Abstract
MicroRNAs (miRNAs) are endogenous noncoding RNAs that negatively regulate gene expression by binding the 3′ noncoding region of the messenger RNA targets inducing their cleavage or blocking the protein translation. They play important roles in multiple biological and metabolic processes, including developmental timing, signal transduction, and cell maintenance and differentiation. Their deregulation can predispose to diseases and cancer. miRNA expression has been demonstrated to be deregulated in many types of human tumors, including thyroid cancers, and could be responsible for tumor initiation and progression. In this paper we reviewed the available data on miRNA deregulation in different thyroid tumors and describe the putative role of miRNA in thyroid cancer development.
1. Introduction
MicroRNAs (miRNAs) are endogenous single-stranded noncoding RNAs of about 22 nucleotides which suppress gene expression by selectively binding to the complementary 3′ untranslated region (3′-UTR) of messenger RNAs (mRNAs) through base-pairing. They play important roles in multiple biological and metabolic processes, such as cell differentiation, proliferation, and survival. Presently, miRNAs are considered one of the most important regulators of gene expression at posttranscriptional level, and it has been estimated that over one third of all human genes could be targeted by miRNAs [1]. Moreover, miRNAs are strongly conserved even among different species, strengthening the hypothesis of their important roles in many essential biological processes.
Recent studies have also supported a role of miRNAs in the initiation and progression of human malignancies. The analysis of global miRNA expression in cancer patients showed different patterns of miRNA overexpression or downregulation in cancer versus normal tissues [2] in several human tumors, such as colorectal neoplasia [3], B cell chronic lymphocytic leukaemia [4, 5], B cell lymphoma [6], lung cancer [7], breast cancer [8], and glioblastoma [9, 10]. The involvement of miRNAs in human cancer is probably due to the fact that >50% of miRNA genes are located at chromosomal fragile sites or common break point sites or within regions of deletion or amplification that are generally altered in human tumors [11]. The deregulation of miRNA expression is suspected to be an important regulator of tumor development and progression in several human tissues. The overexpression of specific miRNAs could lead to the repression of tumor suppressor gene expression, and conversely the downregulation of specific miRNAs could result in an increase of oncogene expression; both these situations induce subsequent malignant effects on cell proliferation, differentiation, and apoptosis that lead to tumor growth and progress.
Experimental evidence demonstrated that the majority of miRNAs present lower expression levels in tumors compared to normal tissues, independent of the cell type. Global miRNA expression is higher in normal tissues compared to their tumoral counterparts or to cancer cell lines. In addition, poorly differentiated tumors present a lower global level of miRNA expression compared to more differentiated tumors [12]. All these data are consistent with the hypothesis that a higher global miRNA expression is associated with cellular differentiation. The reduction of global miRNA expression may reduce cell differentiation that is the hallmark of all human cancers.
2. miRNA Transcription, Maturation, and Mechanisms of Action
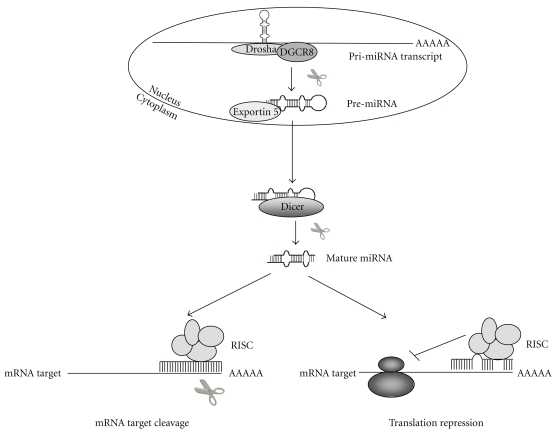
miRNAs are transcribed as long, poly-adenylated, and capped primary transcripts (pri-miRNAs) that are cleaved, at nuclear level, to ~60–70 nucleotide hairpin-shaped intermediates (pre-miRNAs) by the nuclear RNase III Drosha [13]. Following nuclear processing by Drosha, pre-miRNAs are exported to the cytoplasm by the nuclear transport receptor exportin-5 [14]. At cytoplasmic level pre-miRNAs are processed into ~22 nucleotide miRNA duplexes by the cytoplasmic RNase III Dicer [15]. These double-stranded products are unwound by a still unidentified helicase and incorporated as single-stranded RNAs (guide strand) into a ribonucleoprotein complex, known as the RNA-induced silencing complex (RISC) [16]. Which of the two RNA strands is incorporated in the RISC is determined by the stability of the base pairs at the 5′ end of the duplex; the other strand is degraded. The incorporate guide strand leads the RISC to the complementary sequence in the 3′-UTR of target mRNA, negatively regulating gene expression at the posttranscriptional level by targeting the 3′-UTR region of the mRNAs. Nucleotides 2–8 (referred to as “seed”) of the mature miRNAs are evolutionary conserved and result in being crucial in determining target specificity. miRNAs can downregulate gene expression by two distinct posttranscriptional mechanisms: mRNA cleavage or translational repression. The choice of mechanism is determined only by the identity of the mRNA target and the degree of miRNA-mRNA complementarity: miRNA will specify induce the cleavage of mRNA if the target has sufficient complementarity to the miRNA itself, or it will repress translation if the mRNA does not have sufficient complementarity [17, 18] (Figure 1). In the first case miRNA may act as small interfering RNAs (siRNAs) and it cleaves mRNA targets between the nucleotides pairing to positions 10 and 11 of the miRNA [19, 20]; after the cleavage of the target the miRNA remains intact and can guide the recognition and destruction of other mRNAs [17]. Conversely, the reduced complementarity between miRNA and its target mRNA generally creates mismatches and bulges in the central region of the miRNA-mRNA duplex (at positions 10–12 of the mature miRNA sequence) that prevent the siRNA-like cleavage of target mRNA. miRNA-mediated translational repression can be regulated both at initiation level or at postinitiation level of the translation process. In the initiation block the RISC complex inhibits translation by interfering with eIEF4F-cap recognition and 40S small ribosomal subunit recruit or by preventing the assembly of the 60S subunit to form the 80S ribosomal complex. In the postinitial block RISC may inhibit ribosome elongation, inducing ribosome drop-off or facilitating proteolysis of the nascent polypeptides [21].
Figure 1.
Model for miRNA biogenesis and functional mechanisms. miRNA genes are transcribed by RNA polymerase II (pol II) into primary transcripts (pri-miRNAs) that are cleaved by the Drosha-DGCR8 complex to 60–70 nt pre-miRNAs. Pre-miRNAs are then transported to the cytoplasm by exportin-5 and there processed by the endonuclease Dicer to generate a double-stranded mature miRNA of about 21–23 nt. After a strand selection/separation process, the mature miRNA is incorporated into the RISC complex while the other strand is degraded. RISC complex will recognize and mediate cleavage or repression of specific mRNAs.
Recently, it has been hypothesized a role of miRNAs in the upregulation of protein translation. A recent study [22] showed that miR-369-3 linked the AU-rich elements (AREs) of the TNFα mRNA to upregulate translation, through direct base pairing between miR-369-3 “seed” region and complementary ARE regions, under cell cycle arrest conditions. The authors hypothesized that miRNAs may switch between translation repression or activation based on cell cycle status: in proliferating cells they repress translation while under cell cycle arrest they promote translation.
Another study [23] indicated a possible role of miRNAs in positive activation of translation. The authors found that miR-10a interacted with the 5′ untranslated region (5′-UTR) of the mRNAs encoding ribosomal proteins to enhance their translation. Moreover, miR-10a resulted in being capable of both translation repression through 3′-UTR binding or of translation promotion via 5′-UTR binding of different mRNA targets. These data suggest that the same miRNA may exert different effects depending on the site of interaction with its targets.
3. Identification of miRNA Targets
Identification of miRNA targets and of their specific interaction sites is fundamental in the comprehension of miRNA roles in the regulation of biological processes as well as in the development of human malignancies.
Some algorithms, such as miRanda (http://www.microrna.org/microrna/home.do), TargetScan (http://www.targetscan.org/), or PicTar (http://pictar.mdc-berlin.de/), have been developed for the computational prediction of miRNA targets. All these bioinformatic predictions are primarily based, and limited, on conserved interactions involving the miRNA “seed” region and the 3′-UTRs of target mRNAs. A limitation of these prediction programs is that they are not able to reveal novel aspects of miRNA target recognition. False-positive predictions can be eliminated by experimental validation studies but false-negative predictions remain often unsolved.
Experimental approaches to miRNA target identification have mainly focused on analyses of both transcriptome and proteome expression arrays in cells in which a single miRNA has been overexpressed or inhibited. Effects on endogenous target protein levels serve as a good indicator to validate the miRNA-target interaction. However, this approach suffers from a limitation: in fact when a miRNA is introduced, by transfection, in high concentrations into a cell, this may effect the observed effect on target mRNA and generate false-positive results, since the level of translation repression is strongly dependent not only on miRNA and mRNA target complementarity but also on both the amount of target mRNA and the amount of available miRNA in the cell [24].
A direct evidence that a miRNA binds a specific target mRNA can be obtained by formaldehyde cross-linking of the miRNA to its targets [22] or by 4-thiouridine-modified miRNAs [23], both these techniques allow the subsequent mapping of the exact site of binding using primer extension.
4. miRNA Deregulation in Thyroid Tumors
Thyroid tumors represent a good model for studying multistep cancer development in epithelial cells as they comprise a range of lesions with different degrees of malignancy: from benign differentiated and noninvasive adenomas to malignant undifferentiated anaplastic invasive carcinomas. The two most common thyroid tumors are papillary thyroid carcinoma (PTC) and follicular thyroid carcinoma (FTC), accounting, respectively, for about 80% and 15% of all cases, both well differentiated and originating from thyroid follicular cells. PTCs are often multifocal, characterized by classical papillary architecture, and they usually metastasize to the regional lymph nodes. FTCs are often unifocal, encapsulated, and they usually metastasize via the vascular system to lungs and bones. Both these tumors may progress to poorly differentiated carcinoma or to completely nondifferentiated anaplastic carcinoma. Anaplastic thyroid carcinomas (ATCs) are very rare thyroid cancers (2–5% of all cases), extremely undifferentiated and highly aggressive. Medullary thyroid carcinoma (MTC) is a rare thyroid tumor, accounting for less than 5% of all thyroid cancers, that originates from the thyroid intrafollicular C cells.
Several independent studies have analyzed miRNA expression in numerous and different types of thyroid tumors, evidencing a miRNA deregulation in cancer tissues compared to their normal counterparts [25–38]; in thyroid tumors 32% of all known human miRNAs resulted in being upregulated and 38% to be downregulated with more than a 2-fold change as compared to normal tissues [39]. Moreover, the miRNA expression profile presents a significant variability between different kinds of thyroid cancers, even if they originate from the same type of thyroid cells [25, 39]. C-cell-derived MTC has a miRNA expression profile significantly different from those of thyroid tumors originating from follicular cells; but also papillary carcinomas, conventional follicular adenomas and carcinomas, and oncocytic follicular adenomas and carcinomas, all originating from follicular cells, show different and specific miRNA expression profiles. At the moment the exact biological roles of miRNAs in thyroid carcinogenesis remain to be fully elucidated but it seems reasonable that the distinctive pattern of miRNA expression in thyroid tumors compared to normal thyroid tissue may be useful in diagnosis and/or therapy of thyroid neoplasia and that different miRNA expression patterns in different types of thyroid tumors could be useful tools for their classification.
However, the majority of miRNA profiling studies do not provide an estimate of miRNA abundance in normal thyroid tissues and in thyroid tumors. Today, it is recognized that only the most abundantly expressed miRNAs are able to occupy a substantial fraction of their mRNA targets, affecting their translation. The magnitude of translation repression is strongly dependent on the number of miRNA-RISC complexes with respect to the amount of target mRNA molecules. It is hypothesizable that, among all the misregulated miRNAs, only those which are abundantly overexpressed or strongly downregulated are involved in thyroid tumorigenesis. The most abundantly expressed miRNAs in human healthy thyroid gland are listed in Table 1; data are derived from http://www.mirz.unibas.ch/cloningprofiles/ using the “visualization of miRNA expression profiles” tool.
Table 1.
Most abundantly expressed miRNAs in human healthy thyroid gland. The table reports the most abundantly expressed miRNAs in human normal thyroid gland; data are derived from http://www.mirz.unibas.ch/cloningprofiles/. The tool screened a total of 768 human miRNAs. Only miRNAs with an abundance value over 3.0 have been reported in the table.
| miRNA | Abundance value in thyroid gland |
|---|---|
| let-7b | 56.0 |
| let-7a | 52.0 |
| miR-143 | 47.0 |
| miR-126 | 39.0 |
| let-7i | 32.5 |
| let-7c | 29.5 |
| miR-125b | 29.0 |
| miR-16 | 27.0 |
| miR-200c | 24.5 |
| miR-26a | 23.6666666 |
| let-7f | 21.5 |
| miR-23b | 16.0 |
| miR-24 | 14.0 |
| miR-99a | 13.0 |
| miR-29a | 12.0 |
| miR-30d | 12.0 |
| miR-451 | 12.0 |
| miR-23a | 11.0 |
| miR-15a | 9.0 |
| miR-27b | 9.0 |
| miR-30c | 9.0 |
| miR-21 | 8.0 |
| miR-27a | 8.0 |
| miR-30a | 8.0 |
| miR-100 | 8.0 |
| miR-191 | 8.0 |
| let-7e | 7.0 |
| let-7g | 6.0 |
| miR-99b | 6.0 |
| miR-125a-5p | 6.0 |
| miR-145 | 6.0 |
| miR-195 | 5.0 |
| let-7d | 4.0 |
| miR-25 | 4.0 |
| miR-206 | 4.0 |
| miR-10b | 3.0 |
| miR-22 | 3.0 |
| miR-138 | 3.0 |
| miR-152 | 3.0 |
| miR-423-3p | 3.0 |
5. The Role of MicroRNAs in Papillary Thyroid Carcinoma
Some studies analyzed the miRNA expression profile in PTCs [26–32] (Table 2).
Table 2.
Studies of deregulation of miRNA expression profile in thyroid tumors. The table reports a list of published studies on the deregulation of miRNA expression profile in different kinds of thyroid tumors. Type of analyzed thyroid samples, used analysis methods and individuated upregulated or downregulated miRNAs in thyroid tumors are depicted in the table.
| Thyroid tumor type | Analyzed samples | Methods | Tumor upregulated miRNAs | Tumor downregulated miRNAs | Reference |
|---|---|---|---|---|---|
| PTC | 20 fresh PTC tissues versus 20 normal adjacent normal thyroid tissues | Global miRNA microarray, quantitative RT-PCR and Northern blots | miR-146b, miR-221, and miR-222 | — | He et al. [26] |
| PTC | 30 fresh PTC tissues versus 10 normal thyroid tissues | Global miRNA microarray, quantitative RT-PCR, and Northern blots | miR-181b, miR-221, and miR-222 | — | Pallante et al. [27] |
| PTC | 20 formalin-fixed paraffin-embedded PTC tissues versus 20 formalin-fixed paraffin-embedded multinodular goiter | Global miRNA microarray and quantitative RT-PCR | miR-21, miR-31, miR-34a, miR-172, miR-181a, miR-181b, miR-213, miR-221, miR-222, miR-223, and miR-224 | miR-19b-1,2, miR-30a-5p, miR-30c, miR-130b, miR-145sh, miR-218, miR-292-as, miR-300, and miR-345 | Tetzlaff et al. [28] |
| PTC versus non-PTC | 84 formalin-fixed paraffin-embedded tissues and 40 ex vivo aspirate specimens of PTC and non-PTC tumors (follicular adenoma, follicular carcinoma hyperplastic nodules) | Quantitative RT-PCR | miR-146b, miR-221, and miR-222 | — | Chen et al. [29] |
| PTC | 28 BRAF mutated PTC tissues versus 26 BRAF wild-type PTC tissues | Quantitative RT-PCR | No difference between mutated and nonmutated PTC | No difference between mutated and nonmutated PTC | Sheu et al. [31] |
| PTC | 100 PTC tissues versus 16 paired normal thyroid control | Quantitative RT-PCR | miR-146b, miR-221, and miR-222 | — | Chou et al. [32] |
| PTC | 2 PTC cell lines bearing a RET mutation versus normal thyroid cell lines | Global miRNA microarray | miR-34a, miR-96, miR-99a, miR-100, miR-125b, miR-128b, miR-130b, miR-139, miR-141, miR-142-3p, miR-146, miR-148, miR-185, miR-200a, miR-200b, miR-211, miR-213, miR-216, and let-7d | miR-15a, miR-34c, miR-107, miR-127, miR-135b, miR-145, miR-149, miR-154, miR-181a, miR-218, miR-299, miR-302b, miR-302c, miR-323, and miR-370 | Cahill et al. [40] |
| FTC | 22 FTC samples versus 20 FA and 4 normal control thyroid tissues | Global miRNA microarrayand quantitative RT-PCR | miR-192, miR-197, miR-328, and miR-346 | — | Weber et al. [33] |
| FTC PTC, other thyroid tumor variants | A 60-fresh-thyroid tumor and normal samples (23 PTCs, 9 FTCs, 8 FAs, 4 ATCs, 4 poorly differentiated carcinomas, 2 MTCs, 5 hyperplastic nodules, and 5 normal thyroid tissues) and 62 fine-needle aspiration samples | Quantitative RT-PCR | PTC: miR-31, miR-122a, miR-146b, miR-155, miR-187, miR-205, miR-221, miR-222, and miR-224 Conventional FA: miR-190, miR-205, miR-210, miR-224, miR-328, miR-339, and miR-342 Oncocytic FA: miR-31, miR-183, miR-203, miR-221, miR-224, and miR-339 Conventional FTC: miR-146b, miR-155, miR-187, miR-221, miR-222, and miR-224 Oncocytic FTC: miR-183, miR-187, miR-197, miR-221, miR-222, and miR-339 Poorly differentiated carcinomas: miR-129, miR-146b, miR-183, miR-187, miR-221, miR-222, and miR-339 ATC: miR-137, miR-155, miR-187, miR-205, miR-214, miR-221, miR-222, and miR-224 MTC: miR-9, miR-10a, miR-124a, miR-127, miR-129, miR-137, miR-154, miR-224, miR-323, and miR-370 |
— | Nikiforova et al. [25] |
| ATC | ATC tissues versus normal thyroid tissues | Global miRNA microarray, quantitative RT-PCR, Northern blots, and in situ hybridization | — | miR-26a, miR-30a-5p, miR-30d, and miR-125b | Visone et al. [34] |
| ATC | 10 ATC and 5 FTC cell lines, 3 ATC and 8 PTC tissues versus normal thyroid samples | Quantitative RT-PCR | miR-21, miR-146b, miR-221, and miR-222 | miR-26a, miR-138, miR-219, and miR-345 | Mitomo et al. [38] |
| ATC | ATC cell lines and ATC cancer lesions versus normal thyroid tissues | Global miRNA microarray, quantitative RT-PCR, and Northern blots | miR-17-3p, miR-17-5p, miR-18a, miR-19a, miR-19b, miR-20a, and miR-92-1 | — | Takakura et al. [37] |
| ATC | ATC tissues versus PTC and FTC tissues | Global miRNA microarray and quantitative RT-PCR | — | miR-30 and miR-200 | Braun et al. [35] |
Comparing global miRNA expression in human PTCs versus unaffected thyroid tissue He et al. [26] individuated a set of five miRNAs (miR-146, miR-221, miR-222, miR-21, and miR-181a) that were significantly overexpressed in PTCs compared to the adjacent normal tissue. Particularly, three of them, miR-146, miR-221, and miR-222, showed 11- to 19-fold higher level in tumor tissues. The probe sequence of miR-146 used in the miRNA array chip was designed for miR-146a isoform, located on human chromosome 5. Recently a second isoform, named miR-146b, has been described on human chromosome 10. These two miRNAs differ only for two nucleotides in the sequence of their mature forms. Using primers for premature forms of miR-146a and miR-146b, no expression of miR-146a was detected in thyroid tissues by RT-PCR analysis, while a significant overexpression of miR-146b was found in PTC tumor samples as confirmed also by Northern blot for mature miR-146b. Deregulation of miR-146b, miR-221, and miR-222 in the thyroid may be a crucial component of PTC initiation and development. The putative target of these miRNAs was suspected to be KIT, a tyrosine kinase receptor that plays an important role in cell growth and differentiation, acting as an oncogene in many cancers [41, 42]. Nevertheless, neoplastic transformation has been shown to be associated with either overexpression or downregulation of c-KIT in different tissues [43–45]. In PTC tissues in which miR-146b, miR-221, and miR-222 were strongly overexpressed there was a downregulation of KIT transcript and KIT protein. In 50% of cases the reduced expression of KIT was associated with germline single-nucleotide changes in both the two recognition sites of KIT for miR-221 and miR-222 (3′-UTR region of KIT) and for miR-146b (exon 18 of KIT). In conclusion, the upregulation of these five specific miRNAs, and particularly of miR-146b, miR-221, and miR-222, and the subsequent downregulation of KIT seem to be involved in PTC pathogenesis, and sequence changes in miRNA target genes can contribute to their regulation.
c-KIT is frequently expressed in benign thyroid adenomas and goiters, while its expression results in being lowered in about 60% of FTCs and completely absent in PTCs and ATCs. Moreover, the absence of c-KIT expression has been demonstrated also in metastases from primary thyroid tumors, indicating that the modulation of this tyrosine kinase receptor is not dependent on the thyroid microenvironment but is associated to the transformed malignant cell phenotype [46]. To date, the biological significance of loss of c-KIT in thyroid tumors is not elucidated. Surprisingly, the depletion of c-KIT expression in thyroid tumors in contrast with the gain of function of other tyrosine kinase receptors such as c-RET and c-MET or of oncogenes such as c-RAS, suggesting that different tyrosine kinase receptor signaling pathways may exert opposite biological effects in a given cell type, alternatively controlling mitogenesis or cell differentiation. It is hypothesizable that, in thyroid tissue, the c-KIT signaling pathway may control some aspects of the thyrocyte differentiation rather than cell proliferation.
Pallante et al. [27] analyzed the global miRNA expression profile in 30 human PTCs versus 10 normal thyroid tissues using a microarray chip containing 368 human precursor and mature miRNA oligonucleotide probes, accounting for 245 human and mouse miRNA genes and including all the three miR-181 human isoforms (miR-181a, miR-181b, and miR-181c). The analysis revealed an altered miRNA expression profile that distinguished PTCs from normal thyroid tissues: five miRNAs (miR-221, miR-222, miR-213, miR-220, and miR-181b) were overexpressed in the neoplastic tissues. However, a recent work by Chiang et al. [47] strongly suggested that miR-220 is not a miRNA, and miR-213 is not recognized as a miRNA in the miRNA database (http://www.mirbase.org/). The study of Pallante et al. found that miR-221, miR-222, and miR-181b resulted in being abundantly expressed in PTCs, while their expression was only weakly detectable in the healthy thyroid tissues, with the overexpression of miR-221 reaching up to 70-fold increase. overexpression of miR-181b has been also confirmed by quantitative RT-PCR analysis using a couple of primers specifically designed for precursor of the miR-181b isoform to exclude a false-positive result for miR-181b, due to possible cross-hybridization of probes for the miR-181a and miR-181c isoforms in the microarray chip. The authors also demonstrated that miR-221 and miR-222 downregulated the level of c-KIT in PTCs confirming data previously published [41]. In addition, a human thyroid carcinoma cell line transfected with a vector over-expressing miR-221 showed a significant increase of cell growth, and, conversely, the inhibition of miR-221 function by antisense oligonucleotide transfection in the same cell line resulted in a significant reduction of cell growth [27]. The results of these functional studies, together with the previously reported expression data, suggest a critical role of miR-221 in thyroid carcinoma cell growth and, thus, probably in the process of papillary thyroid carcinogenesis.
A study of Tetzlaff et al. [28] analyzed global miRNA expression profile by microarray chip (including known miRNAs from human, mouse, rat, and predicted/candidate human miRNAs) in a series of PTC formalin-fixed paraffin-embedded (FFPE) samples versus benign proliferative multinodular goiters (MNGs). The analysis revealed a set of 13 upregulated miRNAs and a set of 26 downregulated miRNAs. Misregulation has been validated by real-time RT-PCR, in an independent series of tumoral tissues, only for miR-21, miR-31, miR-221, and miR-222, with miR-221 and miR-222 showing a strong upregulation in PTCs compared to independent set of MNGs. Data from this study confirmed miR-221 and miR-222 to be altered in PTCs, as previously described in fresh tissue analyses [26, 27] and, mostly, supported the possibility to use FFPE tumor tissues to analyze miRNA expression when fresh tissues are not available or to perform retrospective studies. The author demonstrated the possibility to extract sufficient miRNA from FFPE tissues using a miRNA labeling method for total RNA extracts (All Total Nucleid Acid Isolation SyStem, Ambion) that bypassed the need for miRNA enrichment and reduced the amount of starting samples, followed by an ammonium acetate/ethanol precipitation.
A recent study by Chen et al. [29] analyzed the expression of a selected set of miRNAs by quantitative RT-PCR in PTCs, non-PTC lesions (follicular adenoma, follicular carcinoma hyperplastic nodules), and normal thyroid tissue. They evidenced an overexpression of miR-146b, miR-221, and miR-222 in PTCs compared to both non-PTC group and healthy controls, but with miR-221 and miR-222 presenting a substantial overlap between different tumor groups. The authors concluded that the expression analysis of miR-221 and miR-222 cannot be resolving in distinguishing PTCs from other tumoral lesions. Conversely, miR-146b resulted in being consistently and specifically overexpressed in classical PTCs, suggesting this miRNA as a possible diagnostic tool to identify PTCs from other thyroid tumors. Putative targets of miR-146b are the nuclear factor-κB (NF-κB), the interleukin-1 receptor-associated kinase 1 (IRAK1), and the tumor necrosis factor receptor-associated factor 6 (TRAF6), whose role in thyroid carcinogenesis has not yet been elucidated.
Mutations of the BRAF gene are implicated in the pathogenesis of PTC through the constitutive activation of the MAPK pathway; classical PTCs are often BRAF mutation positive while follicular PTCs are almost always BRAF mutation negative [48]. Chen et al. [29] found also that miR146b overexpression is common both in classical and follicular variants of PTCs, independently by the BRAF mutation status, suggesting this overexpression as a late event in the PTC progression and probably important for the definition of a complete carcinoma phenotype. This result has been confirmed also by Sheu et al. [31] who analyzed, in a series of 221 PTCs, if the BRAF V600E mutational status was correlated to the miRNA expression profile. They found no difference in the expression of five miRNAs (miR-146b, miR-181b, miR-21, miR-221, and miR-222) between BRAF mutated PTCs and wild-type PTCs. In addition, this study confirmed the overexpression of miR-146b, miR-221, and miR-222 as a useful tool to diagnostic PTCs.
More recently, Chou et al. [32] measured the expression of miR-146b, miR-221, and miR-222 in 100 cases of PTCs finding that their expression levels were associated with extra-thyroidal invasion. In particular miR-146b resulted in being highly expressed in tumors with high risk features and with the BRAF V600E mutation.
Results from all these studies agreed with the fact that miR-221 and miR-222 are both overexpressed in PTCs compared to normal thyroid tissue. Functional analysis on miR-221 function in human PTC-derived cell lines indicated a direct role of this miRNA in PTC carcinogenesis. A vector-induced miR-221 overexpression in these cell lines resulted in a higher number of cell colonies compared to negative control transfected cells; conversely, the block of miR-221 function by antisense oligonucleotide caused a significant reduction in cell proliferation [27]. A subsequent study investigated which pathways or molecular targets were regulated by miR-221 and miR-222 [49]. Some bioinformatic programs suggested the CDKN1B (p27kip1) gene, an important regulator of cell cycle that inhibits the initiation of the S phase, as the putative target of miR-221 and miR-222. This study demonstrated that miR-221 and miR-222 negatively regulate the p27kip1 protein levels in Hela cells and thyroid carcinoma cells by binding to two specific target sites in the 3′-UTR of the p27kip1 gene. Transfection of TPC-1, thyroid papillary carcinoma cell line, with vectors for the overexpression of miR-221 and miR-222 resulted in decreased p27kip1 protein levels. Conversely, inhibition of miR-221 and miR-222 expression by specific 2′-O-Me-221 and 2′-O-Me-222 antisense oligonucleotides increased the p27kip1 protein levels. In both transfection experiments no significant variation of p27kip1 mRNA expression was observed. Results from this study strongly suggested that the overexpression of miR-221 and miR-222 in tumor thyroid cells could be responsible for the posttranscriptional negative regulation of p27kip1 protein expression, inducing cells to enter the S phase of cell cycle and, thus, increasing cell growth.
The RET oncogene mutations occur in about 43% of PTCs [50], constitutively activating the MAPK signal and promoting carcinogenesis. These mutations are responsible for the deregulation of thyroid cell proliferation and differentiation and for cell tumoral transformation. Cahill et al. [40] investigated miRNA expression in two human PTC cell lines bearing a RET mutation, compared to normal thyroid cell lines, finding that 21 miRNAs were significantly overexpressed (miR-34a, miR-96, miR-99a, miR-100, miR-125b, miR-128b, miR-130b, miR-139, miR-141, miR-142-3p, miR-146, miR-148, miR-185, miR-200a, miR-200b, miR-211, miR-213, miR-216 and let-7d) and 14 miRNAs were downregulated (miR-15a, miR-34c, miR-107, miR-127, miR-135b, miR-145, miR-149, miR-154, miR-181a, miR-218, miR-299, miR-302b, miR-302c, miR-323, and miR-370) in tumor cell lines when compared to normal thyroid. These differentially expressed miRNAs potentially regulate genes involved in thyroid functions, and their deregulation could be implicated in thyroid carcinoma progression.
A functional study of Ricarte-Filho et al. [36] investigated the involvement of the let-7f miRNA, recently associated to RAS protein level reduction in lung tumor, in PTC development. The authors found that, in thyroid RET-mutated cell lines, the enhanced expression of RET oncogene reduced the expression of let-7f. The stable transfection of RET-mutated TPC-1 cell line with vector for the overexpression of let-7f inhibited the MAPK activation and reduced cell proliferation. In particular, let-7f increases the expression of thyroid cell differentiation markers such as the TITF1 transcription factor, a key factor in normal thyroid development, and, thus, let-7f is fundamental for the correct regulation of thyroid cell growth and differentiation and the reduced expression of let-7f in RET-mutated thyroid cells is responsible for cell dedifferentiation during PTC malignant progression. Interestingly, let-7f, together with the miRNA let-7 family, is one of the most expressed miRNAs in healthy thyroid gland (Table 1), further suggesting its crucial role in normal thyroid development and functionality. All these data suggested let-7f acting as a tumor suppressor and indicated this miRNA as a potential therapeutic agent in patients with PTCs bearing a RET mutation.
6. The Role of MicroRNAs in Follicular Thyroid Carcinoma
Only two studies have analyzed the miRNA expression alteration in FTCs [25, 33] (Table 2).
In 2006 Weber et al. [33] investigated if miRNAs are differentially expressed between human follicular thyroid carcinomas (FTCs) and follicular adenomas (FAs) testing two high-density miRNA expression arrays on 23 FTCs versus 20 FA samples and 4 normal thyroid controls. Four miRNAs (miR-192, miR-197, miR-328, and miR-346) resulted in being overexpressed in FTCs compared to FAs. None of these miRNAs have previously been associated with other thyroid neoplasia and appear to be specific for FTC phenotype. Two of them, miR-197 and miR-346, have been validated also by quantitative real-time RT-PCR that confirmed their significant overexpression in carcinomas compared to adenomas and healthy tissue. These two miRNAs may participate in the transformation of follicular tumors from benign to malignant status, and they and their target genes may provide novel molecular markers to differentiate malignant (FTCs) from benign (FAs) follicular thyroid neoplasia. The effects of these two deregulated miRNAs have been functionally investigated using two human thyroid cancer cell lines, FTC133 and K5, a human papillary thyroid cancer cell line, NPA87, and a human embryonic kidney cell line, HEK293T as control. The induction of miR-197 and miR-346 overexpression induced cell proliferation in vitro, whereas their inhibition led to cell growth arrest in both FTC133 and K5 cell lines, but not in NPA87 cell line, confirming that the deregulation of miR-197 and miR-346 is a marker of FTC phenotype but not of PTC phenotype. In silico analyses indicated putative targets of miR-197 and miR-346 that have been validated also by in vitro functional analyses. EFEMP2 (fibulin 4), a protein involved in stabilization and organization of extracellular matrix structures that exerts tumor suppressor functions [51, 52], is inhibited by miR-346 overexpression. Activin A receptor type 1 (ACVR1), a potent inhibitor of cell growth in several human cell types including thyroid epithelium [53], and tetraspanin 3 (TSPAN3), whose exact biological role in tumors is still unknown, are both downexpressed as a consequence of miR-197 overexpression. Moreover, the authors performed functional studies also on miR-221 and miR-222 demonstrating that they do not have a role in FTC tumorigenesis.
More recently, Nikiforov et al. [25] compared miRNA expression profiles of principal types of thyroid cancers, finding a distinctive expression pattern associated to follicular thyroid tumors: miR-155, miR-187, miR-221, miR-222, and miR-224 resulted in being highly overexpressed in conventional FTCs, while miR-183, miR-187, miR-197, miR-221, miR-222, and miR-339 were overexpressed in the FTC oncocytic variants.
7. The Role of MicroRNAs in Anaplastic Thyroid Carcinoma
Few studies have analyzed the miRNA expression profiles in ATCs [34, 35, 37, 38] (Table 2).
Visone et al. [34] analyzed the miRNA expression of ATCs using a miRNA microarray chip, finding a significant down-expression of miR-26a, miR-30a-5p, miR-30d, and miR-125b in ATCs compared to normal thyroid samples. These data have been further validated by quantitative RT-PCR, Northern blot analyses, and in situ hybridization. Induced overexpression of miR-26a and miR-125b in two human ATC-derived cell lines resulted in cell growth inhibition, suggesting a role of these two miRNAs in negative cell cycle regulation and that their downregulation could be involved in thyroid tumorigenesis. No effect on cell proliferation was observed after induction of miR-30d and miR-30a-5p overexpression in the same ATC-derived cell lines. miR-26a influences cell cycle progression by negatively regulating the expression of EZH2 oncogene, an epigenetic gene silencer involved in neoplastic development. Recently, Sander et al. [54] suggested miR-26a acting as a potential tumor suppressor in MYC-induced tumors, since overexpression of miR-26a in murine lymphoma cell lines reduced cell proliferation by increasing the percentage of cells in G1 phase. miR-125b resulted in being deregulated in several human tumors, suggesting a role of miR-125b in human carcinogenesis [55–57]. An upregulation of miR-125b expression reduced the proliferation of the CD133-positive glioma cells and arrested the cell cycle at the G1/S transition level [58], through the downregulation of CDK6 and CDC25A, respectively, a cyclin-dependent kinase positively regulating the transition from the G0/G1 phase to the S phase of the cell cycle and a positive regulator of G1/S transition by dephosphorylation and activation of cyclin-CDK complexes. Both CDK6 and CDC25A have been previously reported to modulate the G1/S transition in human embryonic stem cells [59]. Recently, Liang et al. [60] demonstrated that miR-125b suppresses hepatocarcinoma cell proliferation both in vitro and in vivo and that this miRNA increases the expression of p21Cip1/Waf1, arresting cell cycle at the G1 phase.
Another study [38] examined the expression of miRNAs in ATC-derived cell lines versus PTC-derived cell lines as well as ATC tissue samples versus PTC tissue samples, finding that miR-21, miR-146b, miR-221, and miR-222 were overexpressed, while miR-26a, miR-138, miR-219, and miR-345 were downregulated in ATC cell lines and tissues. Moreover, since miR-138 resulted in being the unique miRNA that showed a different expression profile between ATCs and other follicular cell-derived thyroid tumors, the authors investigated its putative role in ATC tumorigenesis, demonstrating that miR-138 could directly target the human telomerase reverse transcriptase (hTERT) gene at posttranscriptional level. hTERT is a catalytic subunit of telomerase that results in being overexpressed in primary ATCs compared to PTCs and associated with cell dedifferentiation and increased metastatic potential [61]. The upregulation of hTERT, subsequent to miR-138 downregulation, could be responsible for malignant progression of well-differentiated PTCs toward undifferentiated ATCs. Since miR-138 downregulation seemed to be strongly associated with the ATC phenotype, this miRNA could be useful as a diagnostic tool for ATC recognition and it may contribute to the development of novel treatment strategy for ATCs.
Takakura et al. [37] reported a group of seven miRNAs (miR-17-3p, miR-17-5p, miR-18a, miR-19a, miR-19b, miR-20a, and miR-92-1), compressively named as miR-17-92 cluster, to be overexpressed in ATC cell lines and ATC cancer lesions compared to adjacent thyroid normal tissues. To investigate functional role of these deregulated miRNAs in ATC tumorigenesis, human ATC cell lines have been induced to silence these miRNAs expression by transfection with specific miRNA antisenses. Inhibition of miR-17-3p expression totally suppressed cell growth and induced apoptosis by the strong activation of caspases 3 and 9. Suppression of miR17-5p or miR-19a caused a strong reduction of cell proliferation, not associated with apoptosis or caspase activation. Moreover, inhibition of miR17-5p, but not of miR-19a, was also responsible for cell senescence. miR-17-5p and miR-19a target, respectively, retinoblastoma 1 (RB1) and phosphatase and tensin homolog (PTEN) genes as confirmed by the fact that the expression of RB1 and PTEN proteins was increased in cells transfected with miR-17-5p and miR-19a inhibitors. Mutations of PTEN gene are associated with Cowden syndrome characterized by breast and thyroid tumors. PTEN acts as a tumor suppressor, negatively regulating cell growth by the repression of the cyclin-dependent kinase inhibitor p27kip1 expression. A reduced expression of PTEN protein is associated with thyroid cancer development [62, 63], and PTEN inactivation is associated with undifferentiated malignant ATCs, rather than other thyroid tumor types [64]. Therefore, the overexpression of miR-19a (and of its isoform miR-19b) might be responsible for PTEN posttranscriptional downregulation and subsequent increased cell growth in ATCs. In addition, downregulation of tumor suppressor activity of RB1 by overexpression of miR-17-5p could further contribute to increased tumor cell proliferation. Selective inhibitors of miR-17-3p, miR-17-5p, and miR-19a demonstrated to significantly reduce cell growth in ATC-derived cell lines, and inhibitor of miR-17-3p also induced cell death. Therefore, these inhibitors could represent valid therapeutic approaches for the treatment of ATCs.
Very recently, Braun et al. [35] identified two miRNAs, miR-30 and miR-200, which were significantly downexpressed in ATCs compared to PTCs and FTCs. overexpression of these two miRNAs in mesenchymal ATC-derived cell lines reduced their invasive and metastatic potential, negatively regulating the expression of the mesenchymal-epithelial transition (MET) proteins. Conversely, inhibition of endogenous miR-200 expression was sufficient to induce the inverse process, the epithelial-mesenchymal transition (EMT) responsible for tumor cell invasiveness and metastatic potential. miR-200 targets the ZEB1 and ZEB2, both repressors of the E-cadherin (CDH1) gene expression. Park et al. [65] demonstrated, in 60 human cell lines conserved at the Nation Cancer Institute, a significant association between miR-200 expression and E-cadherin-vimentin ratio. Induced overexpression of miR-200 caused the upregulation of E-cadherin in tumor cell lines and reduced their motility and invasiveness. Conversely, the inhibition of miR-200 reduced E-cadherin expression, increased expression of vimentin, and induced EMT. All these data suggested miR-200 as a key factor of the epithelial phenotype in cancer cells that could be used as therapeutic agent to reduce the rates of invasive and metastatic thyroid carcinomas.
8. The Role of MicroRNAs in Medullary Thyroid Carcinoma
Only one study [25] has analyzed the miRNA expression in MTCs compared to other thyroid tumors and normal thyroid tissue, finding a set of 10 specific miRNAs (miR-9, miR-10a, miR-124a, miR-127, miR-129, miR-137, miR-154, miR-224, miR-323, and miR-370) upregulated in this tumor type. No functional studies have been yet performed.
9. Conclusion and Future Perspectives
miRNAs are powerful key regulators of gene expression in many fundamental cellular processes such as proliferation, differentiation, and apoptosis. Deregulation of miRNA expression was observed in the initiation, development, and malignant progression of numerous human tumors [2–10]. miRNA expression profiles resulted in being different not only between tumors and healthy tissues but also between different histopathological lesions of the same tissue, between tumors at different stages of malignancy, and between primary tumors and metastases. Therefore, miRNA expression profiles may become useful novel biomarkers for tumor diagnostic and histological characterization. Recent findings suggested that miRNA expression profiles could enable classifying poorly characterized human tumors that can not be accurately classified only by the classical mRNA expression patterns [66]. Moreover, since differences in miRNA expression are, in some cases, associated with the prognosis, analysis of miRNA expression profiles could help also in the therapeutic management of patients.
Very recent data supported the possibility to use circulating miRNAs (plasma, serum, urine, or other body fluids) as a novel class of biomarkers to diagnose tumors, as expression patterns of circulating miRNAs are different between normal tissues and cancers and peculiar miRNA expression profiles are specifically associated with certain types of tumors [67]. In addition, circulating miRNAs present the advantage to be stable molecules with a great resistance to RNase activity that can be easily dosed by noninvasive techniques.
Moreover, since miRNAs regulate cancer cell proliferation, differentiation, apoptosis, and invasiveness, miRNAs and their biological targets could be potential targets of therapeutic genetic strategies in human tumors to interfere with cancer initiation and progression. miRNAs are possible targets for a RNA-based therapy both by positively modulating the expression of specific miRNAs in vivo using expression vectors [68] and/or by inhibiting miRNA expression by transfecting specific 2′-O-methyl-modified antisense RNA (antagomirs) [69].
Acknowledgments
This work was supported by unrestricted grants from Fondazione Ente Cassa di Risparmio di Firenze and Fondazione F.I.R.M.O. Raffaella Becagli to M. L. Brandi.
References
- 1.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120(1):15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 2.Calin GA, Croce CM. MicroRNA-cancer connection: the beginning of a new tale. Cancer Research. 2006;66(15):7390–7394. doi: 10.1158/0008-5472.CAN-06-0800. [DOI] [PubMed] [Google Scholar]
- 3.Michael MZ, O’Connor SM, Van Holst Pellekaan NG, Young GP, James RJ. Reduced accumulation of specific MicroRNAs in colorectal neoplasia. Molecular Cancer Research. 2003;1(12):882–891. [PubMed] [Google Scholar]
- 4.Calin GA, Liu CG, Sevignani C, et al. MicroRNA profiling reveals distinct signatures in B cell chronic lymphocytic leukemias. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(32):11755–11760. doi: 10.1073/pnas.0404432101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Calin GA, Ferracin M, Cimmino A, et al. A microRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. The New England Journal of Medicine. 2005;353(17):1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 6.Eis PS, Tam W, Sun L, et al. Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(10):3627–3632. doi: 10.1073/pnas.0500613102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takamizawa J, Konishi H, Yanagisawa K, et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Research. 2004;64(11):3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- 8.Iorio MV, Ferracin M, Liu CG, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Research. 2005;65(16):7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 9.Ciafrè SA, Galardi S, Mangiola A, et al. Extensive modulation of a set of microRNAs in primary glioblastoma. Biochemical and Biophysical Research Communications. 2005;334(4):1351–1358. doi: 10.1016/j.bbrc.2005.07.030. [DOI] [PubMed] [Google Scholar]
- 10.Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Research. 2005;65(14):6029–6033. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- 11.Calin GA, Sevignani C, Dumitru CD, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(9):2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435(7043):834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 13.Lee Y, Ahn C, Han J, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425(6956):415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 14.Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes and Development. 2003;17(24):3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hutvagner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293(5531):834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- 16.Tang G. siRNA and miRNA: an insight into RISCs. Trends in Biochemical Sciences. 2005;30(2):106–114. doi: 10.1016/j.tibs.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 17.Hutvagner G, Zamore PD. A microRNA in a multiple-turnover RNAi enzyme complex. Science. 2002;297(5589):2056–2060. doi: 10.1126/science.1073827. [DOI] [PubMed] [Google Scholar]
- 18.Zeng Y, Cullen BR. Sequence requirements for micro RNA processing and function in human cells. RNA. 2003;9(1):112–123. doi: 10.1261/rna.2780503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Llave C, Xie Z, Kasschau KD, Carrington JC. Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science. 2002;297(5589):2053–2056. doi: 10.1126/science.1076311. [DOI] [PubMed] [Google Scholar]
- 20.Yekta S, Shih IH, Bartel DP. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304(5670):594–596. doi: 10.1126/science.1097434. [DOI] [PubMed] [Google Scholar]
- 21.Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annual Review of Biochemistry. 2010;79:351–379. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- 22.Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318(5858):1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- 23.Ørom UA, Nielsen FC, Lund AH. MicroRNA-10a binds the 5′UTR of ribosomal protein mRNAs and enhances their translation. Molecular Cell. 2008;30(4):460–471. doi: 10.1016/j.molcel.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 24.Doench JG, Sharp PA. Specificity of microRNA target selection in translational repression. Genes and Development. 2004;18(5):504–511. doi: 10.1101/gad.1184404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nikiforova MN, Tseng GC, Steward D, Diorio D, Nikiforov YE. MicroRNA expression profiling of thyroid tumors: biological significance and diagnostic utility. Journal of Clinical Endocrinology and Metabolism. 2008;93(5):1600–1608. doi: 10.1210/jc.2007-2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He H, Jazdzewski K, Li W, et al. The role of microRNA genes in papillary thyroid carcinoma. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(52):19075–19080. doi: 10.1073/pnas.0509603102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pallante P, Visone R, Ferracin M, et al. MicroRNA deregulation in human thyroid papillary carcinomas. Endocrine-Related Cancer. 2006;13(2):497–508. doi: 10.1677/erc.1.01209. [DOI] [PubMed] [Google Scholar]
- 28.Tetzlaff MT, Liu A, Xu X, et al. Differential expression of miRNAs in papillary thyroid carcinoma compared to multinodular goiter using formalin fixed paraffin embedded tissues. Endocrine Pathology. 2007;18(3):163–173. doi: 10.1007/s12022-007-0023-7. [DOI] [PubMed] [Google Scholar]
- 29.Chen YT, Kitabayashi N, Zhou XK, Fahey TJ, III, Scognamiglio T. MicroRNA analysis as a potential diagnostic tool for papillary thyroid carcinoma. Modern Pathology. 2008;21(9):1139–1146. doi: 10.1038/modpathol.2008.105. [DOI] [PubMed] [Google Scholar]
- 30.Wilson C. Cancer: microRNA expression provides clues about the aggressiveness of papillary thyroid carcinoma. Nature Reviews Endocrinology. 2010;6(8):p. 416. doi: 10.1038/nrendo.2010.97. [DOI] [PubMed] [Google Scholar]
- 31.Sheu SY, Grabellus F, Schwertheim S, Handke S, Worm K, Schmid KW. Lack of correlation between BRAF V600E mutational status and the expression profile of a distinct set of miRNAs in papillary thyroid carcinoma. Hormone and Metabolic Research. 2009;41(6):482–487. doi: 10.1055/s-0029-1215558. [DOI] [PubMed] [Google Scholar]
- 32.Chou CK, Chen RF, Chou FF, et al. MiR-146b is highly expressed in adult papillary thyroid carcinomas with high risk features including extrathyroidal invasion and the BRAF mutation. Thyroid. 2010;20(5):489–494. doi: 10.1089/thy.2009.0027. [DOI] [PubMed] [Google Scholar]
- 33.Weber F, Teresi RE, Broelsch CE, Frilling A, Eng C. A limited set of human MicroRNA is deregulated in follicular thyroid carcinoma. Journal of Clinical Endocrinology and Metabolism. 2006;91(9):3584–3591. doi: 10.1210/jc.2006-0693. [DOI] [PubMed] [Google Scholar]
- 34.Visone R, Pallante P, Vecchione A, et al. Specific microRNAs are downregulated in human thyroid anaplastic carcinomas. Oncogene. 2007;26(54):7590–7595. doi: 10.1038/sj.onc.1210564. [DOI] [PubMed] [Google Scholar]
- 35.Braun J, Hoang-Vu C, Dralle H, Hüttelmaier S. Downregulation of microRNAs directs the EMT and invasive potential of anaplastic thyroid carcinomas. Oncogene. 2010;29(29):4237–4244. doi: 10.1038/onc.2010.169. [DOI] [PubMed] [Google Scholar]
- 36.Ricarte-Filho JCM, Fuziwara CS, Yamashita AS, Rezende E, Da-Silva MJ, Kimura ET. Effects of let-7 microRNA on cell growth and differentiation of papillary thyroid cancer. Translational Oncology. 2009;2(4):236–241. doi: 10.1593/tlo.09151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takakura S, Mitsutake N, Nakashima M, et al. Oncogenic role of miR-17-92 cluster in anaplastic thyroid cancer cells. Cancer Science. 2008;99(6):1147–1154. doi: 10.1111/j.1349-7006.2008.00800.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mitomo S, Maesawa C, Ogasawara S, et al. Downregulation of miR-138 is associated with overexpression of human telomerase reverse transcriptase protein in human anaplastic thyroid carcinoma cell lines. Cancer Science. 2008;99(2):280–286. doi: 10.1111/j.1349-7006.2007.00666.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nikiforova MN, Chiosea SI, Nikiforov YE. MicroRNA expression profiles in thyroid tumors. Endocrine Pathology. 2009;20(2):85–91. doi: 10.1007/s12022-009-9069-z. [DOI] [PubMed] [Google Scholar]
- 40.Cahill S, Smyth P, Finn SP, et al. Effect of ret/PTC 1 rearrangement on transcription and post-transcriptional regulation in a papillary thyroid carcinoma model. Molecular Cancer. 2006;5, article 70 doi: 10.1186/1476-4598-5-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ashman LK. The biology of stem cell factor and its receptor C-kit. International Journal of Biochemistry and Cell Biology. 1999;31(10):1037–1051. doi: 10.1016/s1357-2725(99)00076-x. [DOI] [PubMed] [Google Scholar]
- 42.Kitamura Y, Hirota S. Kit as a human oncogenic tyrosine kinase. Cellular and Molecular Life Sciences. 2004;61(23):2924–2931. doi: 10.1007/s00018-004-4273-y. [DOI] [PubMed] [Google Scholar]
- 43.Natali PG, Nicotra MR, Di Renzo MF, et al. Expression of the c-Met/HGF receptor in human melanocytic neoplasms: demonstration of the relationship to malignant melanoma tumour progression. British Journal of Cancer. 1993;68(4):746–750. doi: 10.1038/bjc.1993.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Natali PG, Nicotra MR, Winkler AB, Cavaliere R, Bigotti A, Ullrich A. Progression of human cutaneous melanoma is associated with loss of expression of c-kit proto-oncogene receptor. International Journal of Cancer. 1992;52(2):197–201. doi: 10.1002/ijc.2910520207. [DOI] [PubMed] [Google Scholar]
- 45.Matsuda R, Takahashi T, Nakamura S, et al. Expression of the c-kit protein in human solid tumors and in corresponding fetal and adult normal tissues. American Journal of Pathology. 1993;142(1):339–346. [PMC free article] [PubMed] [Google Scholar]
- 46.Natali PG, Berlingieri MT, Nicotra MR, et al. Transformation of thyroid epithelium is associated with loss of c-kit receptor. Cancer Research. 1995;55(8):1787–1791. [PubMed] [Google Scholar]
- 47.Chiang HR, Schoenfeld LW, Ruby JG, et al. Mammalian microRNAs: experimental evaluation of novel and previously annotated genes. Genes and Development. 2010;24(10):992–1009. doi: 10.1101/gad.1884710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Adeniran AJ, Zhu Z, Gandhi M, et al. Correlation between genetic alterations and microscopic features, clinical manifestations, and prognostic characteristics of thyroid papillary carcinomas. American Journal of Surgical Pathology. 2006;30(2):216–222. doi: 10.1097/01.pas.0000176432.73455.1b. [DOI] [PubMed] [Google Scholar]
- 49.Visone R, Russo L, Pallante P, et al. MicroRNAs (miR)-221 and miR-222, both overexpressed in human thyroid papillary carcinomas, regulate p27 protein levels and cell cycle. Endocrine-Related Cancer. 2007;14(3):791–798. doi: 10.1677/ERC-07-0129. [DOI] [PubMed] [Google Scholar]
- 50.Fusco A, Santoro M. 20 years of RET/PTC in thyroid cancer: clinico-pathological correlations. Arquivos Brasileiros de Endocrinologia e Metabologia. 2007;51(5):731–735. doi: 10.1590/s0004-27302007000500010. [DOI] [PubMed] [Google Scholar]
- 51.Argraves WS, Greene LM, Cooley MA, Gallagher WM. Fibulins: physiological and disease perspectives. EMBO Reports. 2003;4(12):1127–1131. doi: 10.1038/sj.embor.7400033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gallagher WM, Greene LM, Ryan MP, et al. Human fibulin-4: analysis of its biosynthetic processing and mRNA expression in normal and tumour tissues. FEBS Letters. 2001;489(1):59–66. doi: 10.1016/s0014-5793(00)02389-9. [DOI] [PubMed] [Google Scholar]
- 53.Schulte KM, Jonas C, Krebs R, Röher HD. Activin A and activin receptors in thyroid cancer. Thyroid. 2001;11(1):3–14. doi: 10.1089/10507250150500603. [DOI] [PubMed] [Google Scholar]
- 54.Sander S, Bullinger L, Klapproth K, et al. MYC stimulates EZH2 expression by repression of its negative regulator miR-26a. Blood. 2008;112(10):4202–4212. doi: 10.1182/blood-2008-03-147645. [DOI] [PubMed] [Google Scholar]
- 55.Iorio MV, Ferracin M, Liu CG, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Research. 2005;65(16):7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 56.Henson BJ, Bhattacharjee S, O’Dee DM, Feingold E, Gollin SM. Decreased expression of miR-125b and miR-100 in oral cancer cells contributes to malignancy. Genes Chromosomes and Cancer. 2009;48(7):569–582. doi: 10.1002/gcc.20666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ichimi T, Enokida H, Okuno Y, et al. Identification of novel microRNA targets based on microRNA signatures in bladder cancer. International Journal of Cancer. 2009;125(2):345–352. doi: 10.1002/ijc.24390. [DOI] [PubMed] [Google Scholar]
- 58.Shi L, Zhang J, Pan T, et al. MiR-125b is critical for the suppression of human U251 glioma stem cell proliferation. Brain Research. 2010;1312:120–126. doi: 10.1016/j.brainres.2009.11.056. [DOI] [PubMed] [Google Scholar]
- 59.Zhang X, Neganova I, Przyborski S, et al. A role for NANOG in G1 to S transition in human embryonic stem cells through direct binding of CDK6 and CDC25A. Journal of Cell Biology. 2009;184(1):67–82. doi: 10.1083/jcb.200801009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liang L, Wong C-M, Ying Q, et al. MicroRNA-125b suppressesed human liver cancer cell proliferation and metastasis by directly targeting oncogene LIN28B2. Hepatology. 2010;52(5):1731–1740. doi: 10.1002/hep.23904. [DOI] [PubMed] [Google Scholar]
- 61.Ito Y, Yoshida H, Tomoda C, et al. Telomerase activity in thyroid neoplasms evaluated by the expression of human telomerase reverse transcriptase (hTERT) Anticancer Research. 2005;25(1 B):509–514. [PubMed] [Google Scholar]
- 62.Gimm O, Perren A, Weng LP, et al. Differential nuclear and cytoplasmic expression of PTEN in normal thyroid tissue, and benign and malignant epithelial thyroid tumors. American Journal of Pathology. 2000;156(5):1693–1700. doi: 10.1016/s0002-9440(10)65040-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bruni P, Boccia A, Baldassarre G, et al. PTEN expression is reduced in a subset of sporadic thyroid carcinomas: evidence that PTEN-growth suppressing activity in thyroid cancer cells is mediated by p27(kip1) Oncogene. 2000;19(28):3146–3155. doi: 10.1038/sj.onc.1203633. [DOI] [PubMed] [Google Scholar]
- 64.Frisk T, Foukakis T, Dwight T, et al. Silencing of the PTEN tumor-suppressor gene in anaplastic thyroid cancer. Genes Chromosomes and Cancer. 2002;35(1):74–80. doi: 10.1002/gcc.10098. [DOI] [PubMed] [Google Scholar]
- 65.Park SM, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes and Development. 2008;22(7):894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435(7043):834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 67.Zen K, Zhang CY. Circulating microRNAs: a novel class of biomarkers to diagnose and monitor human cancers. doi: 10.1002/med.20215. Medicinal Research Reviews. In press. [DOI] [PubMed] [Google Scholar]
- 68.Dickins RA, Hemann MT, Zilfou JT, et al. Probing tumor phenotypes using stable and regulated synthetic microRNA precursors. Nature Genetics. 2005;37(11):1289–1295. doi: 10.1038/ng1651. [DOI] [PubMed] [Google Scholar]
- 69.Krützfeldt J, Rajewsky N, Braich R, et al. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438(7068):685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]