. After a period of insulin deprivation, such as after an overnight fast, the forkhead protein is active in liver, and its target genes are fully transcribed. As a result, IRS-2 and PEPCK mRNAs are both abundant. When carbohydrates are ingested, insulin signals through IRS-2, causing forkhead to become phosphorylated and PEPCK transcription to fall. Forkhead phosphorylation also causes IRS-2 transcription to decline and IRS-2 levels fall. Because of the IRS-2 deficiency, the liver becomes insulin-resistant, but this is of no consequence in the normal state because the animal is now postabsorptive and blood sugar and insulin levels have fallen. The fall in insulin allows forkhead to become dephosphorylated. IRS-2 and PEPCK mRNAs rise, and the liver is primed to respond appropriately to the next meal.
The paradox arises in the chronic insulin-resistant state, such as in ob/ob and lipodystrophic mice. When these mice are studied in the fed state, IRS-2 mRNA is low and PEPCK mRNA is high, despite high levels of blood sugar and insulin (2, 7). The high PEPCK indicates that forkhead is dephosphorylated, presumably because of the insulin resistance caused by the deficiency of IRS-2. Why doesn't the dephosphorylated forkhead also activate the IRS-2 gene and reverse the IRS-2 deficiency? It is possible that, in the chronic insulin-resistant state, some other transcription factor becomes altered, and this alteration interferes with IRS-2 transcription even when forkhead is active. Alternatively, it is possible that forkhead actually remains phosphorylated (inactive) in this situation and some other factor is responsible for maintaining high PEPCK levels. It is also possible that different forkhead proteins bind to IRS-2 and PEPCK IREs and that they can be phosphorylated differentially in response to insulin.
A suggestion that different proteins might regulate the IREs of the IRS-2 and PEPCK genes comes from experiments with a cultured line of rat hepatoma cells. These cells are designated CRL-1600 (H4IIE-C3) in the American Type Culture Collection catalogue and H4IIE cells by Hall et al. (9). The addition of 100 nM insulin to the CRL-1600 cells caused a 75–90% decline in PEPCK-driven luciferase activity but no change in IRS-2-driven luciferase activity (J.Z., J.L.G., and M.S.B., unpublished observations). A further understanding of this difference will require detailed studies of the IRS-2 promoter with a delineation of all of the factors that contribute to expression as well as insulin-mediated repression.
Acknowledgments
We thank Dr. Young-Ah Moon for invaluable help with isolation of rat hepatocytes; Dr. Rob Rawson for computer searches; Dr. Ryutaro Komuro for providing IRS-1 cDNA; Dr. Takashi Kadowaki for providing mouse IRS-2 cDNA; Dr. Iichiro Shimomura for helpful comments; Dr. David W. Russell for critical review of the manuscript; and Jeff Cormier and Lorena Avila for expert sequencing of DNA. This research was supported by grants from the National Institutes of Health (HL20948), the Perot Family Foundation, and the W. M. Keck Foundation. J.D.H. is a Pew Scholar in the Biomedical Sciences and is the recipient of an Established Investigator Grant from the American Heart Association.
Abbreviations
- PI3K
phosphatidylinositol 3-kinase
- IGFBP-1
insulin-like growth factor-binding protein-1
- IRE
insulin response element
- PEPCK
phosphoenolpyruvate carboxykinase
- SREBP
sterol regulatory element-binding protein
- CMV
cytomegalovirus
References
- 1.Kahn C R. Diabetes. 1994;43:1066–1084. doi: 10.2337/diab.43.8.1066. [DOI] [PubMed] [Google Scholar]
- 2.Shimomura I, Matsuda M, Hammer R E, Bashmakov Y, Brown M S, Goldstein J L. Mol Cell. 2000;6:77–86. [PubMed] [Google Scholar]
- 3.White M F. Mol Cell Biochem. 1998;182:3–11. [PubMed] [Google Scholar]
- 4.Withers D J, Gutierrez J S, Towery H, Burks D J, Ren J-M, Previs S, Zhang Y, Bernal D, Pons S, Shulman G I, et al. Nature (London) 1998;391:900–904. doi: 10.1038/36116. [DOI] [PubMed] [Google Scholar]
- 5.Previs S F, Withers D J, Ren J-M, White M F, Shulman G I. J Biol Chem. 2000;275:38990–38994. doi: 10.1074/jbc.M006490200. [DOI] [PubMed] [Google Scholar]
- 6.Alessi D R, Andjelkovic M, Caudwell B, Cron P, Morrice N, Cohen P, Hemmings B A. EMBO J. 1996;15:6541–6551. [PMC free article] [PubMed] [Google Scholar]
- 7.Shimomura I, Hammer R E, Ikemoto S, Brown M S, Goldstein J L. Nature (London) 1999;401:73–76. doi: 10.1038/43448. [DOI] [PubMed] [Google Scholar]
- 8.O'Brien R M, Granner D K. Physiol Rev. 1996;76:1109–1161. doi: 10.1152/physrev.1996.76.4.1109. [DOI] [PubMed] [Google Scholar]
- 9.Hall R K, Yamasaki T, Kucera T, Waltner-Law M, O'Brien R, Granner D K. J Biol Chem. 2000;275:30169–30175. doi: 10.1074/jbc.M004898200. [DOI] [PubMed] [Google Scholar]
- 10.Hall R K, Granner D K. J Basic Clin Physiol Pharmacol. 1999;10:119–133. doi: 10.1515/jbcpp.1999.10.2.119. [DOI] [PubMed] [Google Scholar]
- 11.Tang E D, Nuñez G, Barr F G, Guan K-L. J Biol Chem. 1999;274:16741–16746. doi: 10.1074/jbc.274.24.16741. [DOI] [PubMed] [Google Scholar]
- 12.Guo S, Rena G, Cichy S, He X, Cohen P, Unterman T. J Biol Chem. 1999;274:17184–17192. doi: 10.1074/jbc.274.24.17184. [DOI] [PubMed] [Google Scholar]
- 13.Tomizawa M, Kumar A, Perrot V, Nakae J, Accili D, Rechler M M. J Biol Chem. 2000;275:7289–7295. doi: 10.1074/jbc.275.10.7289. [DOI] [PubMed] [Google Scholar]
- 14.Goldstein J L, Basu S K, Brown M S. Methods Enzymol. 1983;98:241–260. doi: 10.1016/0076-6879(83)98152-1. [DOI] [PubMed] [Google Scholar]
- 15.Xu J, Nakamura M T, Cho H P, Clarke S D. J Biol Chem. 1999;274:23577–23583. doi: 10.1074/jbc.274.33.23577. [DOI] [PubMed] [Google Scholar]
- 16.Sambrook J, Russell D W. Molecular Cloning: A Laboratory Manual. 3rd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 2001. [Google Scholar]
- 17.Vaβen L, Wegrzyn W, Klein-Hitpass L. Diabetes. 1999;48:1877–1880. doi: 10.2337/diabetes.48.9.1877. [DOI] [PubMed] [Google Scholar]
- 18.Berry M N, Friend D S. J Cell Biol. 1969;43:506–520. doi: 10.1083/jcb.43.3.506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shimomura I, Bashmakov Y, Ikemoto S, Horton J D, Brown M S, Goldstein J L. Proc Natl Acad Sci USA. 1999;96:13656–13661. doi: 10.1073/pnas.96.24.13656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Repa J J, Liang G, Ou J, Bashmakov Y, Lobaccaro J-M A, Shimomura I, Shan B, Brown M S, Goldstein J L, Mangelsdorf D J. Genes Dev. 2000;14:2819–2830. doi: 10.1101/gad.844900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sakai J, Rawson R B, Espenshade P J, Cheng D, Seegmiller A C, Goldstein J L, Brown M S. Mol Cell. 1998;2:505–514. doi: 10.1016/s1097-2765(00)80150-1. [DOI] [PubMed] [Google Scholar]
- 22.Lakshmanan M R, Nepokroeff C M, Porter J W. Proc Natl Acad Sci USA. 1972;69:3516–3519. doi: 10.1073/pnas.69.12.3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun X J, Pons S, Wang L-M, Zhang Y, Yenush L, Burks D, Myers M G, Jr, Glasheen E, Copeland N G, Jenkins N A, et al. Mol Endocrinol. 1997;11:251–262. doi: 10.1210/mend.11.2.9885. [DOI] [PubMed] [Google Scholar]
- 24.O'Brien R M, Noisin E L, Suwanichkul A, Yamasaki T, Lucas P C, Wang J-C, Powell D R, Granner D K. Mol Cell Biol. 1995;15:1747–1758. doi: 10.1128/mcb.15.3.1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang J-C, Strömstedt P-E, O'Brien R M, Granner D K. Mol Endocrinol. 1996;10:794–800. doi: 10.1210/mend.10.7.8813720. [DOI] [PubMed] [Google Scholar]
- 26.Allander S V, Durham S K, Scheimann A O, Wasserman R M, Suwanichkul A, Powell D R. Endocrinology. 1997;138:4291–4300. doi: 10.1210/endo.138.10.5268. [DOI] [PubMed] [Google Scholar]
- 27.Kimura K D, Tissenbaum H A, Liu Y, Ruvkun G. Science. 1997;277:942–946. doi: 10.1126/science.277.5328.942. [DOI] [PubMed] [Google Scholar]
- 28.Paradis S, Ruvkun G. Genes Dev. 1998;12:2488–2498. doi: 10.1101/gad.12.16.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ogg S, Paradis S, Gottlieb S, Patterson G I, Lee L, Tissenbaum H A, Ruvkun G. Nature (London) 1997;389:994–999. doi: 10.1038/40194. [DOI] [PubMed] [Google Scholar]
- 30.Lin K, Dorman J B, Rodan A, Kenyon C. Science. 1997;278:1319–1322. doi: 10.1126/science.278.5341.1319. [DOI] [PubMed] [Google Scholar]
- 31.Nasrin N, Ogg S, Cahill C M, Biggs W, Nui S, Dore J, Calvo D, Shi Y, Ruvkun G, Alexander-Bridges M C. Proc Natl Acad Sci USA. 2000;97:10412–10417. doi: 10.1073/pnas.190326997. . (First Published September 5, 2000; 10.1073/pnas.190326997) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beale E G, Chrapkiewicz N B, Scoble H A, Metz R J, Quick D P, Noble R L, Donelson J E, Biemann K, Granner D K. J Biol Chem. 1985;260:10748–10760. [PubMed] [Google Scholar]