Abstract
Biomethanation is a mature technology for fuel production. Fourth generation biofuels research will focus on sequestering CO2 and providing carbon-neutral or carbon-negative strategies to cope with dwindling fossil fuel supplies and environmental impact. Formate is an important intermediate in the methanogenic breakdown of complex organic material and serves as an important precursor for biological fuels production in the form of methane, hydrogen, and potentially methanol. Formate is produced by either CoA-dependent cleavage of pyruvate or enzymatic reduction of CO2 in an NADH- or ferredoxin-dependent manner. Formate is consumed through oxidation to CO2 and H2 or can be further reduced via the Wood-Ljungdahl pathway for carbon fixation or industrially for the production of methanol. Here, we review the enzymes involved in the interconversion of formate and discuss potential applications for biofuels production.
1. Introduction
Methane has been recognized as an important fuel source since at least 1778 when Alessandro Volta first identified methane as the primary component of swamp gas.In 1884 Louis Pasteur proposed using methane produced from the anaerobic decay of horse dung to light the streets of Paris. Naturally occurring methane, otherwise known as natural gas, currently provides approximately 20–25% of the US energy demand. The United States' 2010 methane consumption was estimated to be approximately 2.43 × 1016 kJ and demand is anticipated to grow at an annualized rate of 0.4% annually through 2035 to 2.74 × 1016 kJ [1]. Current government support and investment underscores the important role of biomethanation. Several countries, including Japan, the United States, and Sweden, have taken leadership roles in designing the next generation of methane-fueled vehicles [2]. As of 2006, Germany had invested in nearly 3500 biogas production facilities that provided approximately 900 MW of electricity [3]. Denmark and Sweden have made similar investments into biogas production facilities and biomethanation will be one of many tools the Dutch dairy industry relies on to achieve energy self-sufficiency by 2020 [4].
Biomethanation is a complex biological process. It is well established that methanogenic ecosystems require multipart cooperation between at least three trophic guilds. This includes fermentative bacteria, acetogenic bacteria, and methanogens. In short, complex organic molecules are fermented to acetate, hydrogen, formate, and a variety of organic acids (lactate, propionate, and butyrate) and ethanol. Acetogenic bacteria convert these compounds to the methanogenic substrates hydrogen, formate, and acetate. In this cascade, interspecies electron transfer (IET) plays an important role. It results in a syntrophic relationship between acetogenic bacteria and methanogens.
This syntrophic relationship is the rate-limiting step for biological methane production [5] and is essential to proper functioning and maintenance of the overall thermodynamic viability of these systems. The role of interspecies hydrogen transfer is rather well studied, but the role of formate in the methanogenic cascade has received less attention. In anaerobic systems, formate is produced through either formate dehydrogenase (FDH) catalyzed reduction of carbon dioxide or through CoA-dependent cleavage of pyruvate to formate and acetyl-CoA. Conversely, archaeal FDH initially oxidizes formate to H2 and CO2. CO2 can then be reduced by methanogens to form methane. Here, we review the state of our knowledge regarding the production, transfer, and consumption of formate in methanogenic ecosystems by focusing on the properties of the enzymes involved.
2. Formate Production by Pyruvate-Formate Lyase
Pyruvate is the end product of the three major glycolytic pathways: Embden-Meyerhof-Parnas (EMP), Entner-Doudoroff (ED), and Pentose Phosphate Pathways (PPP). Aerobically grown organisms further metabolize pyruvate through NAD-dependent pyruvate dehydrogenase catalyzed oxidation to acetyl-coenzyme A (CoA) and carbon dioxide. In the absence of oxygen, pyruvate is oxidized through either a ferredoxin-dependent oxidoreductase, which catalyzes reduction of ferredoxin coupled to oxidation of pyruvate to acetate and carbon dioxide or through the action of pyruvate formate lyase (PFL). PFL catalyzes CoA-dependent cleavage of pyruvate to form acetyl-CoA without the associated production of NADH or reduced ferredoxin according to the following reaction [6]:
| (1) |
This reaction notably does not result in the production of NADH or reduced ferredoxin and allows for ATP synthesis from acetyl-CoA through the combined action of phosphotransacetylase and acetate kinase [8].
Escherichia coli PFL activity was first demonstrated in 1943 by Kalnitsky and Werkman [9] and was the first glycyl radical enzyme discovered [10, 11]. The forward reaction (1) is energetically favorable and is catalyzed through a “ping-pong” mechanism according to the following reaction scheme [6]:
| (2) |
| (3) |
Mutagenesis experiments showed that the PFL active site consists of three amino acid moieties; the glycyl radical Gly734, Cys418, and Cys419 [12]. Crystal structures of inactive PFL with and without substrate have been solved [13, 14]. These structures reveal that PFL is a α2 homodimer of approximately 170 kDa. The active site residues are contained on opposing hairpin loops with a distance of 4.8 Å between the alpha carbon of Gly734 and Cys419 though this orientation may differ in the activated form of the enzyme [13, 14]. PFL is activated by PFL-activating enzyme (PFL-AE), which mediates hydrogen abstraction from the Gly734 residue to form the glycyl radical [15]. The glycyl radical is thought to be relayed to Cys419 and, possibly, Cys418 forming a thiyl radical, which is responsible for the attack on pyruvate, forming acetylated PFL (2) with associated release of a formyl radical anion (·CO2−) [14].
Two mechanisms for formate formation have been postulated. In the conventional model, the formyl radical is quenched by hydrogen abstraction from Cys419, reforming the Cys419 thiyl radical. Abstraction of hydrogen from CoA facilitates acetyl transfer, releasing acetyl-CoA [14]. Guo and Himo (2004) [16] revisit this reaction mechanism proposing, instead, that the formyl radical is quenched by hydrogen abstraction directly from CoASH without intermediate involvement of a Cys418 thiyl radical [16].
PFL reaction creates a pool of formate that can have diverse fates depending on the environmental conditions in the bioreactor. Under oxygenic- and nitrate-reducing conditions, electrons from the oxidation of formate can be utilized to reduce either oxygen or nitrate. In the absence of nitrate or oxygen, formate can be oxidized directly to CO2 with the reduction of protons to form H2 (discussed later).
3. Interconversion of Formate and H2/CO2
Formate oxidation and CO2 reduction are interconvertible processes that are carried out by two main families of enzymes found in Eubacteria. The first group of enzymes is the iron-sulfur formate dehydrogenase (FDH) enzymes. These enzymes catalyze NAD-independent formate oxidation, have complex quaternary structure, and contain redox active molybdenum (Mo) or tungsten (W) prosthetic groups. The second class of enzymes are the NAD+-dependent FDH enzymes which catalyze the concomitant reduction of NAD+ to NADH and formate oxidation to CO2.
3.1. Structure and Function of Iron-Sulfur-Containing FDH
In enteric bacteria such as E. coli, the fermentative formate dehydrogenase FDH-H together with the hydrogenase Hyd-3 form the energy conserving formate-hydrogen lyase (FHL) complex that oxidizes formate produced via the pyruvate-formate lyase (PFL) system. In this system, FDH-H reduces the Hyd-3 enzyme with electrons extracted from the two-electron oxidation of formate. Reduced Hyd-3 then produces hydrogen gas through the reduction of two protons [17].
Formate dehydrogenase is a member of the dimethylsulfoxide (DMSO) reductase family of enzymes. The first crystal structure obtained was for FDH-H from Escherichia coli; this was found to be a single subunit enzyme with four distinct domains. There is great diversity in subunit composition for these enzymes but the overall topology for all the known crystal structures is very similar; four highly conserved domains are distributed across 1, 2, or 3 subunits. A redox active molybdenum (Mo) or tungsten (W) atom is coordinated in a square-pyramidal manner at the structure's center. Four sulfur atoms distributed on two molybdopterin guanine dinucleotide (MGD) prosthetic groups provide four base ligands. The selenium atom of a selenocysteine residue provides the apical ligand [18, 19].
The active site is approximately 25 Å from the enzyme surface. Formate enters the enzyme through a positively lined funnel-shaped entrance known as the formate cleft. This funnel leads directly to the active site, which is composed of single arginine, histidine, and selenocysteine residues. For the two electron oxidation of formate to carbon dioxide, formate presumably binds directly to Mo[VI] and displaces the SeCys residue. The free selenol is stabilized by an arginine residue. The molybdenum atom likely accepts two electrons from formate with concomitant production of H+ and CO2. A protonable histidine residue accepts the proton from formate. Raaijmakers and colleagues (2002) have identified a putative proton channel in FDH-H and the tungsten-containing formate dehydrogenase of Desulfovibrio gigas. This tunnel, which is oriented perpendicular to the formate cleft, is coated with protonatable glutamic and aspartic acid side chains. Also identified in this study was a hydrophobic channel that may allow the release of CO2 [18, 19].
In addition to the FHL-associated FDH-H enzyme, E. coli expresses two additional FDH enzymes with similar functions, but used under different growth conditions. FDH-N was the first FDH isolated and characterized from E. coli and is expressed when the organism is grown in the presence of nitrate. FDH-N is a heterotrimer composed of an ~110 kDa α-subunit, an ~32 kDa β subunit, and an ~20 kDa γ-subunit. The catalytic α-subunit catalyzes the periplasmic oxidation of formate. Electrons are transferred via the β-subunit and ultimately reduce the cytochrome b-containing γ-subunit; an integral membrane protein which reduces menaquinone to menaquinol. Oxidation of menaquinol by respiratory nitrate reductase Nar results in the translocation of two protons from the cell interior into the periplasm which helps contribute to maintaining proton motive force [17].
FDH-O is topologically similar to FDH-N. FDH-O is expressed when E. coli is grown aerobically and is slightly upregulated in the presence of nitrate. Expression studies led to the postulation that this constitutively expressed enzyme complex acts as a minor formate-to-nitrate respiratory chain ensuring rapid adaptation to nitrate-reducing conditions in the absence of physiologically viable amounts of FDH-N. However conclusive evidence for a physiological role of FDH-O remains enigmatic [20].
3.2. FDH-Catalyzed CO2 Reduction
Carbon dioxide reductase activity has been successfully demonstrated in vitro with FDH enzymes containing a W-MGD cofactor instead of the Mo-MGD cofactor. CO2 reduction is favored because W has a lower redox potential than Mo. It is very likely that CO2 reductase activity is restricted to W-MGD containing FDH enzymes, however it is not yet clear if the inverse relationship is also true: that formate dehydrogenase activity is restricted to Mo-MGD containing enzymes [21].
This activity is essential for carbon fixation by acetogens that reduce CO2 to acetate via the Wood-Ljungdahl pathway. Pyruvate generated during glycolysis is metabolized exclusively via the pyruvate:ferredoxin oxidoreductase system in acetogens. This results in the production of acetate, reduced ferredoxin, and CO2. The first step in acetate synthesis from CO2 involves FDH-catalyzed reduction of CO2 to formate with molecular hydrogen as the physiological electron donor [22–24]. There is high sequence similarity between the formate-producing and formate-consuming FDH enzymes. For example, the acetogen Acetobacterium woodii contains an FDH isoenzyme whose catalytic subunit has 80% identity [25] with the same subunit of E. coli. Given this similarity, it is likely that the catalytic mechanism of carbon dioxide reduction in these organisms is essentially the reverse of the mechanism previously discussed for E. coli FDH-H.
3.3. NAD+-Dependent FDH
The second family of formate oxidizing enzymes is the NAD+-dependent FDH enzymes which are devoid of any metallic prosthetic groups. These enzymes exist as α2 homodimers and catalyze reduction of NAD+ to NADH with concomitant oxidation of formate. This class of enzymes has been studied extensively due to their practical application as a regenerative agent for NADH used in chiral compound synthesis by the pharmaceutical industry. A crystal structure is available [26] and the catalytic mechanisms and structure have been reviewed extensively [27–29]. Recent reports have demonstrated reversibility of these enzymes with NADH-dependent CO2 reduction to form methanol [30–33] (discussed later).
4. Formate to Methane
4.1. Formate Utilization by Methanogens
Biological methane production from formate or hydrogen and carbon dioxide proceeds through a multistep pathway. This pathway has been reviewed at length [34–36] and we wish only to present a basic introduction to the methanogenic pathway. Readers are directed to Methanogenesis edited by Ferry for a more thorough understanding of the methanogens [36].
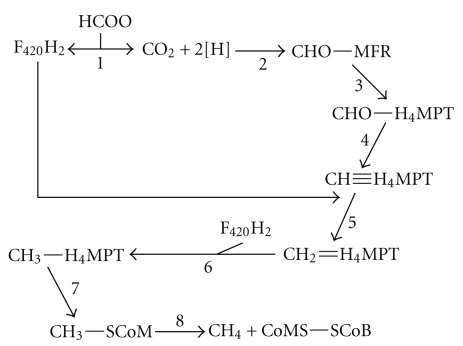
Carbon dioxide is used to form N-formylmethanofuran from CO2 and free methanofuran (MFR) (Figure 1, reaction 2). The formyl group is transferred from formylmethanofuran to tetrahydromethanopterin (H4MPT) by formyl-MFR: H4MPT formyl transferase to form N5-formyl-H4MPT (Figure 1, reaction 3). This undergoes a cyclization reaction catalyzed by N5, N10-methenyl-H4MPT cyclohydrolase to form N5, N10-methenyl-H4MPT (Figure 1, reaction 4). F420-reducing (NiFe) hydrogenase enzymes catalyze two sequential reductions of N5, N10-methenyl-H4MPT; first to N5, N10-methylene-H4MPT (Figure 1, reaction 5) and then to N5, N10-methyl-H4MPT (Figure 1, reaction 6). The methyl group is then transferred to coenzyme M (CoM) (Figure 1, reaction 7) which is reduced to form free methane and oxidized CoM (Figure 1, reaction 8) [36].
Figure 1.
Pathway of hydrogenotrophic methanogenesis from formate. Abbreviations used: HCOO−: formate; F420H2: reduced factor 420; MFR: methanofuran; H4MPT: tetrahydromethanopterin; CoMS: CoEnzyme S; SCoB: CoEnzyme B. For details regarding enzymes and reactions please see accompanying text.
Free formate is not utilized by methanogenic archaea for methanogenesis although there is evidence that formate is needed nutritionally as a formate auxotroph of Methanothermobacter autotrophicus was obtained [37]. Formate is first oxidized to CO2 by formate dehydrogenase with concomitant production of F420H2. F420H2 can then be oxidized by F420 (NiFe) hydrogenase to oxidized F420 and molecular hydrogen. Alternatively, F420H2 can serve as reductant in the sequential reduction of N5, N10-methenyl-H4MPT: first to N5, N10-methylene-H4MPT (Figure 1, reaction 5) and then to N5, N10-methyl-H4MPT (Figure 1, reaction 6). Free hydrogen is not a requirement for reduction of CO2 to methane in organisms growing on formate [36].
There is an important physiological distinction to be made among the hydrogenotrophic methanogens. The presence of cytochromes and the menaquinone analogue methanophenazine seems ubiquitous among members of the Methanosarcinales order, however these features appear restricted to only this group of organisms. Interestingly, cytochrome-containing methanogens demonstrate greater than a twofold higher yield when grown with H2 and CO2 when compared to noncytochrome-containing methanogens, and growth on formate is restricted to methanogens without cytochromes [38].
Costa and colleagues (2010) have proposed another role for formate dehydrogenase. Reduction of methyl-CoM to CoM-S-S-CoB (Figure 1, reaction 8) is the last step in biological methane formation. Cytochrome-containing organisms catalyze this reaction through the action of a membrane-associated, methanophenazine-reducing (NiFe) hydrogenase (Vho) and a methanophenazine-dependent heterodisulfide reductase (Hdr), which oxidizes molecular hydrogen coupled to concomitant reduction of methyl-CoM to form CoM-S-S-CoB and methane. Organisms without cytochromes lack this energy-conserving mechanism and rely instead, evidence suggests, on a cytosolic complex in which electrons flow from formate to a physically associated Hdr. Bifurcation of the electrons allows a coupling of the exergonic reduction of methyl-CoM to CoM-S-S-CoB to the endergonic reduction of ferredoxin. The reduced ferredoxin is then used to reduce CO2 to N-formylmethanofuran in step one of methanogenesis [39].
4.2. Formate as an Interspecies Electron Carrier in Methanogenesis
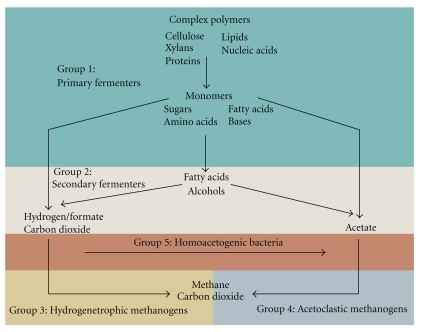
Methanogens are a physiologically specialized group of organisms that use a relatively narrow field of growth substrate: molecular hydrogen and carbon dioxide, formate, acetate, and methanol, and secondary alcohols like 2-propanol or 2-butanol. Primary fermenting organisms (Figure 2, group 1) hydrolyze complex polymers such as polysaccharides, nucleic acids, lipids, and protein to form simple oligomeric and polymeric substrates such as sugars, purines, pyrimidines, fatty acids, glycerol, and amino acids. These monomers are then further fermented to classical fermentation products by the primary fermenting bacteria (Figure 2, group 1): short chain fatty acids such as propionate and butyrate, alcohols such as ethanol, H2 and CO2, and acetate [40–43]. The methanogens rely on substrates which are produced from primary and secondary fermenting organisms (Figure 2, groups 1 & 2). Approximately 70% of the methane produced is through the dismutation of acetate (Table 1) in anaerobic bioreactors (Figure 2, group 3) by acetoclastic methanogens. The remaining portion is produced almost exclusively by hydrogenotrophic methanogens (Figure 2, group 4) [44].
Figure 2.
Carbon flux in methanogenic environments. Primary fermenting organisms (Group 1) degrade complex polymers to monomers, fatty acids, alcohols, hydrogen, carbon dioxide, acetate, and formate. Secondary fermenting organisms (Group 2) degrade fatty acids and alcohols to hydrogen, carbon dioxide, acetate, and formate. Hydrogenotrophic methanogens (Group 3) and acetoclastic methanogens (Group 4) convert carbon dioxide and formate or acetate to methane, respectively. For a more detailed discussion see accompanying text.
Table 1.
Free energy changes of some reactions involved in the degradation of fatty acids and alcohols to methane. Calculations based on Thauer et al. 1977 [45].
| Reaction | ΔG°′ (kJ/mol) |
|---|---|
| ethanol + H2O → acetate− + H+ + 2 H2 | + 9.6 |
| propionate + 2 H2O → acetate− + H+ + CO2 + 3 H2 | + 76 |
| butyrate + 2 H2O → acetate− + 2 H2 | + 48 |
| acetate− + 2 H2O → 2 CO2 + 4 H2 | + 96 |
| 4 H2 + CO2 → CH4 + 2 H2O | −131 |
| 4 formate + 4 H+ → CH4 + 3 CO2 + 2 H2O | −145 |
| CO2 + H2 → formate + H+ | −4.5 |
In well-balanced ecosystems, carbon flows almost exclusively from the monomeric subunits to the formation of H2, CO2, acetate, and formate. If the system becomes unbalanced through the accumulation of short-chain fatty acids, for example, acetate, propionate, and butyrate, the pH will decrease inhibiting methanogenesis. Secondary fermenting organisms are able to metabolize organic acids, but under standard conditions, these fermentation pathways are endergonic. This thermodynamic challenge is solved through syntrophic IET through hydrogenotrophic consumption of hydrogen and/or formate. The classic example of this phenomenon is through hydrogen exchange and was first elucidated with studies of Methanobacillus omelianskii; a coculture containing the S-organism, which fermented ethanol to acetate (Table 1), and the methanogen Methanobacterium bryantii, which consumed hydrogen with concomitant CO2 reduction (Table 1) [46].
IET has been documented with both formate and H2 transfer, and it is unclear whether one mechanism is more physiologically relevant than the other. Metabolic flux calculations [47] coupled with observations that the propionate-oxidizing bacterium Syntrophobacter fumaroxidans catalyzed propionate oxidation when grown together with H2/formate consuming methanogens such as Methanospirillum hungatei and Methanobacterium formicicum, but not when grown with a H2-only-utilizing methanogen Methanobrevibacter arboriphilus [47] have led to speculation that formate is the more important interspecies electron carrier. Experiments that showed that tungsten (W) and molybdenum (Mo) exclusion inhibited syntrophic growth support that formate is important for IET because either element is an essential cofactor for FDH-catalyzed CO2 reduction. However, these results do not conclusively demonstrate a metabolic preference for H2 or formate transfer because Mo/W is also essential for formylmethanofuran dehydrogenase (FMDH) catalyzed reduction of CO2 and formation of N5-formylmethanofuran [48], an indispensable step in methanogenesis [36]. Recent research indicates that multiple hydrogenases and formate dehydrogenases play a key role in syntrophy [49].
5. Perspectives
Biological methane production is a mature technology for providing renewable alternative fuels from readily available complex organic materials. However, for the fourth generation of (bio)fuels, emphasis is on efficient CO2 removal from the Earth's atmosphere. The resulting fuels and gases should not only be renewable, but also be carbon-negative. Carbon negative and carbon-neutral energy sources should also be used to drive the production of reduced carbon compounds to be used as fuels in a regenerative energy economy based on CO2. However, due to its kinetic and thermodynamic stability, strategies to activate and reduce CO2 are required.
Here, we have reviewed the role of formate production and conversion during methanogenic decay of organic material. It is important to recognize that the conversion of formate to hydrogen and CO2 is a reversible process with enzymes capable of both CO2 reduction as well as oxidation identified in the literature. With an oxidation potential close to that of hydrogen, formate is a primary energy source in its own right. Moreover, formate is the first stable intermediate in the biological conversion of CO2 to methanol according the following overall mechanism:
| (4) |
Cofactor-dependent conversion, however, is not economically viable. According to the above equation, one mole of methanol has a theoretical cofactor requirement of three moles of NADH. At current prices (~$500/g NADH) this results in a cost per mole greater than $1,000,000. A recent report demonstrated an efficiency of 127% when in situ biocatalytic cofactor regeneration and enzyme immobilization were employed in bench scale experiments. This would effectively lower the cost per mole methanol to $850,000.
However, as described above, W-MGD-containing iron-sulfur FDH enzymes catalyze CO2 reduction with reductants other than NADH and are physiologically active in homoacetogenic bacteria. In acetogens, hydrogen serves as the physiological electron donor and metabolic engineering pursuits may provide an industrially relevant way to reduce CO2 to formate via W-MGD-containing FDH catalysis. This would provide formate as a precursor molecule for biological methanol formation [30]. However, NADH dependency of downstream enzymes such as formaldehyde dehydrogenase and alcohol dehydrogenase would still pose a considerable economic challenge to cost-effective biological production of methanol.
Potentially of more importance would be developing ways of controlling metabolic flux and directing electrons towards increased formate production and, just as importantly, directing electrons from formate consumption directly into hydrogen or methane production and limiting electron flow down other branches of the metabolic network. Recently, Maeda and colleagues (2008) [50] engineered a strain of E. coli with quadruple mutations that produced 141 times more H2 from formate than the wild-type strain in bench-top experiments. First, hydrogen oxidation by Hyd-1 and Hyd-2 hydrogenase enzymes was inactivated by deleting the corresponding genes (hyaB and hybC, resp.). Next, the gene for the FHL repressor (hycA) was deleted and expression of the FHL inducer encoded by fhlA was increased. These alterations alone contributed to an 80-fold increase in hydrogen production rate compared to wild-type strains. When these mutations were combined with inactivation of the aerobic nitrate dependent FDH-O and FDH-N, a further 1.7-fold increase in hydrogen production rate to 113 (±12) μmol mg-protein−1 hr−1 was observed for a cumulative 141-fold increase in hydrogen production rate compared to the wild-type strain [50]. Thus, streamlining the metabolic flux by channeling electrons from formate directly to the FHL complex rather than down extraneous branches of the metabolic network enhances hydrogen production.
Acknowledgments
This work was supported by a Fulbright Fellowship to B. R. Crable provided by the J. William Fulbright Foundation and the Netherland-America Foundation, underwritten by Merck & Co.
Abbreviations
- ΔG:
Change in Gibb's free energy under conditions specified
- ΔG°′:
Change in Gibb's free energy under standard conditions at pH = 7.0
- CoA:
Coenzyme A
- CoM:
Coenzyme M
- CoB:
Coenzyme B
- DMSO:
Dimethyl sulfoxide
- ED:
Entner Doudoroff pathway of glycolysis
- EMP:
Embden-Meyerhof-Parnas pathway of glycolysis
- FDH:
Formate dehydrogenase
- FHL:
Formate-hydrogen lyase
- FMDH:
Formylmethanofuran dehydrogenase
- H4MPT:
Tetrahydromethanopterin
- IET:
Interspecies electron transfer
- MFR:
Methanofuran
- MGD:
Molybdopterin guanine dinucleotide
- Mo-MGD:
Molybdenum containing MGD
- NAD+:
Nicotinamide adenine dinucleotide
- NADH:
Reduced NAD+
- PFL:
Pyruvate formate lyase
- PFL-AE:
Pyruvate-formate lyase activating enzyme
- PPP:
Pentose phosphate pathway of glycolysis
- W-MGD:
Tungsten containing MGD.
References
- 1.USEIA. Annual Energy Outlook. Washington, DC, USA: Administration USEI; 2010. [Google Scholar]
- 2.Åhman M. Biomethane in the transport sector-an appraisal of the forgotten option. Energy Policy. 2010;38(1):208–217. [Google Scholar]
- 3.Deublein D, Steinhauser A. History and Status to Date in Europe. Biogas from Waste and Renewable Resources. An Introduction. Weinheim, Germany: John Wiley & Sons; 2008. [Google Scholar]
- 4.Gebrezgabher SA, Meuwissen MPM, Lansink AGJMO. Costs of producing biogas at dairy farms in the Netherlands. International Journal on Food System Dynamics. 2010;1(1) [Google Scholar]
- 5.Kim M, Ahn YH, Speece RE. Comparative process stability and efficiency of anaerobic digestion; mesophilic vs. thermophilic. Water Research. 2002;36(17):4369–4385. doi: 10.1016/s0043-1354(02)00147-1. [DOI] [PubMed] [Google Scholar]
- 6.Knappe J, Blaschkowski HP, Groebner P, Schmitt T. Pyruvate formate lyase of Escherichia coli: the acetyl enzyme intermediate. European Journal of Biochemistry. 1974;50(1):253–263. doi: 10.1111/j.1432-1033.1974.tb03894.x. [DOI] [PubMed] [Google Scholar]
- 7.Metzler DE, Metzler CM. Biochemistry: The Chemical Reactions of Living Cells. Burlington, Mass, USA: Harcourt; 2001. [Google Scholar]
- 8.White D. The Physiology and Biochemistry of Prokaryotes. New York, NY, USA: Oxford University Press; 2006. [Google Scholar]
- 9.Kalnitsky G, Werkman CH. The anaerobic dissimilation of pyruvate by a cell-free extract of Escherichia coli. Archives of Biochemistry. 1943;2:113–124. [Google Scholar]
- 10.Knappe J, Neugebauer FA, Blaschkowski HP, Ganzler M. Post-translational activation introduces a free radical into pyruvate formate-lyase. Proceedings of the National Academy of Sciences of the United States of America. 1984;81(3 I):1332–1335. doi: 10.1073/pnas.81.5.1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wagner AFV, Frey M, Neugebauer FA, Schafer W, Knappe J. The free radical in pyruvate formate-lyase is located on glycine-734. Proceedings of the National Academy of Sciences of the United States of America. 1992;89(3):996–1000. doi: 10.1073/pnas.89.3.996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Knappe J, Elbert S, Frey M, Wagner AFV. Pyruvate formate-lyase mechanism involving the protein-based glycyl radical. Biochemical Society Transactions. 1993;21(3, part 3):731–734. doi: 10.1042/bst0210731. [DOI] [PubMed] [Google Scholar]
- 13.Becker A, Fritz-Wolf K, Kabsch W, Knappe J, Schultz S, Volker Wagner AF. Structure and mechanism of the glycyl radical enzyme pyruvate formate- lyase. Nature Structural Biology. 1999;6(10):969–975. doi: 10.1038/13341. [DOI] [PubMed] [Google Scholar]
- 14.Becker A, Kabsch W. X-ray structure of pyruvate formate-lyase in complex with pyruvate and CoA. How the enzyme uses the Cys-418 thiyl radical for pyruvate cleavage. Journal of Biological Chemistry. 2002;277(42):40036–40042. doi: 10.1074/jbc.M205821200. [DOI] [PubMed] [Google Scholar]
- 15.Vey JL, Yang J, Li M, Broderick WE, Broderick JB, Drennan CL. Structural basis for glycyl radical formation by pyruvate formate-lyase activating enzyme. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(42):16137–16141. doi: 10.1073/pnas.0806640105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo JD, Himo F. Catalytic mechanism of pyruvate-formate lyase revisited. Journal of Physical Chemistry B. 2004;108(39):15347–15354. [Google Scholar]
- 17.Sawers G. The hydrogenases and formate dehydrogenases of Escherichia coli. Antonie van Leeuwenhoek. 1994;66(1–3):57–88. doi: 10.1007/BF00871633. [DOI] [PubMed] [Google Scholar]
- 18.Boyington JC, Gladyshev VN, Khangulov SV, Stadtman TC, Sun PD. Crystal structure of formate dehydrogenase H: catalysis involving Mo, molybdopterin, selenocysteine, and an FeS cluster. Science. 1997;275(5304):1305–1308. doi: 10.1126/science.275.5304.1305. [DOI] [PubMed] [Google Scholar]
- 19.Raaijmakers HCA, Romão MJ. Formate-reduced E. coli formate dehydrogenase H: the reinterpretation of the crystal structure suggests a new reaction mechanism. Journal of Biological Inorganic Chemistry. 2006;11(7):849–854. doi: 10.1007/s00775-006-0129-2. [DOI] [PubMed] [Google Scholar]
- 20.Abaibou H, Pommier J, Benoit S, Giordano G, Mandrand-Berthelot MA. Expression and characterization of the Escherichia coli fdo locus and a possible physiological role for aerobic formate dehydrogenase. Journal of Bacteriology. 1995;177(24):7141–7149. doi: 10.1128/jb.177.24.7141-7149.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reda T, Plugge CM, Abram NJ, Hirst J. Reversible interconversion of carbon dioxide and formate by an electroactive enzyme. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(31):10654–10658. doi: 10.1073/pnas.0801290105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Müller V. Energy conservation in acetogenic bacteria. Applied and Environmental Microbiology. 2003;69(11):6345–6353. doi: 10.1128/AEM.69.11.6345-6353.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ragsdale SW. Enzymology of the Wood-Ljungdahl pathway of acetogenesis. Annals of the New York Academy of Sciences. 2008;1125:129–136. doi: 10.1196/annals.1419.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ragsdale SW, Pierce E. Acetogenesis and the Wood-Ljungdahl pathway of CO2 fixation. Biochimica et Biophysica Acta. 2008;1784(12):1873–1898. doi: 10.1016/j.bbapap.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. Journal of Molecular Biology. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 26.Lamzin VS, Aleshin AE, Strokopytov BV, et al. Crystal structure of NAD-dependent formate dehydrogenase. European Journal of Biochemistry. 1992;206(2):441–452. doi: 10.1111/j.1432-1033.1992.tb16945.x. [DOI] [PubMed] [Google Scholar]
- 27.Popov VO, Lamzin VS. NAD-dependent formate dehydrogenase. Biochemical Journal. 1994;301(3):625–643. doi: 10.1042/bj3010625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shabalin IG, Filippova EV, Polyakov KM, et al. Structures of the apo and holo forms of formate dehydrogenase from the bacterium Moraxella sp. C-1: towards understanding the mechanism of the closure of the interdomain cleft. Acta Crystallographica Section D: Biological Crystallography. 2009;65(12):1315–1325. doi: 10.1107/S0907444909040773. [DOI] [PubMed] [Google Scholar]
- 29.Tishkov VI, Popov VO. Catalytic mechanism and application of formate dehydrogenase. Biochemistry. 2004;69(11):1252–1267. doi: 10.1007/s10541-005-0071-x. [DOI] [PubMed] [Google Scholar]
- 30.Baskaya FS, Zhao X, Flickinger MC, Wang P. Thermodynamic feasibility of enzymatic reduction of carbon dioxide to methanol. Applied Biochemistry and Biotechnology. 2010;162(2):391–398. doi: 10.1007/s12010-009-8758-x. [DOI] [PubMed] [Google Scholar]
- 31.El-Zahab B, Donnelly D, Wang P. Particle-tethered NADH for production of methanol from CO2 catalyzed by coimmobilized enzymes. Biotechnology and Bioengineering. 2008;99(3):508–514. doi: 10.1002/bit.21584. [DOI] [PubMed] [Google Scholar]
- 32.Obert R, Dave BC. Enzymatic conversion of carbon dioxide to methanol: enhanced methanol production in silica sol-gel matrices. Journal of the American Chemical Society. 1999;121(51):12192–12193. [Google Scholar]
- 33.Yoshimoto M, Yamashita T, Yamashiro T. Stability and reactivity of liposome-encapsulated formate dehydrogenase and cofactor system in carbon dioxide gas-liquid flow. Biotechnology Progress. 2010;26(4):1047–1053. doi: 10.1002/btpr.409. [DOI] [PubMed] [Google Scholar]
- 34.Deppenmeier U. The unique biochemistry of methanogenesis. Progress in Nucleic Acid Research and Molecular Biology. 2002;71:223–283. doi: 10.1016/s0079-6603(02)71045-3. [DOI] [PubMed] [Google Scholar]
- 35.Deppenmeier U, Müller V. Life close to the thermodynamic limit: how methanogenic archaea conserve energy. Results and Problems in Cell Differentiation. 2008;45:123–152. doi: 10.1007/400_2006_026. [DOI] [PubMed] [Google Scholar]
- 36.Thauer RK, Hedderich R, Fischer R. Reactions and enzymes involved in methanogenesis from CO2 and H2. In: Ferry JG, editor. Methanogenesis: Ecology, Physiology, Biochemistry & Genetics. New York, NY, USA: Chapman & Hall; 1993. pp. 209–295. [Google Scholar]
- 37.Tanner RS, McInerney MJ, Nagle DP. Formate auxotroph of Methanobacterium thermoautotrophicum Marburg. Journal of Bacteriology. 1989;171(12):6534–6538. doi: 10.1128/jb.171.12.6534-6538.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thauer RK, Kaster AK, Seedorf H, Buckel W, Hedderich R. Methanogenic archaea: ecologically relevant differences in energy conservation. Nature Reviews Microbiology. 2008;6(8):579–591. doi: 10.1038/nrmicro1931. [DOI] [PubMed] [Google Scholar]
- 39.Costa KC, Wong PM, Wang T, et al. Protein complexing in a methanogen suggests electron bifurcation and electron delivery from formate to heterodisulfide reductase. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(24):11050–11055. doi: 10.1073/pnas.1003653107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McInerney MJ, Sieber JR, Gunsalus RP. Syntrophy in anaerobic global carbon cycles. Current Opinion in Biotechnology. 2009;20(6):623–632. doi: 10.1016/j.copbio.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McInerney MJ, Struchtemeyer CG, Sieber J, et al. Physiology, ecology, phylogeny, and genomics of microorganisms capable of syntrophic metabolism. Annals of the New York Academy of Sciences. 2008;1125:58–72. doi: 10.1196/annals.1419.005. [DOI] [PubMed] [Google Scholar]
- 42.Schink B, Stams AJM. Syntrophism among prokaryotes. Prokaryotes. 2006;2:309–335. [Google Scholar]
- 43.Worm P, Müller N, Plugge C, Stams AJM, Schink B. Syntrophy in methanogenic degradation. In: Hackstein JHP, editor. (Endo)symbiotic Methanogenic Archaea, Microbiology Monographs. Vol. 19. Berlin, Germany: Springer; 2011. pp. 143–173. [Google Scholar]
- 44.Ferry JG. Methanogenesis: Ecology, Physiology, Biochemistry & Genetics. New York, NY, USA: Chapman & Hall; 1993. [Google Scholar]
- 45.Thauer RK, Jungermann K, Decker K. Energy conservation in chemotrophic anaerobic bacteria. Bacteriological Reviews. 1977;41(1):100–180. doi: 10.1128/br.41.1.100-180.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bryant MP, Wolin EA, Wolin MJ, Wolfe RS. Methanobacillus omelianskii, a symbiotic association of two species of bacteria. Archiv für Mikrobiologie. 1967;59(1–3):20–31. doi: 10.1007/BF00406313. [DOI] [PubMed] [Google Scholar]
- 47.Stams AJM, Dong X. Role of formate and hydrogen in the degradation of propionate and butyrate by defined suspended cocultures of acetogenic and methanogenic bacteria. Antonie van Leeuwenhoek. 1995;68(4):281–284. doi: 10.1007/BF00874137. [DOI] [PubMed] [Google Scholar]
- 48.Plugge CM, Jiang BO, De Bok FAM, Tsai C, Stams AJM. Effect of tungsten and molybdenum on growth of a syntrophic coculture of Syntrophobacter fumaroxidans and Methanospirillum hungatei. Archives of Microbiology. 2009;191(1):55–61. doi: 10.1007/s00203-008-0428-9. [DOI] [PubMed] [Google Scholar]
- 49.Worm P, Stams AJM, Cheng X, Plugge CM. Growth- and substrate-dependent transcription of formate dehydrogenase and hydrogenase coding genes in Syntrophobacter fumaroxidans and Methanospirillum hungatei. Microbiology. 2011;157(1):280–289. doi: 10.1099/mic.0.043927-0. [DOI] [PubMed] [Google Scholar]
- 50.Maeda T, Sanchez-Torres V, Wood TK. Metabolic engineering to enhance bacterial hydrogen production. Microbial Biotechnology. 2008;1(1):30–39. doi: 10.1111/j.1751-7915.2007.00003.x. [DOI] [PMC free article] [PubMed] [Google Scholar]