Abstract
The yeast cytosol contains multiple homologs of the DnaK and DnaJ chaperone family. Our current understanding of which homologs functionally interact is incomplete. Zuotin is a DnaJ homolog bound to the yeast ribosome. We have now identified the DnaK homolog Ssz1p/Pdr13p as zuotin's partner chaperone. Zuotin and Ssz1p form a ribosome-associated complex (RAC) that is bound to the ribosome via the zuotin subunit. RAC is unique among the eukaryotic DnaK-DnaJ systems, as the 1:1 complex is stable, even in the presence of ATP or ADP. In vitro, RAC stimulates the translocation of a ribosome-bound mitochondrial precursor protein into mitochondria, providing evidence for its chaperone-like effect on nascent chains. In agreement with the existence of a functional complex, deletion of each RAC subunit resulted in a similar phenotype in vivo. However, overexpression of zuotin partly rescued the growth defect of the Δssz1 strain, whereas overexpression of Ssz1p did not affect the Δzuo1 strain, suggesting a pivotal function for the DnaJ homolog.
Proteins synthesized on cytosolic ribosomes either fold or translocate to their final location cotranslationally or posttranslationally. Translocation and folding require chaperone-like proteins that often serve multiple and overlapping functions. The complex chaperone network in the eukaryotic cell is currently only poorly understood (1–3).
Most posttranslational translocation events require the translocating proteins to be unfolded. This requirement is at least partly ensured by binding of cytosolic chaperones to newly translated proteins. The yeast DnaK/Hsp70 homolog Ssa1/2p and its partner protein, the DnaJ homolog Ydj1p, are involved in protein translocation into a variety of compartments (4). Besides their role in translocation, Ssa1/2p and Ydj1p are involved in cytosolic protein folding, most likely in a posttranslational manner (5–7). Other chaperones assisting posttranslational folding in the eukaryotic cytosol are the chaperonin CCT and Hsp90 (2, 8, 9).
Some chaperones interact cotranslationally with polypeptides. In both eukaryotes and prokaryotes, soluble DnaK and DnaJ homologs bind to nascent chains and subsequently assist their folding (2, 10, 11). The ribosome-bound DnaK homolog Ssb1/2p can be crosslinked to nascent chains, providing evidence for a functionally important interaction (12).
There is long-standing circumstantial evidence of cotranslational import into mitochondria (13). More recent data suggest that some precursor proteins even require a cotranslational mechanism to be imported into mitochondria (14–16). However, no specialized component of a mitochondrial cotranslational translocation system, comparable to a signal recognition particle or signal recognition particle receptor, has been identified (17).
In a previous study we introduced an in vitro mitochondrial protein import assay for the identification of cytosolic components involved in either cotranslational translocation or interacting with nascent precursor proteins in a chaperone-like manner. The assay, based on the translocation of ribosome-bound nascent chains into mitochondria, led to the identification of nascent polypeptide-associated complex (NAC) as a factor stimulating mitochondrial protein import in vitro (18). NAC has also been found to affect the delivery of artificial precursor proteins to mitochondria in vivo (19) and is involved in import into the endoplasmic reticulum (20, 21). However, deletion of the genes encoding NAC in yeast does not significantly affect growth, suggesting functional redundancy (19, 22). To identify factors that can functionally replace NAC we made use of a yeast strain carrying disruptions in EGD1 and EGD2 encoding the two NAC subunits (ΔNAC). From this strain we have now purified a ribosome-associated complex (termed RAC) that stimulates translocation into mitochondria in vitro. The complex is a heterodimer composed of the DnaJ homolog zuotin and the DnaK homolog Ssz1p/Pdr13p, with an essentially quantitative engagement of both chaperones in complex formation. The individual subunits have previously been described by others; however, their stable interaction had not been detected (23–26). Our in vivo data indicate that the two subunits of RAC have overlapping but nonidentical functions and serve multiple roles in the cell.
Materials and Methods
Yeast Strains and Plasmids.
Standard yeast genetic techniques were used (27). MH272–3f a/α (ura3/ura3, leu2/leu2, his3/his3, trp1/trp1, ade2/ade2), the parental wild-type strain of all derivatives in this study, is the ade− variant of JK9–3d a/α (19, 28). YRG16 MATa (ura3, leu2, his3, trp1, ade2, egd2∷ADE2, egd1∷URA3), lacking the αNAC (EGD2) and βNAC (EGD1) subunits, was used for the purification of RAC. YRG16 is derived from YRLG3 (ura3, leu2, his3, trp1, ade2, egd2∷ADE2) (19) by deletion of the EGD1 gene (R. George and T.L., unpublished observations). IDA1 (MATa, ura3, leu2, his3, trp1, ade2, zuo1∷TRP1) and IDA2 (MATa, ura3, leu2, his3, trp1, ade2, ssz1∷LEU2, and IDA12 (MATα, ura3, leu2, his3, trp1, ade2, zuo1∷TRP1 ssz1∷LEU2) were generated by disruption of the respective genes in the diploid strain MH272-3f a/α followed by tetrad analysis. ZUO1 (YRG285C) was disrupted by replacing the 1.2-kb PinAI/PshAI fragment of the coding region with TRP1. SSZ1 (YHR064C) was disrupted by replacing the 1.2-kb BglII/HindIII fragment of the coding region with LEU2. For overexpression in yeast, SSZ1 was cloned into pRS423 (2μ, HIS3) (29) and ZUO1 was cloned into YEplac195 (2μ, URA3) (30), resulting in the plasmids 2μ-ZUO1 and 2μ-SSZ1, respectively. The plasmids were transformed into IDA12, resulting in the strains IDA12–2μ-SSZ1 (MATα, ura3, leu2, his3, trp1, ade2, zuo1∷TRP1 ssz1∷LEU2, 2μ-SSZ1) and IDA12–2μ-ZUO1 (MATα, ura3, leu2, his3, trp1, ade2, zuo1∷TRP1 ssz1∷LEU2, 2μ-ZUO1), and into MH272–3fα, resulting in wt-2μ-SSZ1 (MATα, ura3, leu2, his3, trp1, ade2, 2μ-SSZ1) and wt-2μ-ZUO1 (MATα, ura3, leu2, his3, trp1, ade2, 2μ-ZUO1).
Purification of RAC.
Cytosol prepared as described (18) from a 10-liter culture of YRG16 was layered on top of 1 vol of buffer S1 (25% sucrose/20 mM Hepes⋅KOH, pH 7.4/120 mM Kacetate, pH 7.4/5 mM MgAcetate/2 mM DTT/0.5 mM PMSF) and centrifuged for 2.5 h at 160,000 × g. After the ribosomal pellet was suspended in buffer S1 containing 0.72 M Kacetate, the suspension was centrifuged for 1 h at 200,000 × g. The ribosome-free supernatant, termed ribosomal salt-wash, was diluted with 6 vol of buffer S1 lacking sucrose, Kacetate, and DTT and loaded onto a ResourceQ anion-exchange column (Amersham Pharmacia). Bound proteins were eluted with a 100–800 mM 30-ml linear Kacetate gradient in 40 mM Hepes⋅KOH, pH 7.4. RAC eluted at a concentration of 500–600 mM Kacetate. RAC-containing fractions were pooled, concentrated in Centricon-30 devices (Sartorius) to a final volume of 2 ml, and loaded onto a Hi-Load 16/60 Supedex200 prep grade gel filtration column (Amersham Pharmacia) preequilibrated with 40 mM Hepes⋅KOH, 300 mM Kacetate, pH 7.4. RAC-containing fractions were collected and applied to a MonoQ anion-exchange column (Amersham Pharmacia). Bound proteins were eluted with a 300–1,200 mM 25 ml linear Kacetate gradient in 40 mM Hepes⋅KOH (pH 7.4). RAC eluted at 800–900 mM Kacetate. RAC-containing fractions were pooled, washed with 20 mM Hepes⋅KOH, 120 mM Kacetate, and 5 mM Mgacetate (pH 7.4) with the use of Centricon-30 devices, concentrated, frozen in liquid nitrogen, and stored at −80°C. Yield from a 10-liter culture was ≈0.3 mg RAC.
Generation of Antibodies.
Antibodies were generated against purified αNAC, βNAC, Ssz1p, zuotin, and Rpl16a. Immunization of rabbits was performed by Eurogentec (Brussels). Each of the antibodies recognizes a specific band in total yeast extracts (see Figs. 4 and 5 and data not shown). In particular, αSsz1p did not recognize zuotin and vice versa.
Figure 4.
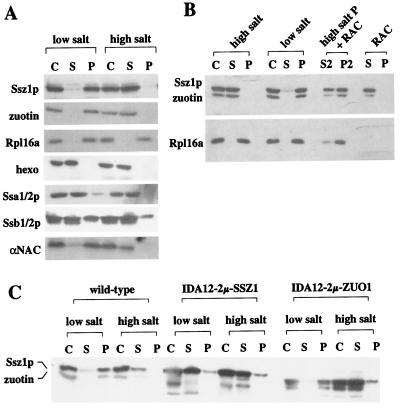
Binding of RAC to ribosomes. (A) The major fraction of RAC is bound to ribosomes in a salt-sensitive manner. Translation-competent cytosol (C) was separated into a postribosomal supernatant (S) and a ribosomal pellet (P) in the presence of either 120 mM Kacetate (low salt) or 700 mM Kacetate (high salt). (B) RAC is reversibly released by high concentrations of salt. After release of endogenous RAC from the ribosome (high-salt P), the ribosomal pellet was resuspended under low-salt conditions, and purified RAC was added to a final concentration of 150 nM (high-salt P + RAC). After incubation for 5 min on ice the ribosomes (P2) were separated from the supernatant (S2) for a second time. As a control, RAC was treated the same, but in the absence of ribosomes (RAC). (C) Binding of RAC to ribosomes requires zuotin. Cytosol of MH272–3f (wild type), IDA12–2μ-SSZ1, and IDA12–2μ-ZUO1 were separated into postribosomal supernatant and ribosomal pellet in the presence of 120 mM Kacetate (low salt) or 700 mM Kacetate (high salt). Note that both Ssz1p and zuotin partly degrade during extract preparation in the absence of their partner protein. (A–C) Corresponding amounts of cytosol, supernatant, and ribosomal pellet were separated by SDS/PAGE and analyzed by immunodecoration, with the use of antibodies specifically recognizing Ssz1p, zuotin, Rpl16a, hexokinase (hexo), Ssa1/2p, Ssb1/2p, and αNAC.
Figure 5.
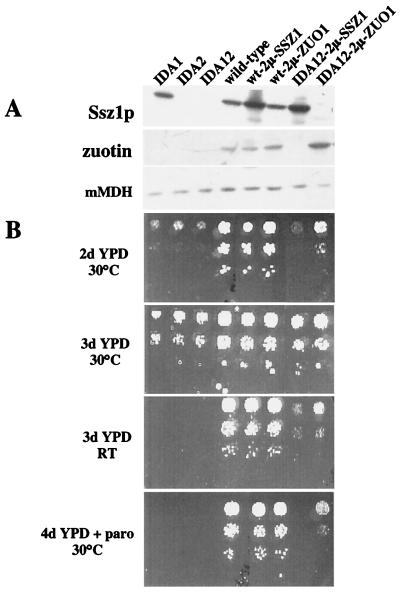
Deletions in SSZ1 and ZUO1 display a similar but nonidentical phenotype. Yeast strains IDA1 (Δzuo), IDA2 (Δssz1), IDA12 (Δzuo1Δssz1), wild type (MH272–3fα), wt-2μ-SSZ1 (MH272–3fα overexpressing SSZ1), wt-2μ-ZUO1 (MH272–3fα overexpressing ZUO1), IDA12–2μ-SSZ1 (IDA12 overexpressing SSZ1), and IDA12–2μ-ZUO1 (IDA12 overexpressing ZUO1) were grown to early log phase at 30°C on minimal glucose medium. (A) Equal amounts of total yeast protein were separated on SDS/PAGE followed by immunodecoration with antibodies directed against Ssz1p, zuotin, and, as a control for loading of equal amounts, malate dehydrogenase (mMDH). (B) Serial 5-fold dilutions of early log-phase cultures. Dilutions containing the same number of cells were spotted from top to bottom onto rich glucose-containing medium (YPD, yeast extract/peptone/dextrose). The time and temperature of incubation are given. RT, room temperature; d, days; paro, 75 μg/ml paromomycin.
In Vitro Translation, Preparation of Ribosome-Nascent Chain Complexes (RNCs), and Translocation Assays.
Yeast translation extract was prepared as described (31) from either JK9-3dα (28) or YRG16. Translation of yeast mitochondrial malate dehydrogenase (Mdh1p) was performed as described (18). Mitochondria were isolated from JK9–3dα grown on lactate-based medium and purified as described (32). Translocation reactions contained 0.8 mg/ml mitochondria in import buffer (20 mM Hepes⋅KOH, pH 7.4/120 mM Kacetate, pH 7.4/5 mM Mgacetate/0.6 M sorbitol/0.05 units/μl RNase inhibitor/2 mM DTT/2 mM ATP/2 mM NADH/2 mM KPi). Mitochondria were premixed with import buffer, and the reaction was started by the addition of 12.5% (vol/vol) of RNCs at 20°C. Volumes of the assay varied from 100 to 500 μl. After 12 min 100-μl samples were withdrawn, and mitochondria were reisolated by centrifugation. The supernatants obtained after reisolation of the mitochondria contained the fraction of RNCs that had not bound to the mitochondria (data not shown). When the membrane potential was destroyed by the addition of 1 μg/ml valinomycin in control reactions, translocation was abolished (data not shown). Translocation efficiency varied by a factor of 2, depending on the RNC preparation. However, the relative translocation efficiency within the experiments was highly reproducible.
Immunoprecipitation Assay.
Immunoprecipitations were performed with protein A-Sepharose beads (CL-4B; Amersham Pharmacia) coated with IgGs against Ssz1p or zuotin or with preimmune IgGs as described (33) in 20 mM Hepes-KOH, 240 mM Kacetate, 5 mM Mgacetate, 200 mM sucrose, 1 mM EDTA, 1 mM PMSF, 0.1% Triton X-100, pH 7.4.
Ribosome Binding Assay.
In a typical experiment 60 μl yeast cytosol was diluted by 1:2 with either low-salt dilution buffer (20 mM Hepes⋅KOH/120 mM Kacetate/2 mM DTT/5 mM Mgacetate/1 mM PMSF, pH 7.4) or high-salt dilution buffer (20 mM Hepes⋅KOH/1.4 M Kacetate/2 mM DTT/5 mM Mgacetate/1 mM PMSF, pH 7.4). After removal of an aliquot (total), 40 μl of the samples was layered on top of a 100-μl low-salt sucrose cushion (20 mM Hepes⋅KOH/25% sucrose/120 mM Kacetate/2 mM DTT/5 mM Mgacetate/1 mM PMSF, pH 7.4) or a 100-μl high-salt sucrose cushion (20 mM Hepes⋅KOH, 7.4/25% sucrose/700 mM Kacetate/2 mM DTT/5 mM Mgacetate/1 mM PMSF, pH 7.4). After centrifugation at 200,000 × g in a RC M120 GX ultracentrifuge (Sorvall) for 180 min at 4°C, samples were split into supernatant and pellet, and corresponding amounts were analyzed by SDS/PAGE, followed by immunoblotting.
Analytical Ultracentrifugation.
RAC was analyzed at initial protein concentrations of 80–200 μg/ml in 20 mM Hepes⋅KOH (pH 7.4), 120 mM Kacetate, 5 mM Mgacetate with a Beckman Optima XL-A centrifuge and a 50Ti rotor. Sedimentation equilibrium measurements (absorption at 230 and 280 nm) were carried out in double sector cells at 10,000 rpm and 20°C. Data were analyzed with the software provided by Beckman Instruments (Palo Alto, CA).
Miscellaneous.
Total yeast protein for immunoblot analysis was prepared by the method of Yaffe and Schatz (34). Protein concentrations were determined by the Bradford assay (Bio-Rad), with BSA as a standard. 125I-labeled protein A was used to develop the immunoblots (35). Quantification of 125I and 35S on immunoblots or SDS gels, respectively was made with an Aida ImageAnalyzer (Raytest, Isotopenmessgeräte GmbH, Straubenhardt, Germany). Identification of Ssz1p and zuotin was by sequencing after transfer to polyvinylidene fluoride membrane. A stock solution of 2.4 M Kacetate (pH 7.4) was prepared by mixing potassium acetate and acetic acid according to the Henderson–Hasselbalch equation.
Results
Identification of a RAC.
When energized mitochondria are incubated with the precursor of mitochondrial malate dehydrogenase (Mdh1p) attached to the ribosome as a nascent chain, the N terminus of the precursor reaches the matrix and its presequence is cleaved off. However, because Mdh1p cannot leave the RNC, translocation is not completed and a two-membrane-spanning intermediate is generated. Translocation of nascent Mdh1p depends on ribosome-associated proteins that can be removed by treatment with high concentrations of salt. The major translocation-stimulating factor bound to wild-type RNCs is NAC (18). When compared with RNCs prepared from wild-type yeast, the translocation efficiency of ΔNAC RNCs was reduced; however, translocation efficiency could be further reduced by high-salt treatment (data not shown).
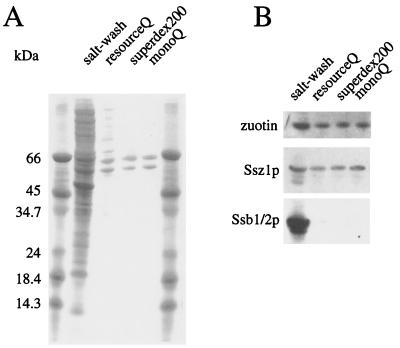
We have used the translocation assay as a functional test to purify the ribosome-associated stimulating factor from the ΔNAC strain. As judged by Coomassie staining, two proteins with apparent molecular masses of 60 kDa and 55 kDa, respectively, made up more than 90% of the active preparation (Fig. 1A). The band of higher molecular mass was identified as the DnaK homolog Pdr13p (24) (termed Ssz1p in this study), and the band of lower molecular mass was identified as the DnaJ homolog zuotin (23, 25, 36). The ratio between Ssz1p and zuotin was constant during purification, suggesting that the two proteins formed a ribosome-associated complex. RAC was free of the ribosome-bound DnaK homologs Ssb1/2p (Fig. 1B).
Figure 1.
Purification of RAC. (A) Yeast RAC was purified as described in Materials and Methods. The purification was monitored by SDS/PAGE and staining with Coomassie blue. Lanes represent proteins released from ribosomes by treatment with 700 mM Kacetate (salt wash); the pool of the active fractions after separation of the salt wash on a ResourceQ (resourceQ); the pool of the active fractions after separation on Superdex200 (superdex200); and the pool of the active fractions after separation on MonoQ (monoQ). Molecular mass standards are indicated on the left. (B) Purified RAC is free of Ssb1/2p. Aliquots of the samples shown in A were analyzed by immunoblotting with antibodies specific for Ssz1p, zuotin, and Ssb1/2p.
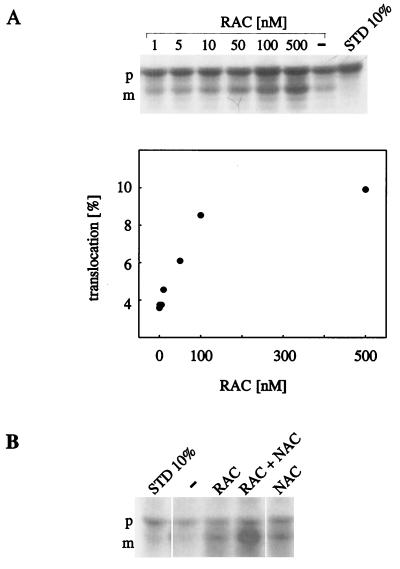
RAC stimulated translocation of high-salt-washed RNCs into mitochondria in a concentration-dependent manner (Fig. 2A). The concentration of ribosomes in a standard translocation assay was ≈100 nM, as determined by quantitative immunoblotting with the purified ribosomal protein Rpl16a as a standard. Although this estimate might vary by a factor of 2–5, it suggests that stimulation by RAC reaches a plateau when the ratio of RAC to ribosomes is ≈1:1 (Fig. 2A and data not shown).
Figure 2.
RAC stimulates translocation of RNCs into mitochondria. Ribosome-bound Mdh1p was generated in the presence of [35S]methionine in a yeast translation extract derived from the ΔNAC strain YRG16. RNCs were isolated under high-salt conditions, with factors stimulating translocation removed, and subsequently incubated with isolated mitochondria. After the reaction mitochondria were reisolated, equal aliquots of mitochondrial pellets, containing mitochondria-bound RNCs, were resuspended, precipitated with trichloroacetic acid, and analyzed by SDS/PAGE, followed by fluorography. STD 10%, 10% of the RNCs added per assay. p, Mdh1p precursor; m, mature Mdh1p. (A) Purified RAC stimulates translocation in a dose-dependent manner. High-salt-washed ΔNAC-RNCs were preincubated in the absence (−) or in the presence of purified RAC. The concentration of RAC is given as the final concentration in the assay. Translocation efficiency is given as the percentage of the total amount of RNCs added to the assay (Lower). (B) RAC and NAC synergistically stimulate translocation. The translocation assay was performed as in A, except that high-salt-washed RNCs were preincubated in the absence (−) or in the presence of purified RAC (RAC), a combination of RAC and NAC (RAC + NAC), or NAC (NAC). The final concentrations in the translocation assay were 100 nM for RAC and 40 nM for NAC, respectively.
A comparison of RAC and NAC revealed that the two proteins effectively stimulated translocation of high-salt-washed RNCs at similar concentrations (Fig. 2B and data not shown). In the experiment shown in Fig. 2B, translocation efficiency in the presence of either RAC or NAC alone was about 20%. Increasing the concentration of RAC or NAC, respectively, did not further increase translocation efficiency (data not shown). However, when the two complexes were combined, translocation efficiency increased to 38%, suggesting that the two complexes did not compete in binding to the nascent chain but rather acted synergistically (Fig. 2B).
The Complex of Ssz1p and Zuotin Is a Stable Heterodimer.
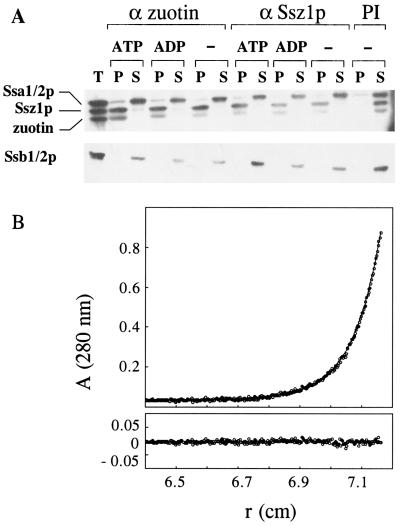
IgGs specific for either Ssz1p or zuotin immunoprecipitated RAC efficiently from the protein mixture released by salt wash from ribosomes. Coimmunoprecipitation of the two subunits was unaffected by the addition of 2 mM ATP or ADP (Fig. 3A). Stability of the complex in the presence of adenine nucleotides strongly suggests that zuotin does not bind to Ssz1p in a substrate-like manner. The DnaK homologs Ssa1/2p and Ssb1/2p contained in the ribosomal salt wash remained unbound to IgGs directed against Ssz1p or zuotin, respectively (Fig. 3A). The small fraction of Ssa1/2p recovered in the pellet was also detected in the preimmune control, suggesting that Ssa1/2p was unspecifically bound to the beads (Fig. 3A, PI).
Figure 3.
RAC is a stable heterodimer. (A) RAC is stable in the presence of adenine nucleotides. Proteins released from ribosomes by 700 mM Kacetate were used for immunoprecipitations in the presence of 2 mM ATP (ATP) or 2 mM ADP (ADP) or in the absence of nucleotide (−) with antisera directed against zuotin (α zuotin) or Ssz1p (α Ssz1p), or with preimmune serum (PI). After incubation for 3 h at 4°C, bound proteins were recovered in the pellet (P) and unbound proteins in the supernatant (S), and both fractions were analyzed by SDS/PAGE and immunoblotting with antibodies specific for Ssa1/2p, Ssz1p, zuotin, or Ssb1/2p as indicated. The total of the ribosomal salt wash added to each single reaction is shown on the left (T). (B) Molecular mass determination of RAC was performed by sedimentation equilibrium measurements at 10,000 rpm and 20°C in an analytical ultracentrifuge. The experimental data could be fitted to a homogeneous population of particles with an apparent mass of 126 kDa. (Upper) Experimental data and fit (−). (Lower) Deviation of fit and experimental data.
The stoichiometry between Ssz1p and zuotin in the complex was determined by analytical ultracentrifugation. The predicted molecular masses of zuotin and Ssz1p are 49 kDa and 62 kDa, respectively. At equilibrium, RAC behaved like a uniform population of particles with a molecular mass of 126,000 Da (Fig. 3B). No higher oligomers or aggregates were detected in sedimentation velocity experiments (data not shown). The results were independent of the concentration the range of 0.7 to 1.8 μM RAC and are consistent with the presence of a stable heterodimeric complex.
Zuotin Anchors Ssz1p to Cytosolic Ribosomes.
To determine the fraction of Ssz1p complexed with zuotin and to analyze the distribution of the proteins in relation to cytosolic ribosomes, yeast cytosol was separated into a postribosomal supernatant and a ribosomal pellet in the presence of low or high concentrations of salt. Independently of the salt concentration the core ribosomal protein Rpl16a was recovered in the pellet, and hexokinase, a cytosolic marker, was recovered in the supernatant. The distribution of Ssa1/2p, Ssb1/2p, and NAC under high- and low-salt conditions was in agreement with published data (12, 19, 22, 37) (Fig. 4A). RAC was almost exclusively recovered in the ribosomal pellet at low concentrations of salt. At high concentrations of salt, RAC was released from the ribosomes and recovered in the supernatant (Fig. 4). Release was reversible: when high-salt-washed ribosomes were incubated with purified RAC at low salt concentrations and the ribosomes were subsequently reisolated, RAC was recovered in the ribosomal pellet (Fig. 4B). In a control reaction, RAC was recovered in the supernatant in the absence of ribosomes. We conclude that the bulk of RAC is bound to ribosomes and can efficiently rebind to high-salt-washed ribosomes.
Cytosol containing either Ssz1p or zuotin, but not the other subunit, was prepared from yeast strains lacking either Ssz1p or zuotin. In the absence of Ssz1p the major fraction of zuotin was recovered in the ribosomal pellet, and high-salt treatment caused its release to the supernatant (Fig. 4C, IDA12–2μ-ZUO1). In the absence of zuotin, however, Ssz1p was recovered in the postribosomal supernatant even under low-salt conditions (Fig. 4C, IDA12–2μ-SSZ1). The combination of data indicates that stable binding of Ssz1p to ribosomes requires its partner protein zuotin.
Deletion of Either Ssz1p or Zuotin Results in Similar but Nonidentical Phenotypes.
We have generated the single deletions of either ZUO1 (IDA1) or SSZ1 (IDA2), the double deletion of ZUO1 and SSZ1 (IDA12), and strains overexpressing either the individual or both subunits. Immunoblotting confirmed the predicted levels of Ssz1p and zuotin in the different strains; the level of zuotin in IDA2 was significantly reduced when compared with wild type, suggesting that zuotin is destabilized in the absence of Ssz1p (Fig. 5A and data not shown). (For a characterization of the single deletion strains also compare refs. 23 and 24.)
IDA1, IDA2, and IDA12 displayed slow growth (compare growth after 2 days and 3 days, respectively), cold sensitivity, and hypersensitivity to the protein synthesis inhibitor paromomycin (Fig. 5B). This combination of growth defects has been connected with ribosome-bound factors thought to aid the passage of nascent chains through the ribosome channel into the cytosol (23, 38).
The phenotypic similarity of IDA1, IDA2, and IDA12 is in agreement with a single functional complex formed by Ssz1p and zuotin. However, overexpression of zuotin partly rescued slow growth, cold sensitivity, and paromomycin hypersensitivity of IDA12, whereas overexpression of Ssz1p did not (Fig. 5B). The data suggest that deletion of ZUO1 has a more severe effect on the biogenesis of newly synthesized proteins than deletion of SSZ1 and that ZUO1 is a multicopy suppressor of Δssz1 (compare also the Discussion).
Discussion
Physical Interaction Between Ssz1p and Zuotin.
Zuotin was previously found to be a ribosome-associated protein (23), and binding of a fraction of Ssz1p to the ribosome has been suggested recently (26, 39). Here we show by a variety of methods that virtually all Ssz1p and zuotin contained in the yeast cytosol forms a stable, ribosome-associated complex that is a heterodimer in solution.
It is well established that DnaK and DnaJ homologs interact with each other (40, 41). However, the functionally important interaction between the N-terminal ATPase domain of DnaK and the conserved J-domain is highly transient. Recent findings suggest that previously reported stable interaction between DnaJ and DnaK is probably due to substrate-like binding of DnaJ to DnaK (42–44). The stable complex between Ssz1p and zuotin is thus unusual, but not without precedence. Thermus thermophilus DnaK and DnaJ form a stable 3:3 complex that contains an additional three subunits of the 8-kDa protein Daf (45).
We can only speculate about which domains mediate the stable interaction between Ssz1p and zuotin. The transient interaction of the ATPase domains of DnaK homologs with J-domains might be stable in this particular case. Such stability, however, would imply significant mechanistic differences between RAC and other DnaK-DnaJ partners (40). We regard this possibility as unlikely and favor a model in which Ssz1p and zuotin stably interact via yet uncharacterized domains, facilitating the transient functional interaction of the Ssz1p ATPase and the zuotin J-domain in the presence of substrate protein. In this context a deletion mutant of zuotin is of interest. In this mutant the very N terminus, up to the J-domain, results in a nonfunctional protein that is still able to bind to the ribosome (23). This N-terminal segment, which bears no homology to any other known J-protein, is a good candidate domain for binding to Ssz1p.
Two Ribosome-Bound DnaK Systems Are Involved in Cytosolic Translation.
A function of RAC in translation is suggested by the finding that yeast strains lacking functional RAC are hypersensitive to nonsense-suppression-inducing aminoglycosides like paromomycin and G418 (Fig. 5B and data not shown). The very same phenotype has previously been reported for the ribosome-bound DnaK homolog Ssb1/2p, which is implicated in translation (23, 38). Although the DnaK homologs Ssb1/2p and Ssz1p function during the early stages of protein biogenesis, they differ in a number of mechanistically important properties: Ssz1p is a subunit of a heterodimeric complex, whereas Ssb1/2p is not. RAC (Ssz1p and zuotin) binds to ribosomes almost quantitatively, whereas only a fraction of Ssb1/2p copurifies with ribosomes. The stoichiometry between Ssz1p and ribosomes is ≈1:1 (data not shown), whereas the steady-state level of Ssb1/2p is 2- to 4-fold higher than that of ribosomes. Ssz1p binds to ribosomes independently of a nascent chain, whereas Ssb1/2p binds more stably to translating ribosomes. Finally, Ssz1p is bound to the ribosome only in the presence of zuotin, whereas Ssb1/2p binds to the ribosome directly (compare the Results section of this study and refs. 12, 25, and 38). It will be most interesting to learn whether the two ribosome-bound DnaK systems have overlapping functions, act sequentially on nascent chains, or differ in their substrate specificity.
The in Vivo Functions of Ssz1p and Zuotin Are Similar but Not Identical.
Although the phenotypes of the IDA1, IDA2, and IDA12 strains are similar, their differences have implications for the function of zuotin and Ssz1p: (i) zuotin is a multicopy suppressor of Δssz1, whereas overexpression of SSZ1 does not rescue the Δzuo1 phenotype; (ii) the steady-state level of zuotin is significantly reduced in the Δssz1 strain, suggesting that zuotin is destabilized in the absence of its partner protein. Two different models might account for this combination of results. In the first model the observed phenotype is primarily caused by a lack of zuotin. Δssz1 results in the same phenotype because, as a consequence, the steady-state level of zuotin is decreased. According to this model zuotin might be functional without its partner protein. This hypothesis is in line with results suggesting that DnaJ homologs do not merely function as cochaperones of DnaK but also interact with substrate proteins by themselves (10, 46–50). In the second model the function of Ssz1p can be partially replaced by one (or more) DnaK homologs, whereas zuotin's function is nonredundant. In the absence of Ssz1p, overexpression of zuotin would lead to an increased level of the protein and as a consequence permit the binding of a yet unidentified DnaK homolog to the ribosome. In this model zuotin serves a dual function in recruiting its partner to the ribosome and priming it for the interaction with its natural substrates (40, 44).
Does RAC Also Function in Targeting to the Mitochondria?
We have initially identified RAC in an attempt to find cytosolic components stimulating protein translocation into mitochondria. RAC shares the ability to stimulate mitochondrial protein translocation with NAC (18). Although our in vitro data suggest that RAC and NAC are involved in cotranslational protein translocation into mitochondria, the respective knockout strains did not display typical mitochondrion-specific defects: (i) In the absence of RAC yeast did not lose the ability to respire (data not shown); furthermore, we were unable to detect accumulated mitochondrial precursor proteins in the cytosol at steady state (data not shown). (ii) The quadruple deletion strain Δegd1/Δegd2/Δssz1/Δzuo1, lacking both NAC and RAC-, displayed a phenotype no more severe than that of the Δssz1/Δzuo1 double deletion strain. (iii) The steady-state level of RAC was not increased in the ΔNAC strain (data not shown). The in vitro translocation assay might thus not specifically identify components involved in cotranslational translocation into mitochondria. However, the combination of data is still compatible with a function of RAC in mitochondrial translocation. Factors involved in cotranslational protein translocation into mitochondria might be highly redundant, such that even in the absence of NAC and RAC, functional replacement is possible. In this context it is interesting that the salt wash of ΔNAC ribosomes contained at least one additional factor besides RAC-stimulating translocation of high-salt-washed RNCs (data not shown). Alternatively, mitochondrial precursor proteins translocated via a cotranslational route might switch to posttranslational translocation in the absence of RAC and/or NAC. A comparable situation exists for translocation into the yeast endoplasmic reticulum. Yeast lacking functional signal recognition particles is viable; protein translocation into the endoplasmic reticulum under these conditions must occur posttranslationally (51).
Whether RAC, NAC, and potential other cytosolic proteins are involved in cotranslational translocation into mitochondria has yet to be determined. Regardless of the outcome, the translocation assay used in this study represents a useful and reliable tool for the identification of ribosome-bound proteins affecting nascent chains in a chaperone-like manner.
Acknowledgments
The Ssb1/2p antibody was a kind gift of Elisabeth Craig (University of Wisconsin). The strains MH272–3f and JK9–3d were from Mike Hall (University of Basel). We thank Rebecca George for critical comments on the manuscript and help with the yeast strain YRG16 and Rudi Glockshuber for giving M.G. the opportunity to work in his laboratory. Most helpful during all stages of the project were discussions with Yves Dubaquié. This study was supported by a grant from the Swiss National Science Foundation and by the Fonds der Chemischen Industrie (to S.R.).
Abbreviations
- NAC
nascent polypeptide-associated complex
- RAC
ribosome-associated complex consisting of Ssz1p and zuotin
- RNC
ribosome-nascent chain complex
- Mdh1p
mitochondrial malate dehydrogenase
References
- 1.Agashe V R, Hartl F U. Semin Cell Dev Biol. 2000;11:15–25. doi: 10.1006/scdb.1999.0347. [DOI] [PubMed] [Google Scholar]
- 2.Bukau B, Deuerling E, Pfund C, Craig E A. Cell. 2000;101:119–122. doi: 10.1016/S0092-8674(00)80806-5. [DOI] [PubMed] [Google Scholar]
- 3.Schatz G, Dobberstein B. Science. 1996;271:1519–1526. doi: 10.1126/science.271.5255.1519. [DOI] [PubMed] [Google Scholar]
- 4.Rassow J, Voos W, Pfanner N. Trends Cell Biol. 1995;5:207–212. doi: 10.1016/s0962-8924(00)89001-7. [DOI] [PubMed] [Google Scholar]
- 5.Kim S, Schilke B, Craig E A, Horwich A L. Proc Natl Acad Sci USA. 1998;95:12860–12865. doi: 10.1073/pnas.95.22.12860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bush G L, Meyer D I. J Cell Biol. 1996;135:1229–1237. doi: 10.1083/jcb.135.5.1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu Z, Cyr D M. J Biol Chem. 1998;273:27824–27830. doi: 10.1074/jbc.273.43.27824. [DOI] [PubMed] [Google Scholar]
- 8.Leroux M R, Hartl F U. Curr Biol. 2000;10:260–264. doi: 10.1016/s0960-9822(00)00432-2. [DOI] [PubMed] [Google Scholar]
- 9.Johnson J L, Craig E A. Cell. 1997;90:201–204. doi: 10.1016/s0092-8674(00)80327-x. [DOI] [PubMed] [Google Scholar]
- 10.Hendrick J P, Langer T, Davis T A, Hartl F U, Wiedmann M. Proc Natl Acad Sci USA. 1993;90:10216–10220. doi: 10.1073/pnas.90.21.10216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beckmann R P, Mizzen L A, Welch W J. Science. 1990;248:850–854. doi: 10.1126/science.2188360. [DOI] [PubMed] [Google Scholar]
- 12.Pfund C, Lopez-Hoyo N, Ziegelhoffer T, Schilke B A, Lopez-Buesa P, Walter W A, Wiedmann M, Craig E A. EMBO J. 1998;17:3981–3989. doi: 10.1093/emboj/17.14.3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Verner K. Trends Biochem Sci. 1993;18:366–371. doi: 10.1016/0968-0004(93)90090-a. [DOI] [PubMed] [Google Scholar]
- 14.Corral-Debrinski M, Belgareh N, Blugeon C, Claros M G, Doye V, Jacq C. Mol Microbiol. 1999;31:1499–1511. doi: 10.1046/j.1365-2958.1999.01295.x. [DOI] [PubMed] [Google Scholar]
- 15.Ni L, Heard T S, Weiner H. J Biol Chem. 1999;274:12685–12691. doi: 10.1074/jbc.274.18.12685. [DOI] [PubMed] [Google Scholar]
- 16.Knox C, Sass E, Neupert W, Pines O. J Biol Chem. 1998;273:25587–25593. doi: 10.1074/jbc.273.40.25587. [DOI] [PubMed] [Google Scholar]
- 17.Stroud R M, Walter P. Curr Opin Struct Biol. 1999;9:754–759. doi: 10.1016/s0959-440x(99)00040-8. [DOI] [PubMed] [Google Scholar]
- 18.Fünfschilling U, Rospert S. Mol Biol Cell. 1999;10:3289–3299. doi: 10.1091/mbc.10.10.3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.George R, Beddoe T, Landl K, Lithgow T. Proc Natl Acad Sci USA. 1998;95:2296–2301. doi: 10.1073/pnas.95.5.2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wiedmann B, Prehn S. FEBS Lett. 1999;458:51–54. doi: 10.1016/s0014-5793(99)01118-7. [DOI] [PubMed] [Google Scholar]
- 21.Wiedmann B, Sakai H, Davis T A, Wiedmann M. Nature (London) 1994;370:434–440. doi: 10.1038/370434a0. [DOI] [PubMed] [Google Scholar]
- 22.Reimann B, Bradsher J, Franke J, Hartmann E, Wiedmann M, Prehn S, Wiedmann B. Yeast. 1999;15:397–407. doi: 10.1002/(SICI)1097-0061(19990330)15:5<397::AID-YEA384>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 23.Yan W, Schilke B, Pfund C, Walter W, Kim S, Craig E A. EMBO J. 1998;17:4809–4817. doi: 10.1093/emboj/17.16.4809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hallstrom T C, Katzmann D J, Torres R J, Sharp W J, Moye-Rowley W S. Mol Cell Biol. 1998;18:1147–1155. doi: 10.1128/mcb.18.3.1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang S, Lockshin C, Herbert A, Winter E, Rich A. EMBO J. 1992;11:3787–3796. doi: 10.1002/j.1460-2075.1992.tb05464.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Michimoto T, Aoki T, Toh-e A, Kikuchi Y. Gene. 2000;257:131–137. doi: 10.1016/s0378-1119(00)00381-4. [DOI] [PubMed] [Google Scholar]
- 27.Sherman F, Fink G R, Hicks J B. Methods in Yeast Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1986. [Google Scholar]
- 28.Heitmann J, Movva N R, Hiestand P C, Hall M N. Proc Natl Acad Sci USA. 1991;88:1948–1952. doi: 10.1073/pnas.88.5.1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Christianson T W, Sikorski R S, Dante M, Shero J H, Hieter P. Gene. 1992;110:119–122. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
- 30.Gietz R D, Sugino A. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- 31.Garcia P D, Hansen W, Walter P. Methods Enzymol. 1991;194:675–682. doi: 10.1016/0076-6879(91)94049-i. [DOI] [PubMed] [Google Scholar]
- 32.Glick B S, Pon L A. Methods Enzymol. 1995;260:213–223. doi: 10.1016/0076-6879(95)60139-2. [DOI] [PubMed] [Google Scholar]
- 33.Rospert S, Hallberg R L. Methods Enzymol. 1995;260:287–292. doi: 10.1016/0076-6879(95)60145-7. [DOI] [PubMed] [Google Scholar]
- 34.Yaffe M P, Schatz G. Proc Natl Acad Sci USA. 1984;81:4819–4823. doi: 10.1073/pnas.81.15.4819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Haid A, Suissa M. Methods Enzymol. 1983;96:192–205. doi: 10.1016/s0076-6879(83)96017-2. [DOI] [PubMed] [Google Scholar]
- 36.Wilhelm M L, Reinbolt J, Gangloff J, Dirheimer G, Wilhelm F X. FEBS Lett. 1994;349:260–264. doi: 10.1016/0014-5793(94)00683-0. [DOI] [PubMed] [Google Scholar]
- 37.James P, Pfund C, Craig E A. Science. 1997;275:387–389. doi: 10.1126/science.275.5298.387. [DOI] [PubMed] [Google Scholar]
- 38.Nelson R J, Ziegelhoffer T, Nicolet C, Werner-Washburne M, Craig E A. Cell. 1992;71:97–105. doi: 10.1016/0092-8674(92)90269-i. [DOI] [PubMed] [Google Scholar]
- 39.Hallstrom T C, Moye-Rowley W S. Mol Microbiol. 2000;36:402–413. doi: 10.1046/j.1365-2958.2000.01858.x. [DOI] [PubMed] [Google Scholar]
- 40.Bukau B, Horwich A L. Cell. 1998;92:351–366. doi: 10.1016/s0092-8674(00)80928-9. [DOI] [PubMed] [Google Scholar]
- 41.Kelley W L. Trends Biochem Sci. 1998;23:222–227. doi: 10.1016/s0968-0004(98)01215-8. [DOI] [PubMed] [Google Scholar]
- 42.Mayer M P, Laufen T, Paal K, McCarty J S, Bukau B. J Mol Biol. 1999;289:1131–1144. doi: 10.1006/jmbi.1999.2844. [DOI] [PubMed] [Google Scholar]
- 43.Greene M K, Maskos K, Landry S J. Proc Natl Acad Sci USA. 1998;95:6108–6113. doi: 10.1073/pnas.95.11.6108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Misselwitz B, Staeck O, Rapoport T A. Mol Cell. 1998;2:593–603. doi: 10.1016/s1097-2765(00)80158-6. [DOI] [PubMed] [Google Scholar]
- 45.Motohashi K, Yohda M, Endo I, Yoshida M. J Biol Chem. 1996;271:17343–17348. doi: 10.1074/jbc.271.29.17343. [DOI] [PubMed] [Google Scholar]
- 46.Szabo A, Korszun R, Hartl F U, Flanagan J. EMBO J. 1996;15:408–417. [PMC free article] [PubMed] [Google Scholar]
- 47.Ungewickell E, Ungewickell H, Holstein S E, Lindner R, Prasad K, Barouch W, Martin B, Greene L E, Eisenberg E. Nature (London) 1995;378:632–635. doi: 10.1038/378632a0. [DOI] [PubMed] [Google Scholar]
- 48.Langer T, Lu C, Echols H, Flanagan J, Hayer M K, Hartl F U. Nature (London) 1992;356:683–689. doi: 10.1038/356683a0. [DOI] [PubMed] [Google Scholar]
- 49.Schröder H, Langer T, Hartl F-U, Bukau B. EMBO J. 1993;12:4137–4144. doi: 10.1002/j.1460-2075.1993.tb06097.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gamer J, Bujard H, Bukau B. Cell. 1992;69:833–842. doi: 10.1016/0092-8674(92)90294-m. [DOI] [PubMed] [Google Scholar]
- 51.Hann B C, Walter P. Cell. 1991;67:131–144. doi: 10.1016/0092-8674(91)90577-l. [DOI] [PubMed] [Google Scholar]