Abstract
Polyamines are essential for Trypanosoma cruzi, the causative agent of Chagas disease. As T. cruzi behaves as a natural auxotrophic organism, it relies on host polyamines biosynthesis. In this paper we obtained a double-transfected T. cruzi parasite that expresses the green fluorescent protein (GFP) and a heterologous ornithine decarboxylase (ODC), used itself as a novel selectable marker. These autotrophic and fluorescent parasites were characterized; the ODC presented an apparent Km for ornithine of 0.51 ± 0.16 mM and an estimated Vmax value of 476.2 nmoles/h/mg of protein. These expressing ODC parasites showed higher metacyclogenesis capacity than the auxotrophic counterpart, supporting the idea that polyamines are engaged in this process. This double-transfected T. cruzi parasite results in a powerful tool—easy to follow by its fluorescence—to study the role of polyamines in Chagas disease pathology and in related processes such as parasite survival, invasion, proliferation, metacyclogenesis, and tissue spreading.
1. Introduction
Protozoan parasites cause disease, death, and severe social and economic losses, particularly in developing countries. Trypanosoma cruzi is the causative agent of Chagas disease, which affects 16–18 million people mainly in Latin America [1, 2].
T. cruzi is a digenetic parasite whose life cycle involves an insect vector (from Reduviidae family) and a vertebrate host. The parasite undergoes deep morphological, biochemical, and physiological changes as an adaptive response to the extreme environmental variations that it faces around the life cycle. The epimastigote form (E) is a proliferative and noninfective extracellular stage, present in the insect gut. At the insect rectum, it differentiates to metacyclic trypomastigote (MT), an infective stage present in the contaminated insect faeces, which enter the vertebrate host through the insect wound or via mucosal membranes. Once inside, the trypomastigotes invade different kinds of cells, including macrophages, fibroblasts, and muscle cells [3]. There, they transform to amastigote stage and replicate to reach a critical number of parasites per cell, then they differentiate to blood stream trypomastigotes (BT), host cells burst, and release trypomastigotes, which either infect other cells or get sucked up by a bug vector [4–6]. The recent construction of stable green fluorescent protein (GFP) expressing T. cruzi strains has resulted in a useful tool to follow processes as cell invasion and infection progression [7, 8].
Polyamines are ubiquitous cell compounds essential for macromolecular biosynthesis [9, 10] and normal cell survival, proliferation and differentiation [11–13]. Several studies related to polyamine metabolism in pathogenic protozoa have been carried out with the aim to affect the proliferation, infectivity, or differentiation of these organisms and find new therapeutic targets against parasitic diseases [1, 14, 15]. With this kind of approach it was discovered that the acute infections of Trypanosoma brucei brucei in mice as well as the human African sleeping sickness, caused by T. brucei gambiense, can be treated by α-difluoromethylornithine (DFMO), the specific and irreversible ornithine decarboxylase (ODC) inhibitor [16–18].
The naturally occurring polyamines in most cell types, including mammalian cells, are putrescine (actually a diamine precursor), spermidine and spermine, while in trypanosomes this last polyamine was only found in trace amounts [11, 19, 20]. There are some differences related to polyamine biosynthetic enzymes between mammalian cells and parasites; however, polyamines are essential for both kinds of organisms. In fact, only two kinds of wild-type cells, Halobacteriales and Methanobacteriales, have been described with the ability to grow in the absence of a detectable polyamine concentration [21, 22].
The main route of putrescine biosynthesis is through the ornithine decarboxylation using the enzyme ornithine decarboxylase (ODC; EC 4.1.1.17), key enzyme capable of regulating cell growth and differentiation [22–24]. Many plants, bacteria and some specific mammalian tissues possess an alternative pathway by arginine decarboxylase (ADC; EC 4.1.1.19) where arginine is converted to putrescine via the intermediate agmatine [25–29].
The next step in polyamine biosynthesis is catalyzed by spermidine synthase (SpdS; EC 2.5.1.16) that transfers an aminopropyl group from the donor decarboxylated S-adenosylmethionine (dcAdoMet) to putrescine, producing spermidine. Spermine synthase (SpmS; EC 2.5.1.22) transfers a second aminopropyl group from dcAdoMet producing spermine. This enzyme is generally absent in the protozoan parasites; however, T. cruzi would be an exception since its genome presents two aminopropyltransferase genes, one of them a putative SpmS. This function has been proposed in silico but not functionally validated yet [1, 30].
The biosynthesis of polyamines is irreversible, but there is a route for polyamine back-conversion involving two steps: a spermidine/spermine N-acetyltransferase (EC 2.3.1.57) and polyamine oxidase (PAO; EC 1.5.3.13) [31, 32].
In trypanosomatids, spermidine plays a central role because it is the precursor of the derivative N1, N8-bis-glutathionyl-spermidine (trypanothione). This compound, unique in these protozoan organisms, is essential to maintain the redox equilibrium in the parasite cell and plays the metabolic and antioxidant functions of glutathione [33–35]. For this and other reasons, it is not surprising that polyamines are essential for survival and proliferation of T. cruzi [36–40]. However, T. cruzi is one of the extremely few eukaryotic organisms auxotrophic for polyamines. We have previously demonstrated that T. cruzi epimastigotes are unable to synthesize putrescine de novo because they contain neither ODC nor ADC enzymatic activity, due to the absence of ODC and ADC genes [38, 41, 42]. This conclusion is also supported by data from the T. cruzi genome project [30]. Polyamines seem to be crucial for the other T. cruzi stages, particularly in trypomastigote internalization process in, at least, some cellular types, and infection progression as well as for other related parasites infection [6, 43–46]. Since parasites live inside other organisms, numerous essential metabolites can be obtained directly from the host without the need to synthesize them [47]. It seems to be the case for polyamines, where its uptake systems from the parasite together with the ODC enzymatic activity from the host are a concerted and critical system for T. cruzi survival and host infection.
In this work we obtained a double-transfected T. cruzi strain that expresses the green fluorescent protein (GFP) and a heterologous ODC enzyme. The latter is encoded by the ODC gene, an auxotrophy rescuing gene used here as a selectable marker. Then we selected a Y-GFP-ODC clone with high expression of both GFP [7] and ODC activity [38]. These autotrophic and fluorescent parasites were characterized as well as the heterologous ODC enzyme activity. As T. cruzi is a naturally auxotrophic organism for polyamines, the autotrophy acquisition could provide information about the role of polyamines in processes such as parasite survival, invasion, proliferation, metacyclogenesis and tissue spreading, under different conditions of polyamines availability natural or induced with polyamine inhibitors; the green fluorescent protein allows to easily follow those processes microscopically. Therefore, the double-transfected T. cruzi parasite is a powerful new tool to study the role of polyamines, naturally provided by the host ODC, in the parasite cell cycle. This would help to better understand Chagas disease pathology and to find targets for new therapies.
2. Materials and Methods
2.1. Chemicals
Brain heart infusion, tryptose, and yeast extract were obtained from Difco Laboratories (Detroit, MI, USA). Minimal essential medium (SMEM), heat-inactivated fetal calf serum (FCS), and amino acids were from GIBCO BRL (Gaithersburg, MD, USA); vitamins, bases, haemin, polyamines, the ODC cofactor pyridoxal 5′-phosphate (PLP), protease inhibitors, HEPES buffer, antibiotics, cycloheximide and Rhodamine-conjugated phalloidin were purchased from Sigma (St. Louis, MO, USA). L-[1-14C]ornithine (57.1 Ci/mol) was from NEN Life Science Product, Inc (Boston, MA). The inhibitor α-difluoromethylornithine (DFMO) was a gift from Merrell Dow Research Institute (Cincinnati, OH).
2.2. T. cruzi Epimastigote Cultures
Epimastigote culture of Y strain previously transfected with the green fluorescent protein (GFP) gene (kindly provided by Dr S. Schenkman, Departamento de Microbiologia, Imunologia e Parasitologia, Universidade Federal de São Paulo (São Paulo, SP, Brazil)) (Y-GFP) was maintained at 28°C in rich media BHT, while the derived culture, Y-GFP retransfected with ornithine decarboxylase (ODC) gene, Y-GFP-ODC population (Y-GFP-ODC strain), and the selected clone (Y-GFP-ODC clone) were cultured at 28°C in a semisynthetic medium (SDM79) which contains only traces of polyamines [38]. Haemin (20 mg/L), 10% heat-inactivated fetal calf serum and the antibiotics streptomycin (100 μg/mL), penicillin (100 U/mL) and the selective drug G418 (250 μg/mL) were added to all cultures.
To determine the growth kinetics of each strain, a part of Y-GFP culture was maintained in BHT while others were placed in SDM79 medium in absence or presence of 1 mM putrescine or in Diamond in absence or presence of 2 mg/mL glucose. Y-GFP-ODC strain and the derived clone were maintained in SDM79 medium without or with the addition of 5 mM α-difluoromethylornithine (DFMO). Parasite growth was followed by cell counting. Cultures were diluted to 10–20 × 106 cells/mL every 48–72 h, when the stationary phase was reached.
2.3. Parasite Transfection
Epimastigote forms of Y strain had been previously transfected with pROCKGFPNeo and characterized [7, 48]. This recombinant plasmid contains the GFP coding region and sequences that allow a stable DNA integration at the β-tubulin loci [7]. A selected clone from this Y-GFP strain was retransfected with the recombinant vector pODC7, as previously described [38]. pODC7 contains the complete open reading frame (ORF) of ODC gene from C. fasciculata [49] cloned into the expression vector pRIBOTEX [50] that integrates at the ribosomal genes loci [38, 51].
After transfection, parasite culture was diluted with rich medium and incubated for 72 h at 28°C to allow its recovery before adding the restrictive culture conditions, consisting in 500 μg/mL Geneticin (G 418) (selection by antibiotic resistance) and semisynthetic medium SDM79 (selection by polyamine autotrophy development). Subsequent growth was followed by cell counting. Samples of T. cruzi cultures before and after transfection were taken to analyze cellular morphology, fluorescence, and ODC activity. Drug selection was completed 50 days after electroporation as indicated by the mock-transfected parasites (transfected with empty p-RIBOTEX) and the appearance of ODC activity. This time was slightly longer than that seen in each of the previous single-transfection cases (the T. cruzi transfection with GFP [7] and with ODC [38], in both cases selected by resistance to 250 μg/mL G418).
2.4. Selection of T. cruzi Y-GFP-ODC Clones
After selection of the double-recombinant Y-GFP-ODC T. cruzi strain, cloned subpopulations were obtained by diluting part of the culture to a final density of 0.1 cells/0.2 mL of SDM79 medium supplemented with 10% FCS and 250 μg/mL of G418 and plated in 96-well plates. They were incubated at 28°C in humid chamber for 4-5 weeks and analyzed with a fluorescence or confocal microscope. GFP-expressing clones were selected and transferred into 1 mL of the same medium in culture bottles and incubated at 28°C. Once they reached the plateau the cultures were expanded to 5 mL and ODC activity was assayed (see Section 3).
2.5. Preparation of Cell Extracts and ODC Enzymatic Assays
Cell extracts were obtained and the enzymatic activity was measured by the release of 14CO2 from L-[1-14C]ornithine as previously described [52]. Briefly, parasites were collected at the exponential phase of growth. They were washed and lysated by two cycles of freeze-thawing and sonication, in buffer A (50 mM Hepes, pH 7.4, 1 mM DTT, 0.5 mM EDTA and 0.1 mM pyridoxal 5′-phosphate) supplemented with protease inhibitors and 0.5% Nonidet P40. Cell lysates were centrifuged for 10 min at 10000 ×g. Supernatant fractions were tested for ODC activity by incubation at 37°C in presence of 2 mM L-[1-14C] ornithine (5 uCi/mL). The enzymatic reaction was performed for the indicated times and stopped by the addition of 0.5 N perchloric acid. The radioactive CO2 released during the reaction was trapped on a piece of Whatman 3 MM paper soaked with 2 M KOH and then measured in a scintillation counter. Protein concentration was measured, using BSA as standard, according to Bradford [53].
2.6. Kinetic Parameters of Heterologous ODC
Ornithine decarboxylation was carried out at different times, substrate and inhibitor concentrations. The determination of the kinetic constants apparent Km and Vmax values for ornithine of the enzyme expressed in Y-GFP-ODC were calculated from Lineweaver-Burk plots. Studies of inactivation by DFMO were carried out as previously described [38, 54].
2.7. Trypanosoma cruzi Metacyclogenesis
For in vitro differentiation of T. cruzi epimastigotes, we have used a method previously described [55, 56] with some modifications. Briefly, 25 × 106 epimastigotes from 15-day-old cultures from Y-GFP and Y-GFP-ODC strain and clone grown in Diamond or SDM79 media, respectively, were harvested by centrifugation at 600 ×g for 15 min at 15°C. The parasites were then incubated for 2 h at 37°C in 50 μL of triatomine artificial urine (TAU) medium (190 mM NaCl, 17 mM KCl, 2 mM MgCl2, 2 mM CaCl2, 8 mM phosphate buffer, pH 6.0). After that, parasites were diluted (1 : 100 dilution) in TAU3AAG medium (TAU supplemented with 10 mM L-proline, 50 mM L-sodium glutamate, 2 mM L-sodium aspartate, and 10 mM D-glucose) and maintained for 24 or 48 h in culture flasks at 28°C [55, 56]. Epimastigotes treated with α-difluoromethylornithine (DFMO) or Putrescine (Put) during metacyclogenesis were obtained by addition of 5 mM DFMO or 0,2 mM Put in both TAU and TAUAAG media at the same conditions as controls.
Mixed parasitic forms corresponding to epimastigotes and metacyclic trypomastigotes (E-MT suspensions) were harvested by centrifugation at 600 ×g for 15 min and used to infect Vero cells monolayers using 24-well plates containing glass coverslips in D-MEM supplemented with 3% FBS and antibiotics (infection medium). 24 h after infection, cells were washed, fixed, and stained with Rhodamine-phalloidin for the fluorescence microscopy analysis (see below). The metacyclogenesis capacity of these suspensions was estimated by counting the percentage of infected cells. Parasites were directly localized as green structures inside cells, whereas host cell extension was visualized in red, as indicate in the following.
2.8. Fluorescence Microscopy
For Rhodamine-phalloidin staining, Vero cells were washed three times with PBS to remove free extracellular parasites, fixed with 3% paraformaldehyde solution in PBS for 10 min at RT, washed with PBS, and blocked with 50 mM NH4Cl in PBS. Subsequently, cells were permeabilized with 0.05% saponin in PBS containing 0.5% BSA and then incubated with Rhodamine-conjugated phalloidin for 1 h at 37°C to label F-actin. Cells were washed three times with PBS and mounted with Mowiol before fluorescence microscopy analysis. Confocal images were obtained with an Olympus FV1000 confocal microscope and the FV 10-ASW 1.7 program (Olympus, Japan). Images were processed using Adobe 7.0 software (Adobe Systems).
3. Results
3.1. Growth Kinetics of Y-GFP and Y-GFP-ODC Strain and Clone
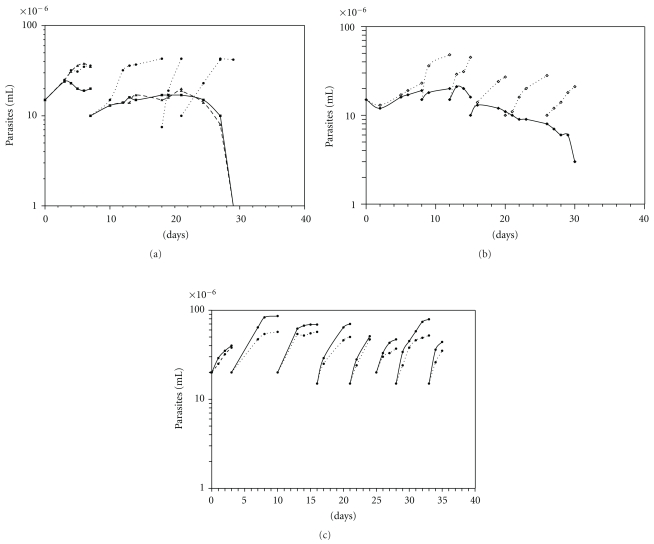
Y-GFP strain cultured in the rich medium BHT, supplemented with G418, maintained the proliferative capacity for long periods of time having a doubling time of 2-3 days and a constant maximal cell density of ~40 × 106 parasites/mL (Figure 1(a)). On the other hand, Y-GFP cultured in Diamond, the rich medium habitually used for maintaining this strain [7, 8], grew slowly with a doubling time of approximately 7 days. The maximal cell density (~37 × 106 parasites/mL) was scarcely reached at the first-second passage (Figure 1(a)). To discard that the low proliferation rate was due to low availability of glucose, an important source of energy for T. cruzi [57], we followed the Y-GFP growth in Diamond supplemented with 2 mg/mL glucose. There were no differences in the kinetic characteristics between both conditions. When Y-GFP strain was cultured in the semisynthetic medium SDM79 (containing only traces of polyamines), the kinetics of growth (Figure 1(b)) was similar to that observed in Diamond. After 10–15 days of culture under these three conditions, Y-GFP parasites progressively stopped growth, started decreasing, and finally were lysed. However, when Y-GFP was cultured in SDM79 supplemented with 1 mM putrescine the culture could recover the proliferation rate (Figure 1(b)).
Figure 1.
Growth kinetics of Y-GFP and Y-GFP-ODC clone cultured at different conditions. Cultures carried out under the indicated conditions were diluted to 10–20 × 106 cells/mL when the stationary phase was reached. (a) Y-GFP cultured in rich media BHT (- -⬤- -), Diamond (—■—), and in Diamond supplemented with 2 mg/mL glucose (- -△- -). (b) Y-GFP cultured in the semisynthetic medium SDM79 without (—◆ —) or with (- -◊- -) 1 mM putrescine. (c) Y-GFP-ODC clone cultured in SDM79 (—⬤—) or in SDM79 supplemented with 5 mM α-difluoromethylornithine (DFMO) (- -⬤- -). All cultures were continuously maintained in presence of 250 μg/mL G418.
The double-transfected Y-GFP-ODC clone grew continuously in SDM79, even better than Y-GFP in BHT, with a doubling time of 2-3 days and a maximal cell density of ~80 × 106 parasites/mL (Figure 1(c)). Similar results were obtained for the Y-GFP-ODC strain (not shown). The continuous proliferation of the double-transfected Y-GFP-ODC strain and clone in absence of polyamines indicates that the original auxotrophy for diamines of T. cruzi was reversed after transfection with the heterologous ODC gene, as it was previously described for monotransfected strains [38].
α-difluoromethylornithine (DFMO), the specific and irreversible inhibitor of ODC, altered the growth rate of Y-GFP-ODC clone, particularly the maximal cell density (lower than 55 × 106 parasites/mL) (Figure 1(c)). The partial effect of DFMO on the proliferative rate could be explained in different ways [58, 59] that will be discussed later.
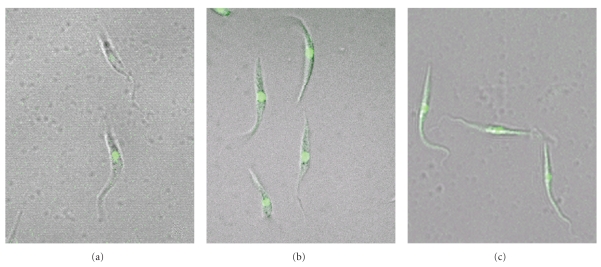
The three cultures were analyzed by confocal microscopy. There were ∼80% of parasites expressing GFP in Y-GFP strain, while the Y-GFP-ODC double-transfected strain and the selected clone presented values of ∼95%, similar to those described by Pires et al. [7]. The cell morphology of the three cultures was similar (Figure 2).
Figure 2.
Fluorescence microscopy of Y-GFP (a) and Y-GFP-ODC strain (b) and the selected Y-GFP-ODC clone epimastigotes (c) (1000x).
3.2. ODC Enzymatic Activity in Double-Transfected T. cruzi Epimastigotes
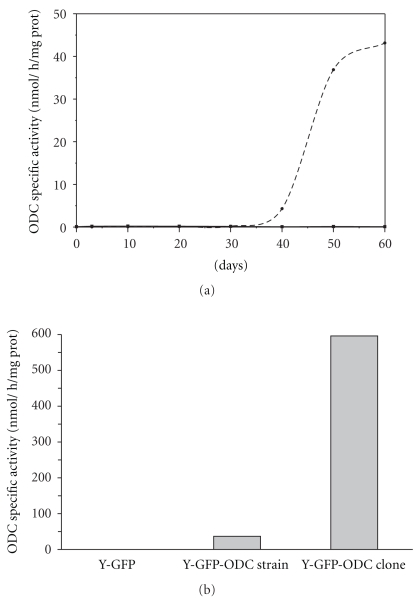
The ODC activity was measured at 72 h and 10, 20, 30, 40, and 50 days after transfection of Y-GFP T. cruzi epimastigotes with the heterologous ODC. We have found that double-transfected Y-GFP-ODC cells expressed high levels of ODC activity after 50 days following transfection (Figure 3(a)). It was different from previous reports, where a transient expression (between 48 h and 7 days) was detectable in ODC single-transfected parasites; that expression decreased markedly during the following weeks and recovered at 30–40 days after transfection [38]. This difference could be related to the cell machinery readaptation to transcribe-translate a new foreign and active gene, doing this already with GFP and the neomycin phosphotransferase (neomycin resistance).
Figure 3.
Ornithine decarboxylase (ODC) specific activity in Y-GFP and Y-GFP-ODC cell extracts. (a) ODC specific activity was measured in cell extracts obtained at different times after transfection; (—■—) Y-GFP strain; (- -⬤- -) Y-GFP-ODC strain. (b) ODC activity in extracts of exponentially growing cultures at 60 days after transfection and clone selection. All values are the average of at least three experiments.
The ODC specific activity in the double-transfected Y-GFP-ODC was around 39 ± 4 nmol/h/mg prot. Once the Y-GFP-ODC clone was selected, it showed an ODC activity with little variation during the course of the experiment reaching values of 590 nmol/h/mg prot (Figure 3(b)). The ODC activity in the Y-GFP strain was, as expected, negligible in all assays.
3.3. Properties of the Heterologous ODC Expressed in the Selected Y-GFP-ODC Clone
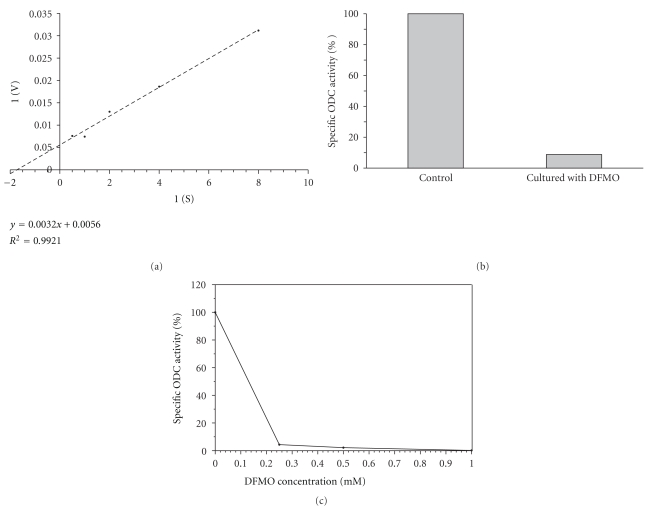
The kinetic parameters of ODC expressed in the Y-GFP-ODC clone were calculated from a Lineweaver-Burk plot (Figure 4(a)). The estimated Vmax value was 476.2 nmoles/h/mg of protein, and the apparent Km for ornithine was 0.51 ± 0.16 mM. While the Km value was similar to those obtained previously for other transfected T. cruzi strains [38] and for C. fasciculata [60], the Vmax was higher than those obtained in previous assays of single-transfection with ODC gene in DFMO-sensitive strains. Instead, the Vmax value obtained in Y-GFP-ODC clone was similar to that observed for ODC-transfected strains after reaching DFMO resistance [38, 58, 59].
Figure 4.
ODC activity characterization. (a) Plot of Lineweaver-Burk. ODC activity was assayed in vitro at different concentrations of the substrate ornithine to estimate apparent Km and Vmax. (b) The Y-GFP-ODC clone was cultured in absence or presence of 5 mM α-difluoromethylornithine (DFMO) for 24 hs, then cells were harvested, washed and ODC activity was measured. (c) ODC activity was carried out in the presence of DFMO at different concentrations. Values in B and C are given as percentages of maximal activity. All other details as indicated in Materials and Methods. These values are the average of two independent experiments.
The assays of in vivo ODC turnover (determined by stopping protein synthesis with 50 μg/mL cycloheximide and measuring the remaining enzymatic activity at different times) showed that the ODC expressed in Y-GFP-ODC clone was very stable, with a half-life longer than 6 h (data not shown), remarkably longer than the half-life of the ODC expressed in Crithidia fasciculata, its natural cellular context [60]. This notorious change in stability has already been observed in experiments of single transfection in other strains of T. cruzi [38].
When Y-GFP-ODC clone was cultured in presence of DFMO, the inhibitor was effectively internalized into the cell causing a notable ODC activity inhibition (Figure 4(b)). Additional in vitro assays showed that DFMO inhibits the ODC reaction in a dose-response way, with an IC50 lower than 0.13 mM (Figure 4(c)).
Based on these observations, the best explanation of why DFMO caused only a moderated effect on Y-GFP-ODC clone proliferation (Figure 1(c)) seems to be the high Vmax of ODC activity that correlates with a high number of active ODC molecules, making Y-GFP-ODC clone resistant to DFMO.
3.4. T. cruzi Metacyclogenesis in Y-GFP and Y-GFP-ODC Parasites
We next compared the capacity of Y-GFP-ODC (strain, and clone) and the auxotrophic Y-GFP to differentiate from epimastigotes (E) to metacyclic trypomastigotes (MT) forms using an in vitro protocol previously described [55, 56]. Y-GFP, Y-GFP-ODC strain and Y-GFP-ODC clone were incubated with TAU medium for 2 h and subsequently with TAU3AAG medium for 48 hrs to induce T. cruzi differentiation (TAU treatment). The MT generated for each cell culture was estimated using the E-MT suspensions to infect Vero cell monolayers during 24 h and comparing with their controls, in Diamond or SDM79 media for Y-GFP or Y-GFP-ODC, respectively.
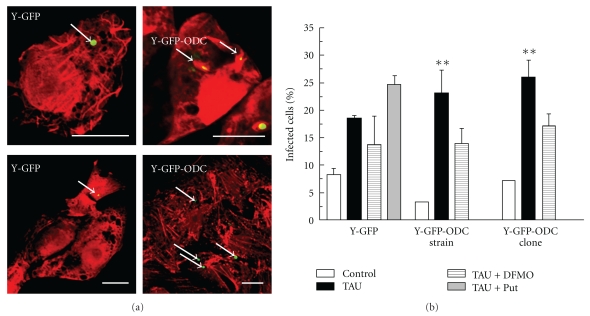
After the infection time, the cells were washed, fixed and incubated with Rhodamine-phalloidin to localize actin cytoskeleton of host cell by fluorescence microscopy, process that allows the clear observation of cellular limits, whereas T. cruzi parasites were observed directly by the presence of GFP protein. Figure 5(a) shows, at two differential amplification levels, confocal images of Vero cells infected by MT generated by epimastigotes of Y-GFP or Y-GFP-ODC strain under TAU treatment. Quantification studies indicated that the percentage of infected cells increased when comparing parasites obtained from Y-GFP-ODC metacyclogenesis with the Y-GFP control (Figure 5(b), black bars); even more, in some cases it was possible to observe more than one Y-GFP-ODC parasite per cell. Although the magnitude of this increment was not as high as to be statistically significant, the tendency was constant. Supporting this result, we observed that the presence of DFMO in the metacyclogenesis assay (see details in Materials and Methods) impaired the infection of Y-GFP-ODC strain and clone, while it has no evident effect over Y-GFP (Figure 5(b), dashed bars) because the inhibitor target is not present in this strain. Conversely, the percentage of infected cells was increased when Put was added to Y-GFP parasites during the differentiation process (Figure 5(b), grey bars) showing similar values to those found in Y-GFP-ODC parasites. Those results indicate that polyamines supply favours the E to MT conversion.
Figure 5.
Metacyclogenesis of Y-GFP and Y-GFP-ODC strain and clone. After metacyclogenesis protocol (see details in Materials and Methods), the total mixed suspensions of E and MT from each condition (Control, TAU, or TAU + DFMO) were used to infect Vero cells for 24 h followed by detection of the host cell actin cytoskeleton as indicated under Materials and Methods. (a) Confocal images showing Vero cells infected for 24 h with T. cruzi Y-GFP and Y-GFP-ODC clone obtained from TAU medium. (b) Percentage of infected Vero cells with T. cruzi Y-GFP, T. cruzi Y-GFP-ODC strain, and T. cruzi Y-GFP-ODC clone at indicated conditions. Data represent the mean ± SEM of at least two independent experiments (number of counted cells ≈100). Significantly different from control: **P < .01, Bars: 10 μm.
There is an important number of morphological and biochemical events that occur during metacyclogenesis process. Although the molecular causes of those changes are largely unknown, these experiments clearly show the participation of polyamines in that process. Recently published results showed that spermidine can regulate the transcription of cell cycle and autophagy genes in numerous organisms [13]. Our results show that the availability of polyamines during T. cruzi differentiation favours the global process, increasing the infective capacity of E-MT suspensions. As our results were obtained by indirect infection of Vero cells, the major infective capacity, reached in the presence of endogenous or exogenous source of polyamines, could be explained in different ways. Polyamines could, for example, activate the expression of genes that participate in differentiation from E to MT or those that code to membrane molecules necessary for host cell interaction and invasion, improving the quantity or quality of the process, respectively.
4. Concluding Remarks
Polyamines availability shows to be an important requirement for T. cruzi proliferation, invasion of mammalian host cells, intracellular multiplication, and infection progression, but the way that they act is still uncertain [1, 44, 61–63]. Unlike other trypanosomatid parasites, T. cruzi does not synthesize polyamines by an endogenous ODC and relies on the polyamine availability from the parasite external milieu [38, 41]. However, some evidence suggests that polyamines are not as free and available as it could be predicted by polyamine abundance in the host; polyamines could be conjugated or stored in reservoirs so they are not homogeneously distributed and equally available around all host cells and fluids or under different treatments [37, 46]. The polyamine content in the host relies on diet and, specially, in its ODC enzyme, whose activity is highly regulated at multiple levels [64]. Different treatments that alter the host ODC activity modulate, in turn, polyamine intracellular content—ergo its availability for T. cruzi intracellular stages—as much as polyamine concentration in extracellular fluids, the natural medium for bloodstream stages. In this context, the host ODC as well as the parasite uptake system would be potential targets, as by self or as a complementary target increasing the effect of antitrypanosomal therapies. As there has been some discrepancies about the possible presence of arginine decarboxylase (ADC) activity under specific metabolic conditions [41, 42, 65–68] as well as there are clear evidence about the presence of ADC in at least some kind of cells that could be infected by T. cruzi [27–29], this other possible way to obtain polyamines should be taken into account when the role of polyamines among the T. cruzi biological cycle is studied. In any case, the growth curves and the results obtained in metacyclogenesis assays support the idea that polyamines are engaged in parasite proliferation and in differentiation process. Therefore, this double transfected T. cruzi becomes a powerful and easy-to-follow tool to study the role of polyamines in Chagas disease pathology. In particular, the expression of GFP gene in this double-mutant (or in new mutants for other genes), which allows a simple visualization of T. cruzi cellular limits, represents an advantage compared to the detection techniques frequently used (such as T. cruzi nuclear and kinetoplast DNA Hoechst staining or indirect immunofluorescence) to localize and characterize the host cell parasitophorous vacuole (TcPV) as well as the transit and escape of T. cruzi from the TcPV. Further experiments of infection in cell monolayers and animal models will be done to confirm and extend the knowledge about the role of polyamines in host cell invasion and infection progression.
Acknowledgments
The authors are grateful to Dr. S. Schenkman and Esteban Lozano who gave them the Y-GFP strain. They also wish to thank Dr. NS González for helpful advice and discussions along the work and Dr. ID Algranati for support and critical reading of this manuscript. This work was partially supported by PIP 2010-2012 GI from the Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET, Argentina) and PICT 2005-33431 from Agencia Nacional de Promoción Científica y Tecnológica (ANPCyT, Argentina) to C. Carrillo and Comisión Nacional Salud Investiga, Ministerio de Salud de la Nación and Fundación Bunge y Born to P. S. Romano. J. J. Barclay, L. G. Morosi, and M. C. Vanrell contributed equally to this work.
References
- 1.Heby O, Persson L, Rentala M. Targeting the polyamine biosynthetic enzymes: a promising approach to therapy of African sleeping sickness, Chagas’ disease, and leishmaniasis. Amino Acids. 2007;33(2):359–366. doi: 10.1007/s00726-007-0537-9. [DOI] [PubMed] [Google Scholar]
- 2.Schofield CJ, Maudlin I. Trypanosomiasis control. International Journal for Parasitology. 2001;31(5-6):615–620. doi: 10.1016/s0020-7519(01)00162-x. [DOI] [PubMed] [Google Scholar]
- 3.Andrade LO, Andrews NW. Opinion: the Trypanosoma cruzi—host-cell interplay: location, invasion, retention. Nature Reviews Microbiology. 2005;3(10):819–823. doi: 10.1038/nrmicro1249. [DOI] [PubMed] [Google Scholar]
- 4.Tyler KM, Engman DM. The life cycle of Trypanosoma cruzi revisited. International Journal for Parasitology. 2001;31(5-6):472–481. doi: 10.1016/s0020-7519(01)00153-9. [DOI] [PubMed] [Google Scholar]
- 5.Kohl L, Robinson D, Bastin P. Novel roles for the flagellum in cell morphogenesis and cytokinesis of trypanosomes. The EMBO Journal. 2003;22(20):5336–5346. doi: 10.1093/emboj/cdg518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Romano PS, Arboit MA, Vázquez CL, Colombo MI. The autophagic pathway is a key component in the lysosomal dependent entry of Trypanosoma cruzi into the host cell. Autophagy. 2009;5(1):6–18. doi: 10.4161/auto.5.1.7160. [DOI] [PubMed] [Google Scholar]
- 7.Pires SF, DaRocha WD, Freitas JM, et al. Cell culture and animal infection with distinct Trypanosoma cruzi strains expressing red and green fluorescent proteins. International Journal for Parasitology. 2008;38(3-4):289–297. doi: 10.1016/j.ijpara.2007.08.013. [DOI] [PubMed] [Google Scholar]
- 8.Ramirez MI, Yamauchi LM, De Freitas LHG, Uemura H, Schenkman S. The use of the green fluorescent protein to monitor and improve transfection in Trypanosoma cruzi. Molecular and Biochemical Parasitology. 2000;111(1):235–240. doi: 10.1016/s0166-6851(00)00309-1. [DOI] [PubMed] [Google Scholar]
- 9.Geiger LE, Morris DR. Stimulation of deoxyribonucleic acid replication fork movement by spermidine analogs in polyamine-deficient Escherichia coli. Journal of Bacteriology. 1980;141(3):1192–1198. doi: 10.1128/jb.141.3.1192-1198.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Algranati ID, Goldemberg SH. Initiation, elongation and termination of polypeptide synthesis in cell-free systems from polyamine-deficient bacteria. Biochemical and Biophysical Research Communications. 1981;103(1):8–15. doi: 10.1016/0006-291x(81)91653-3. [DOI] [PubMed] [Google Scholar]
- 11.Tabor CW, Tabor H. Polyamines. Annual Review of Biochemistry. 1984;53:749–790. doi: 10.1146/annurev.bi.53.070184.003533. [DOI] [PubMed] [Google Scholar]
- 12.Wallace HM, Fraser AV, Hughes A. A perspective of polyamine metabolism. Biochemical Journal. 2003;376(1):1–14. doi: 10.1042/BJ20031327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eisenberg T, Knauer H, Schauer A, et al. Induction of autophagy by spermidine promotes longevity. Nature Cell Biology. 2009;11(11):1305–1314. doi: 10.1038/ncb1975. [DOI] [PubMed] [Google Scholar]
- 14.Bacchi CJ, McCann PP. Inhibition of polyamine metabolism: biological significance and bases for new therapies. In: McCann PP, Pegg AE, Sjoerdsma A, editors. Polyamines. Orlando, Fla, USA: Academic Press; 1987. pp. 317–344. [Google Scholar]
- 15.Algranati ID. Polyamine metabolism in Trypanosoma cruzi: studies on the expression and regulation of heterologous genes involved in polyamine biosynthesis. Amino Acids. 2010;38(2):645–651. doi: 10.1007/s00726-009-0425-6. [DOI] [PubMed] [Google Scholar]
- 16.Bacchi CJ, Nathan HC, Hutner SH. Polyamine metabolism: a potential therapeutic target in trypanosomes. Science. 1980;210(4467):332–334. doi: 10.1126/science.6775372. [DOI] [PubMed] [Google Scholar]
- 17.Milord F, Pépin J, Loko L, Ethier L, Mpia B. Efficacy and toxicity of eflornithine for treatment of Trypanosoma brucei gambiense sleeping sickness. The Lancet. 1992;340(8820):652–655. doi: 10.1016/0140-6736(92)92180-n. [DOI] [PubMed] [Google Scholar]
- 18.Sjoerdsma A, Schechter PJ. Eflornithine for African sleeping sickness. The Lancet. 1999;354(9174):p. 254. doi: 10.1016/S0140-6736(05)66324-6. [DOI] [PubMed] [Google Scholar]
- 19.Pegg AE. Recent advances in the biochemistry of polyamines in eukaryocytes. Biochemical Journal. 1986;234(2):249–262. doi: 10.1042/bj2340249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ariyanayagam MR, Oza SL, Mehlert A, Fairlamb AH. Bis(glutathionyl)spermine and other novel trypanothione analogues in Trypanosoma cruzi. Journal of Biological Chemistry. 2003;278(30):27612–27619. doi: 10.1074/jbc.M302750200. [DOI] [PubMed] [Google Scholar]
- 21.Hamana K, Matsuzaki S. Polyamines as a chemotaxonomic marker in bacterial systematics. Critical Reviews in Microbiology. 1992;18(4):261–283. doi: 10.3109/10408419209113518. [DOI] [PubMed] [Google Scholar]
- 22.Marton LJ, Pegg AE. Polyamines as targets for therapeutic intervention. Annual Review of Pharmacology and Toxicology. 1995;35:55–91. doi: 10.1146/annurev.pa.35.040195.000415. [DOI] [PubMed] [Google Scholar]
- 23.Janne J, Poso H, Raina A. Polyamines in rapid growth and cancer. Biochimica et Biophysica Acta. 1978;473(3-4):241–293. doi: 10.1016/0304-419x(78)90015-x. [DOI] [PubMed] [Google Scholar]
- 24.Pegg AE. Polyamine metabolism and its importance in neoplastic growth and as a target for chemotherapy. Cancer Research. 1988;48(4):759–774. [PubMed] [Google Scholar]
- 25.Cunin R, Glansdorff N, Pierard A, Stalon V. Biosynthesis and metabolism of arginine in bacteria. Microbiological Reviews. 1986;50(3):314–352. doi: 10.1128/mr.50.3.314-352.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith TA. L-arginine carboxy-lyase of higher plants and its relation to potassium nutrition. Phytochemistry. 1963;2(3):241–252. [Google Scholar]
- 27.Iyo AH, Zhu MY, Ordway GA, Regunathan S. Expression of arginine decarboxylase in brain regions and neuronal cells. Journal of Neurochemistry. 2006;96(4):1042–1050. doi: 10.1111/j.1471-4159.2005.03544.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhu MY, Iyo A, Piletz JE, Regunathan S. Expression of human arginine decarboxylase, the biosynthetic enzyme for agmatine. Biochimica et Biophysica Acta. 2004;1670(2):156–164. doi: 10.1016/j.bbagen.2003.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mella C, Martínez F, De Los Angeles García M, et al. Expression and localization of an agmatinase-like protein in the rat brain. Histochemistry and Cell Biology. 2010;134(2):137–144. doi: 10.1007/s00418-010-0720-z. [DOI] [PubMed] [Google Scholar]
- 30.El-Sayed NM, Myler PJ, Bartholomeu DC, et al. The genome sequence of Trypanosoma cruzi, etiologic agent of chagas disease. Science. 2005;309(5733):409–435. doi: 10.1126/science.1112631. [DOI] [PubMed] [Google Scholar]
- 31.Della Ragione F, Pegg AE. Purification and characterization of spermidine/spermine N-acetyltransferase from rat liver. Biochemistry. 1982;21(24):6152–6158. doi: 10.1021/bi00267a020. [DOI] [PubMed] [Google Scholar]
- 32.Vujcic S, Liang P, Diegelman P, Kramer DL, Porter CW. Genomic identification and biochemical characterization of the mammalian polyamine oxidase involved in polyamine back-conversion. Biochemical Journal. 2003;370(1):19–28. doi: 10.1042/BJ20021779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fairlamb AH, Cerami A. Metabolism and functions of trypanothione in the kinetoplastida. Annual Review of Microbiology. 1992;46:695–729. doi: 10.1146/annurev.mi.46.100192.003403. [DOI] [PubMed] [Google Scholar]
- 34.Fairlamb AH, Blackburn P, Ulrich P, Chait BT, Cerami A. Trypanothione: a novel bis(glutathionyl)spermidine cofactor for glutathione reductase in trypanosomatids. Science. 1985;227(4693):1485–1487. doi: 10.1126/science.3883489. [DOI] [PubMed] [Google Scholar]
- 35.Fairlamb AH, Cerami A. Identification of a novel, thiol-containing co-factor essential for glutathione reductase enzyme activity in trypanosomatids. Molecular and Biochemical Parasitology. 1985;14(2):187–198. doi: 10.1016/0166-6851(85)90037-4. [DOI] [PubMed] [Google Scholar]
- 36.González NS, Huber A, Algranati ID. Spermidine is essential for normal proliferation of trypanosomatid protozoa. FEBS Letters. 2001;508(3):323–326. doi: 10.1016/s0014-5793(01)03091-5. [DOI] [PubMed] [Google Scholar]
- 37.Igarashi K, Kashiwagi K. Polyamines: mysterious modulators of cellular functions. Biochemical and Biophysical Research Communications. 2000;271(3):559–564. doi: 10.1006/bbrc.2000.2601. [DOI] [PubMed] [Google Scholar]
- 38.Carrillo C, Cejas S, González NS, Algranati ID. Trypanosoma cruzi epimastigotes lack ornithine decarboxylase but can express a foreign gene encoding this enzyme. FEBS Letters. 1999;454(3):192–196. doi: 10.1016/s0014-5793(99)00804-2. [DOI] [PubMed] [Google Scholar]
- 39.Algranati ID, Sánchez C, González NS. Polyamines in Trypanosoma cruzi and Leishmania mexicana. In: Goldemberg SH, Algranati ID, editors. The Biology and Chemistry of Polyamines. Oxford, UK: Oxford University Press; 1990. pp. 137–146. [Google Scholar]
- 40.Ariyanayagam MR, Fairlamb AH. Diamine auxotrophy may be a universal feature of Trypanosoma cruzi epimastigotes. Molecular and Biochemical Parasitology. 1997;84(1):111–121. doi: 10.1016/s0166-6851(96)02788-0. [DOI] [PubMed] [Google Scholar]
- 41.Carrillo C, Cejas S, Huber A, González NS, Algranati ID. Lack of arginine decarboxylase in Trypanosoma cruzi epimastigotes. Journal of Eukaryotic Microbiology. 2003;50(5):312–316. doi: 10.1111/j.1550-7408.2003.tb00141.x. [DOI] [PubMed] [Google Scholar]
- 42.Carrillo C, Serra MP, Pereira CA, Huber A, González NS, Algranati ID. Heterologous expression of a plant arginine decarboxylase gene in Trypanosoma cruzi. Biochimica et Biophysica Acta. 2004;1674(3):223–230. doi: 10.1016/j.bbagen.2004.06.017. [DOI] [PubMed] [Google Scholar]
- 43.Yakubu MA, Majumder S, Kierszenbaum F. Inhibition of S-adenosyl-L-methionine (AdoMet) decarboxylase by the decarboxylated AdoMet analog 5’-{[(Z)-4-amino-2-butenyl]methylamino}-5’-deoxyadenosine (MDL 73811) decreases the capacities of Trypanosoma cruzi to infect and multiply within a mammalian host cell. Journal of Parasitology. 1993;79(4):525–532. [PubMed] [Google Scholar]
- 44.Kierszenbaum F, Wirth JJ, McCann PP, Sjoerdsma A. Arginine decarboxylase inhibitors reduce the capacity of Trypanosoma cruzi to infect and multiply in mammalian host cells. Proceedings of the National Academy of Sciences of the United States of America. 1987;84(12):4278–4282. doi: 10.1073/pnas.84.12.4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kierszenbaum F, Wirth JJ, McCann PP, Sjoerdsma A. Impairment of macrophage function by inhibitors of ornithine decarboxylase activity. Infection and Immunity. 1987;55(10):2461–2464. doi: 10.1128/iai.55.10.2461-2464.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Olenyik T, Gilroy C, Ullman B. Oral putrescine restores virulence of ornithine decarboxylase-deficient Leishmania donovani in mice. Molecular and Biochemical Parasitology. 2011;176(2):109–111. doi: 10.1016/j.molbiopara.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fairlamb AH. Metabolic pathway analysis in trypanosomes and malaria parasites. Philosophical Transactions of the Royal Society B. 2002;357(1417):101–107. doi: 10.1098/rstb.2001.1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.DaRocha WD, Silva RA, Bartholomeu DC, et al. Expression of exogenous genes in Trypanosoma cruzi: improving vectors and electroporation protocols. Parasitology Research. 2004;92(2):113–120. doi: 10.1007/s00436-003-1004-5. [DOI] [PubMed] [Google Scholar]
- 49.Svensson F, Ceriani C, Wallström EL, et al. Cloning of a trypanosomatid gene coding for an ornithine decarboxylase that is metabolically unstable even though it lacks the C-terminal degradation domain. Proceedings of the National Academy of Sciences of the United States of America. 1997;94(2):397–402. doi: 10.1073/pnas.94.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Martínez-Calvillo S, López I, Hernández R. pRIBOTEX expression vector: a pTEX derivative for a rapid selection of Trypanosoma cruzi transfectants. Gene. 1997;199(1-2):71–76. doi: 10.1016/s0378-1119(97)00348-x. [DOI] [PubMed] [Google Scholar]
- 51.Lorenzi HA, Vazquez MP, Levin MJ. Integration of expression vectors into the ribosomal locus of Trypanosoma cruzi. Gene. 2003;310(1-2):91–99. doi: 10.1016/s0378-1119(03)00502-x. [DOI] [PubMed] [Google Scholar]
- 52.Marcora MS, Cejas S, González NS, Carrillo C, Algranati ID. Polyamine biosynthesis in Phytomonas: biochemical characterisation of a very unstable ornithine decarboxylase. International Journal for Parasitology. 2010;40:1389–1394. doi: 10.1016/j.ijpara.2010.04.003. [DOI] [PubMed] [Google Scholar]
- 53.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein dye binding. Analytical Biochemistry. 1976;72(1-2):248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 54.Sánchez CP, Mucci J, González NS, Ochoa A, Zakin MM, Algranati ID. α-difluoromethylornithine-resistant cell lines obtained after one-step selection of Leishmania mexicana promastigote cultures. Biochemical Journal. 1997;324(3):847–853. doi: 10.1042/bj3240847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Contreras VT, Morel CM, Goldenberg S. Stage specific gene expression precedes morphological changes during Trypanosoma cruzi metacyclogenesis. Molecular and Biochemical Parasitology. 1985;14(1):83–96. doi: 10.1016/0166-6851(85)90108-2. [DOI] [PubMed] [Google Scholar]
- 56.Ferreira LRP, Silva AM, Michailowsky V, Reis LFL, Gazzinelli RT. Expression of serum amyloid A3 mRNA by inflammatory macrophages exposed to membrane glycoconjugates from Trypanosoma cruzi. Journal of Leukocyte Biology. 1999;66(4):593–600. doi: 10.1002/jlb.66.4.593. [DOI] [PubMed] [Google Scholar]
- 57.Opperdoes FR, Michels PAM. Enzymes of carbohydrate metabolism as potential drug targets. International Journal for Parasitology. 2001;31(5-6):482–490. doi: 10.1016/s0020-7519(01)00155-2. [DOI] [PubMed] [Google Scholar]
- 58.Carrillo C, González NS, Algranati ID. Trypanosoma cruzi as a model system to study the expression of exogenous genes coding for polyamine biosynthetic enzymes. Induction of DFMO resistance in transgenic parasites. Biochimica et Biophysica Acta. 2007;1770(12):1605–1611. doi: 10.1016/j.bbagen.2007.08.013. [DOI] [PubMed] [Google Scholar]
- 59.Carrillo C, Cejas S, Cortés M, et al. Sensitivity of trypanosomatid protozoa to DFMO and metabolic turnover of ornithine decarboxylase. Biochemical and Biophysical Research Communications. 2000;279(2):663–668. doi: 10.1006/bbrc.2000.3996. [DOI] [PubMed] [Google Scholar]
- 60.Ceriani C, González NS, Algranati ID. Ornithine decarboxylase from Crithidia fasciculata is metabolically unstable and resistant to polyamine down-regulation. FEBS Letters. 1992;301(3):261–264. doi: 10.1016/0014-5793(92)80253-d. [DOI] [PubMed] [Google Scholar]
- 61.Stempin CC, Dulgerian LR, Garrido VV, Cerban FM. Arginase in parasitic infections: macrophage activation, immunosuppression, and intracellular signals. Journal of Biomedicine and Biotechnology. 2010;2010:10 pages. doi: 10.1155/2010/683485. Article ID 683485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Peluffo G, Piacenza L, Irigoín F, Alvarez MN, Radi R. L-arginine metabolism during interaction of Trypanosoma cruzi with host cells. Trends in Parasitology. 2004;20(8):363–369. doi: 10.1016/j.pt.2004.05.010. [DOI] [PubMed] [Google Scholar]
- 63.Bacchi CJ, Braunstein VL, Rattendi D, Yarlett N, Wittner M, Tanowitz HB. Stage-specific polyamine metabolism in Trypanosoma cruzi. Journal of Eukaryotic Microbiology. 2001;48:201S–202S. doi: 10.1111/j.1550-7408.2001.tb00519.x. [DOI] [PubMed] [Google Scholar]
- 64.Pegg AE. Regulation of ornithine decarboxylase. Journal of Biological Chemistry. 2006;281(21):14529–14532. doi: 10.1074/jbc.R500031200. [DOI] [PubMed] [Google Scholar]
- 65.Hernández S, Schwarcz de Tarlovsky S. Arginine decarboxylase in Trypanosoma cruzi, characteristics and kinetic properties. Cellular and Molecular Biology. 1999;45(4):383–391. [PubMed] [Google Scholar]
- 66.Piacenza L, Peluffo G, Radi R. L-arginine-dependent suppression of apoptosis in Trypanosoma cruzi: contribution of the nitric oxide and polyamine pathways. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(13):7301–7306. doi: 10.1073/pnas.121520398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Le Quesne SA, Fairlamb AH. Regulation of a high-affinity diamine transport system in Trypanosoma cruzi epimastigotes. Biochemical Journal. 1996;316(2):481–486. doi: 10.1042/bj3160481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Majumder S, Wirth JJ, Bitonti AJ, McCann PP, Kierszenbaum F. Biochemical evidence for the presence of arginine decarboxylase activity in Trypanosoma cruzi. Journal of Parasitology. 1992;78(2):371–374. [PubMed] [Google Scholar]