Abstract
We have explored the organization of the axonal lobes in Drosophila mushroom bodies by using a panel of immunohistochemical markers. These markers consist of antibodies to eight proteins expressed preferentially in the mushroom bodies: DAMB, DCO, DRK, FASII, LEO, OAMB, PKA RII, and RUT. Previous to this work, four axonal lobes, two projecting dorsally (α and α′) and two medially (β and γ), had been described in Drosophila mushroom bodies. However, our analysis of immunohistochemically stained frontal and sagittal sections of the brain revealed three medially projecting lobes. The newly distinguished lobe, which we term β′, lies along the dorsal surface of β, just posterior to γ. In addition to resolving a fifth lobe, our studies revealed that there are specific lobe sets defined by equivalent marker expression levels. These sets are (1) the α and β lobes, (2) the α′ and β′ lobes, and (3) the γ lobe and heel (a lateral projection formed by a hairpin turn of some of the peduncle fibers). All of the markers we have examined are consistent with these three sets. Previous Golgi studies demonstrate that each mushroom body cell projects one axon that branches into a dorsal lobe and a medial lobe, or one unbranched axon that projects medially. Taken together with the lobe sets listed above, we propose that there are three major projection configurations of mushroom body cell axons: (1) one branch in the α and one in the β lobe, (2) one branch in the α′ and one in the β′ lobe, and (3) one unbranched axon projecting to the heel and the γ lobe. The fact that these neuron types exhibit differential expression levels of a number of mushroom body genes suggests that they may have corresponding functional differences. These functions may be conserved in the larvae, as several of these genes were expressed in larval and embryonic mushroom bodies as well. The basic mushroom body structure, including the denritic calyx, peduncle, and lobes, was already visible by the late stages of embryogenesis. With new insights into mushroom body organization, and the characterization of markers for developing mushroom bodies, we are beginning to understand how these structures form and function.
Mushroom bodies are conserved brain structures that receive inputs from several sensory systems. Electrophysiological experiments show that mushroom body neurons are responsive to olfactory, visual, tactile, and gustatory stimuli (Erber 1978; Erber et al. 1980; Gronenberg 1987). The prominent antennoglomerular tract and anterior superior optic tract convey olfactory and visual information to the mushroom body calyces, whereas additional afferents relay mechanosensory information (Strausfeld 1976; Mobbs 1982; Rybak and Menzel 1993). This convergence suggests that the mushroom bodies may be sites of sensory integration, an essential component to associative learning. Behavioral experiments correspondingly show that mushroom bodies are required for spatial memory in the cockroach (Mizunami et al. 1993), and for olfactory learning in Drosophila (de Belle and Heisenberg 1994) and the honeybee (Erber et al. 1980; Hammer and Menzel 1995).
Evidence of how mushroom bodies encode the identity of sensory information and how their architecture facilitates sensory integration are just beginning to emerge. As shown in the locust, synchronized cell firing may encode odor identity (MacLeod and Laurent 1996). Perhaps information from other sensory modalities is encoded in mushroom body cells by the same mechanism, and overlapping assemblies of synchronized cells mediate sensory integration. More clues to how cell organization might allow integration came from resolving the mushroom body architecture in the honeybee. The honeybee calyx is subdivided into three anatomically discrete regions that receive afferents from different sensory systems (Mobbs 1982). Therefore, communication between these calycal regions may result in sensory associations. Integrated information might then be forwarded to mushroom body efferents through the lobes, which are subdivided into distinct fiber bundles. This segregation of the lobes provides a neuroanatomical basis for discrete output signals. Indeed, in honeybee and cockroaches, different lobes may mediate separate behaviors (Erber et al. 1980; Mizunami et al. 1993). To test these models, it is crucial to be able to identify the cells comprising specific assemblies.
The genetics and molecular biology developed in Drosophila make it a powerful model to study the relationship between the organization and function of mushroom body neurons. As in all insects, the mushroom bodies of Drosophila consist of posterior clusters of Kenyon cells, which extend processes that form a dendritic calyx, and a fasciculated axonal tract (the peduncle), which branches into distinct lobes. Four lobes (α, α′, β, γ) and a heel have been described in Drosophila (Heisenberg 1980; Ito et al. 1997) To distinguish these lobes and determine how the structure of the mushroom body enables its function, a battery of markers would be highly valuable. Therefore, we have compiled the expression patterns for eight gene markers, including three members of the cAMP cascade—adenylyl cyclase (RUT) [RUTABAGA], and the catalytic and regulatory subunits of protein kinase A (PKA) (DCO and RII), two members of the mitogen-activated protein (MAP) kinase cascade [(DRK) Drosophila receptor kinase and 14-3-3 (LEONARDO, LEO)], the biogenic dopamine and octopamine amine receptors (DAMB and OAMB), and the cell adhesion molecule, fasciclin II (FASII). With the exception of DRK, the mushroom body expression of all of these genes has been documented previously (Han et al. 1992, 1996, 1998; Skoulakis et al. 1993; Skoulakis and Davis 1996; Muller 1997). However, it is the systematic comparison of expression patterns of all these genes that has provided insight to the organization of the lobes.
The eight markers exhibited five patterns of expression in the mushroom body lobes. Interestingly, there were sets of lobes that exhibited equal levels of expression with any marker that we used. These sets are: (1) the α and β lobes, (2) the α′ and β′ lobes, and (3) the γ lobe and heel (Fig. 1, below). The identification of these three sets of lobes is interesting in light of Golgi studies, that suggest that Kenyon cells extend either one axon that branches to form a dorsal projection and a medial projection, or one unbranched axon that projects medially (Mobbs 1982; Yang et al. 1995; M. Heisenberg, pers. comm.). In conjuction with our model of mushroom body lobe organization, these studies suggest that the majority of Kenyon cells may be classified into one of three projection configurations, corresponding to the three lobe sets described above. In addition to describing the adult expression patterns for these markers, we have followed several of them throughout development. The concentration of their expression in mushroom bodies was conserved in larvae; therefore, these markers may serve not only to dissect the role of mushroom bodies in adults, but in larvae as well. The markers FASII, DCO, RII, and LEO were also expressed in embryonic mushroom bodies. These are important resources, given the paucity of gene markers for embryonic mushroom bodies. With the markers defined in this paper, we now have tools to dissect the development, neuroanatomy, and ultimately the function of the Drosophila mushroom bodies.
Figure 1.
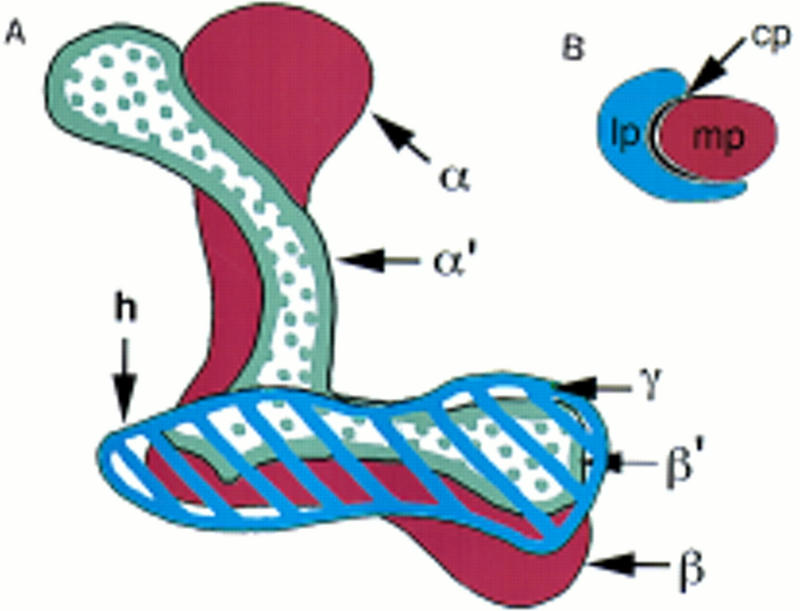
(A) Cartoon of the mushroom body lobes depicted from an anterior viewpoint. Although the mushroom bodies are bilateral, this diagram depicts only the left lobe structure. Dorsal is up; medial is to the right. The peduncle would extend behind the plane of paper toward the Kenyon cells. The most anterior lobe, γ, is shown striped in blue, and is continuous with the heel (h). Just behind the γ lobe are the α′ and β′ collateral lobes, stippled in gray. The β lobe, ventral to the β′ lobe, and its collateral α, are in brown. (B) Cartoon of a cross section through the peduncle at the level of the fan-shaped body. The lateral peduncle is in blue, the central peduncle in black, and the medial peduncle in brown, corresponding to the coloration of the lobes to which they project.
Materials and Methods
Drosophila melanogaster flies were raised at room temperature on standard sucrose and cornmeal media. Flies used for immunohistochemical experiments were either Canton-S or adh cn pr.
PARAFFIN EMBEDDING
Flies were fixed in Carnoy’s solution (60% vol/vol ethanol, 30% vol/vol chloroform, 10% vol/vol glacial acetic acid) for 4 hr at room temperature. After four 100% ethanol washes of 30 min each, the tissue was immersed in methyl benzoate overnight and then transferred to a 1:1 solution of methyl benzoate/paraffin for 1.5 hr at 56°C. The flies were then immersed in pure paraffin, four times for 1 hr each at 56°C, and finally embedded in paraffin at room temperature. The paraffin blocks were cut into 3-μm floating sections that were mounted on gelatinized glass slides. The tissue was deparaffinized with a 3-min wash in xylene, and rehydrated through an ethanol series and PBHT [0.02 m NaPO4, 0.5 m NaCl, 0.2% Triton X-100 (pH 7.4)]. For anti-DAMB immunohistochemistry, cryosections were used as described in Han et al. (1996). Parafilm was used to cover the sections during all antibody incubations. For the primary antibody reaction, the samples were blocked with 1.5% goat serum in PBHT for 1 hr at room temperature, followed by incubation with antibody in blocking solution at room temperature for 12–14 hr. The sections were washed in PBHT, and a 1:400 dilution of the secondary antibody (Vector Labs) in blocking solution was applied at room temperature for 3 hr. Slides were washed in PBHT and exposed to HRP conjugated to streptavidin at a dilution of 1:400 in PBHT. After a final PBHT wash, the HRP was reacted with a substrate solution of 1 mg/ml diaminobenzidine and 0.03% H2O2 in PBHT. The unreacted substrate was washed away with water, two times for 5 min each, and the slides were mounted with Glycergel (DAKO). The larvae were fixed and processed whole, essentially like adults. For the embryos, the first fixation was done using 4% formaldehyde, as described below. This was followed by devitellinization in methanol, and a final fixation in Carnoy’s solution. Paraffin impregnation and embedding were then carried out as described for adults. Antigen retrieval was required to detect RUT. After deparaffinization and rehydration, the slides were immersed in 200 ml of H2O in an 18-cm square plastic dish, and microwaved for 10 min at high power. Slides were then transferred to PBHT and processed as above.
PLASTIC EMBEDDING OF EMBRYONIC BRAINS
Canton-S or adh cn pr flies were kept in the dark at 25°C in chambers containing grape juice agar plates. Egg-laden plates were collected from a 2-hr laying period and aged for 18–19 hr at 25°C. Approximately 60% of the eggs have hatched by this time, therefore larvae were removed from the plates using 10% sucrose and water washes; the plates were checked to insure that no larvae remained. Embryos were collected and dechorionated using standard methods for immunohistochemistry. The embryos were fixed with 4% formaldehyde in [180 mm PIPES (pH 6.9), 4 mm MgSO4, 2 mm EGTA] heptane solution (1:1) for 20 min, and then washed three times with 5 mm EGTA in cold 95% methanol, and three times in 100% methanol. The methanol was replaced by PBHT, and the central nervous system was dissected from the embryo by holding the anterior tip of the animal with forceps, and ripping the body wall away with a sharp tungsten needle. The dissected central nervous system was then placed in a 0.6-ml thin-walled PCR tube (a clear tube allows visibility) for immunohistochemistry. Antibody incubations were carried out at room temperature on a Labquake rocker, with the same solutions and durations as described for paraffin sections. After removal of the DAB substrate, the tissue was dehydrated through an ethanol series and embedded in methacrylate according to the manufacturer’s instructions (Polysciences, Inc). One-micron sections were cut with a dry glass knife and mounted on uncoated glass slides with Glycergel (DAKO).
Results
MUSHROOM BODY LOBE CONFIGURATION
Drosophila mushroom bodies are paired structures with their perikarya located on the dorsoposterior surface of the brain. Each cell projects a neurite that extends ventroanteriorly into the dendritic calyx, and an axonal peduncle that terminates in lobes at the anterior face of the brain. The peduncle branches into five lobes (Figs. 1 and 2), two projecting dorsally, α and α′ (Fig. 3, 17–24), and three medially, β, β′, and γ (Fig. 3, 25–32). At the lobe branchpoint, some peduncle fibers fan out laterally to form the “heel” (Fig. 3, 17–24). Previous to this work, only two medially projecting lobes were described in Drosophila, β and γ. However, by examining frontal, sagittal, and horizontal (not shown) brain sections stained with mushroom body markers, it is clear that the medially projecting fibers comprise three distinct bundles. As labeled in Figures 3, 27, and 4g, we refer to the ventral most lobe as β, the middle lobe as β′, and the dorsal lobe as γ. The γ lobe is the most anterior lobe, being the only lobe visible in anterior frontal sections (Fig. 3, 33–40). As one moves posteriorly, β′ and β become visible below γ, such that β′ is sandwiched between the other two (Fig. 3, 25–32). In more posterior sections, γ disappears, and β′ appears to be the most dorsal lobe, lying along the dorsal surface of β (Fig. 3, 19). In the past, the interleaved nature of the medial projections has contributed to difficulties of lobe identification. For example, the γ lobe has been described as wrapping almost entirely around the β lobe in Musca (Fig. 6.5 in Strausfeld 1976). By this criterion, the interior fiber bundle observed in sagittal sections (β′ in Fig. 4g) might be defined as the β lobe (Han et al. 1996). However, the more ventral lobe (β in Fig. 3, 27, and Fig. 4g) has been referred to as β in published frontal sections (Heisenberg 1980; Nighorn et al. 1991; Yang et al. 1995; Ito et al. 1997; Muller 1997), so we conformed to that designation in this paper. The spatial relationship of the α and α′ lobes has been described more consistently (Heisenberg 1980; Ito et al. 1997). We found that α′ wraps closely around α, such that its tip appears lateral to α in frontal sections (Fig. 3, 17–24), and posterior to α in sagittal sections (Fig. 4d–f). This wrapping is particularly clear in Figure 4e. This description of the mushroom body structure reflects the entwined relationship of the lobes. Uncovering their arrangement has necessitated the examination of frontal, sagittal, and horizontal brain sections stained with mushroom body markers. Not only were our markers useful for generally highlighting the mushroom body morphology, but their differential distribution allowed us to distinguish more easily the lobes from one another. For example, FASII was expressed more intensely in the β lobe (Figs. 3, 25, and 4i) than the β′ or γ lobe, which facilitated our consistent identification of these lobes between frontal and sagittal sections. In fact, five different patterns of expression among the lobes were observed with our markers (Fig. 3; Table 1), and they were all consistent with the lobe configuration described above.
Figure 2.
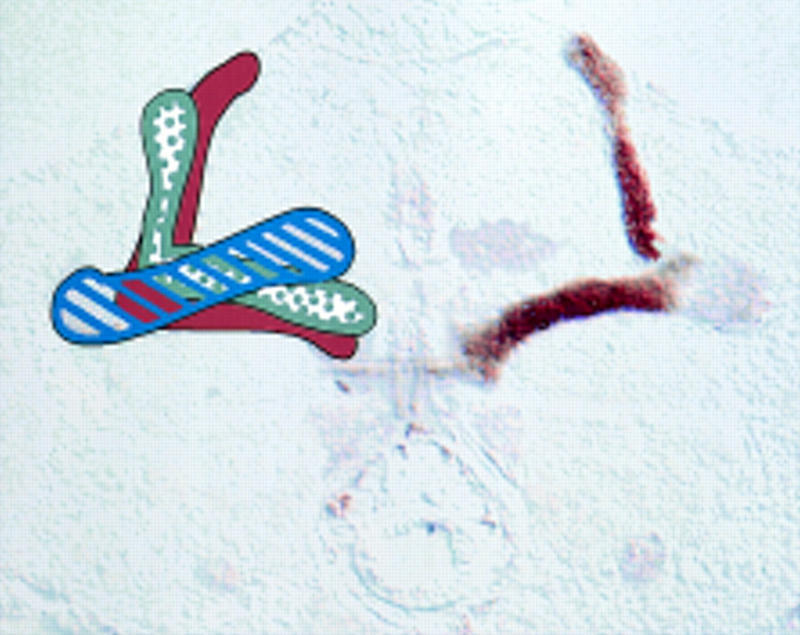
Superimposition of the frontal sections in Fig. 3, 17 and 25, to display all five lobes and the heel in one plane. These sections show the bilateral nature of the mushroom bodies, with the right lobe structure stained with FASII and the left lobe structure colored as in Fig. 1. The heel/γ lobe are striped in blue, the α/β collateral lobes are in solid brown, and the α′/β′ collateral lobes are stippled in gray. Correspondingly, FASII shows three levels of staining: Weak in the heel and γ lobe, strong in the α/β lobes, and absent in the α′/β′ lobes. Because this photograph shows sections through the middle of the brain, at the level of the α/β lobes, only the posterior portion of the γ lobe is visible.
Figure 3.
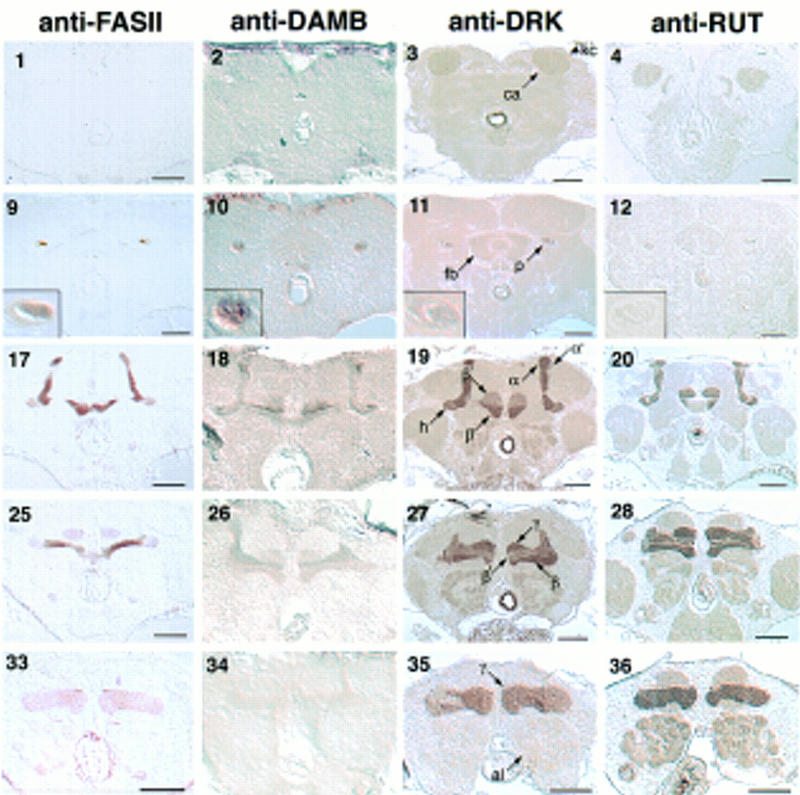
Frontal sections through the adult brain stained for mushroom body markers. Columns are labeled with the antibody used. Each row shows a different plane through the mushroom bodies, advancing from posterior to anterior. (1–8) Calyces (ca) and Kenyon cells (kc); (9–16) peduncles (p) and fan-shaped body (fb). Insets show a magnification of the peduncle to resolve its lateral, central, and medial portions, as drawn in Fig. 1B. Anti-FASII staining demarcates these three fiber tracts by differential staining; the medial peduncle is strongly stained, the narrow central peduncle is unstained, and the lateral peduncle is weakly stained. (17–24) α lobe, α′ lobe, and posterior portions of the β and β′ lobes. The heel (h) is delineated by the weak staining of anti-FASII. (25–32) β′ lobe interleaved between the β lobe and the γ lobe. (33–40) γ lobe above the antennal lobes (al). Bar, 50 μm.
Figure 4.
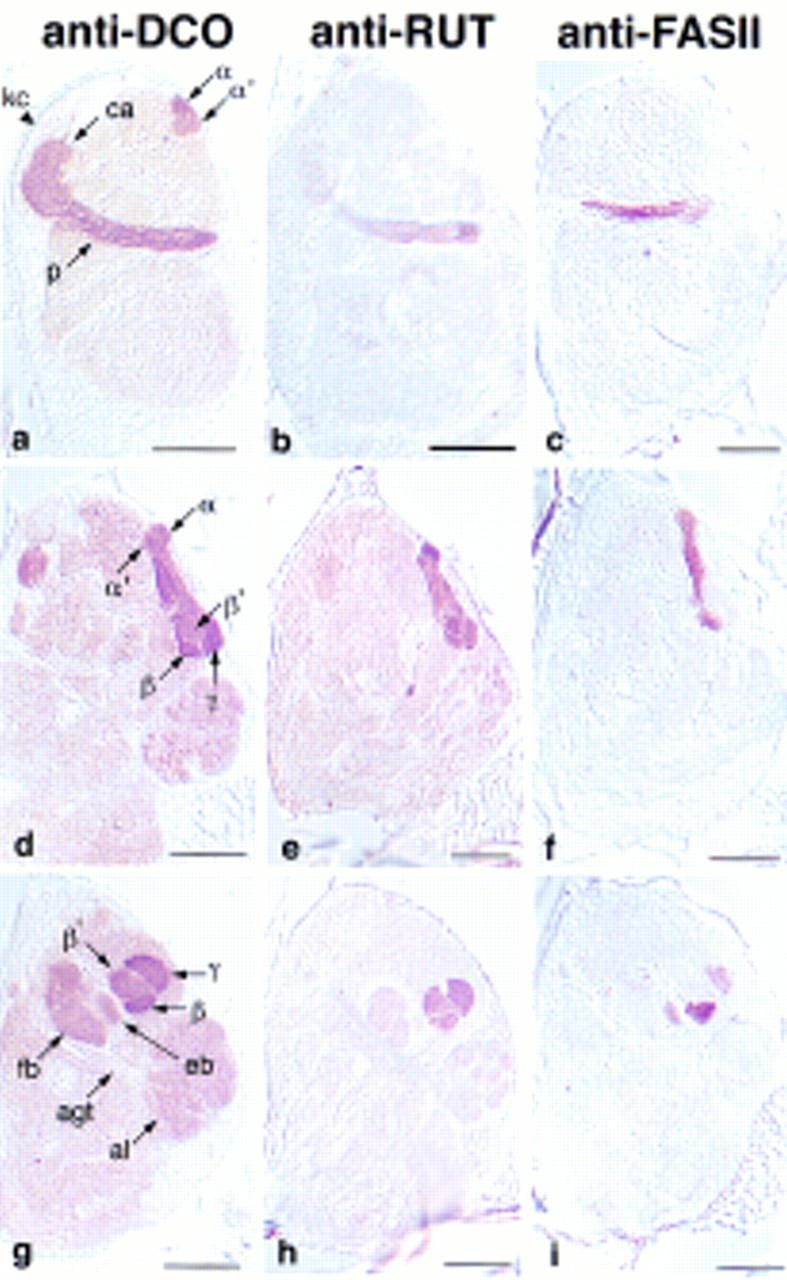
Sagittal sections through the adult brain. Anterior is to the right. Sections advance from lateral in the top row, to medial in the bottom row. (a–c) Kenyon cells, calyx, and peduncles. The tips of the α and α′ lobes are visible with anti-DCO. (d–f) α′ lobe wrapping around the a lobe; d is from a lateral perspective; e and f are from a medial perspective. (g–i) γ lobe covering the anterior face of the β′ lobe; antennal glomerular tract (agt) extending to the mushroom body calyx. Bar, 50 μm. Abbreviations are as in Fig. 3.
Table 1.
Marker expression patterns in mushroom bodies
| FASII | DAMB | DCO | DRK | OAMB | RII | RUT | LEO | |
|---|---|---|---|---|---|---|---|---|
| α′ and β′ | none | medium | medium | weak | strong | medium | strong | medium |
| α and β | strong | weak | strong | strong | weak | strong | strong | medium |
| γ and heel | weak | weak | strong | strong | strong | strong | strong | medium |
| Lat. peduncle | weak | weak | strong | strong | strong | strong | weak | medium |
| Cent. peduncle | none | weak | medium | weak | variable | weak | weak | medium |
| Med. peduncle | strong | weak | strong | strong | weak | strong | weak | medium |
| Calyx | none | none | strong | weak | medium | strong | weak | medium |
| Perikarya | none | none | medium | none | none | medium | none | medium |
For all antibodies, the description refers to how preferential the staining is relative to background. The identification of the lateral, central, and medial peduncle was made in frontal sections at the level of the fan-shaped body.
MARKER EXPRESSION PATTERNS
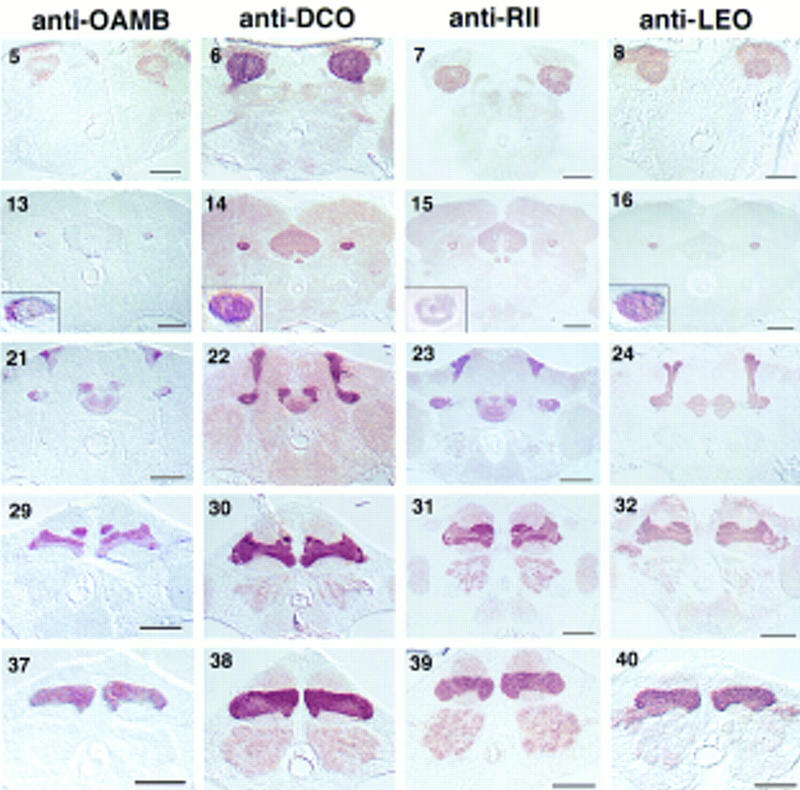
Six of the markers that we examined showed differential expression among the lobes. FASII exhibited the most complex staining pattern, being detected at high levels in the α and β lobes, low levels in the heel and γ lobe, and not at all in the α′ and β′ lobes. DCO, RII, and DRK were all highly expressed in the heel, and α, β, and γ lobes, with somewhat reduced expression in the α′ and β′ lobes. The contrast was slight for DCO, intermediate for RII, and dramatic for DRK. A complementary pattern was found for DAMB. DAMB antibodies highlighted the α′ and β′ lobes, with relatively weak staining in the heel and the α, β, and γ lobes. OAMB displayed yet another pattern, being detected at high levels in the heel and α′, β′, and γ lobes, with intermediate staining of the α and β lobes. Finally, RUT and LEO were present at approximately equal levels in all five lobes. The differential gene expression and discrete morphology of the individual lobes suggests that they constitute functional units of the mushroom bodies.
The markers may also be classified by their localization at the subcellular level to dendrites, axons, or somata. The catalytic and regulatory subunits of PKA (DCO and RII), LEO, and OAMB, were detected in the calyx, lobes, and, with the exception of OAMB, the somata. Because these proteins are found in mushroom body dendrites, they may serve a role in the receipt of sensory information by mushroom bodies. Antibodies to the markers FASII, DAMB, DRK, and RUT preferentially decorated the lobes. The lobes have both afferent and efferent connections, so that these proteins may be serving in either a presynaptic or a postsynaptic capacity of mushroom body neurons. The mechanism by which these proteins are excluded from the calyx is unknown; however, FASII may be localized to the axons by its interaction with DISCS-LARGE-1 (DLG), a PDZ protein responsible for localizing FASII to the neuromuscular junction (Zito et. al. 1997). In summary, the differential distribution of the markers within single neurons, and between different neurons, yielded six different mushroom body expression patterns: (1) ubiquitous; (2) preferential to the calyx, heel, α, β, and γ lobes; (3) preferential to the heel, α, β, and γ lobes (not calyx); (4) specific to the heel, α, β, and γ lobes; (5) preferential to the calyx, α′, β′, and γ lobes; and (6) preferential to the α′ and β′ lobes.
MARKER EXPRESSION PATTERN REVEAL PEDUNCLE FIBER ARRANGEMENT
How are the mushroom body axons organized into lobes? In cross sections through the peduncle, a medial fiber tract and a lateral tract have been defined (Heisenberg 1980; Nighorn et al. 1991; Yang et al. 1995; Ito et al. 1997). The medial tract appears round in frontal sections, and the lateral tract forms a crescent around it (Figs. 1 and 3, 9–16). The medial tract branches to form the α and β lobes, whereas the lateral tract fans out to form the heel, and then turns medially to form the γ lobe (Yang et al. 1995; Ito et al. 1997; J. Crittenden et al., unpubl.). The expression patterns of our markers are accordant with this organization: anti-FASII strongly stains the medial peduncle, which corresponds to the intense staining of the α and β lobes, whereas the weakly staining lateral peduncle corresponds to the low expression in the γ lobe and the heel (Table 1 and Fig. 3, 9–16 insets). Likewise, DRK is expressed strongly in both the medial and lateral peduncle, which parallels its high expression in the α, β, and γ lobes, and heel (Fig. 3 insets). In contrast, anti-OAMB exhibits low staining in the medial peduncle, α, and β lobes, and high staining in the lateral peduncle, the γ lobe, and the heel. In addition to confirming the continuity of the medial tract with the α and β lobes, and the lateral tract with the heel and γ lobe, our markers define a third peduncle tract corresponding to the α′ and β′ lobes. This tract forms a crescent between the medial and lateral tracts, which we refer to as the central peduncle (Fig. 1). This tract is not stained by anti-FASII (Fig. 3, 9 inset), which corresponds to the lack of FASII expression in the α′ and β′ lobes. Similarly, anti-DRK and anti-RII only weakly stain the central peduncle (Fig. 3, 11 and 15 insets) and the α′ and β′ lobes. Uniform staining of the peduncle was observed with anti-LEO (Fig. 3, 16 inset), which equally stains all of the lobes and the heel. Conclusions culd not be drawn from DAMB, OAMB, or RUT, as their expression in the peduncle was weak or variable.
MARKERS DEFINE COLLATERAL MUSHROOM BODY LOBES
Golgi stains reveal that some mushroom body axons branch to form collaterals in two lobes, one projecting dorsally and one projecting medially (Yang et al. 1995). By this criterion, Kenyon cells may have six collateral configurations: α–β, α–β′, α–γ, α′–β, α′–β′, and α′–γ. However, our analysis of marker expression patterns is consistent with only two of these collateral configurations and one unbranched configuration (projecting to only one lobe). This follows from reasoning that equivalent expression of a marker in any two lobes is consistent with their being collateral (assuming even distribution of markers between branches). For example, anti-FASII demarcated three potential fiber bundles by three levels of expression (Fig. 3, 9, 17, and 25): (1) the medial peduncle, α lobe, and β lobe had a high level of expression, (2) the lateral peduncle, heel, and γ lobe had a low level of expression; and (3) the central peduncle, α′ lobe, and β′ lobe had no expression. Remarkably, all of the markers were expressed in patterns consistent with this tripartite arrangement. DRK was expressed at high levels in the α/β lobes and the heel/γ lobe, whereas lower levels were observed in the α′/β′ lobes (Fig. 3, 11, 19, and 27). DCO and RII were expressed in the same pattern but with less specificity (Fig. 3, 14, 15, 22, 23, 30, and 31). A key complementary pattern was exhibited by DAMB, which was expressed strongly in the α′/β′ lobes, and weakly in the α/β lobes and heel/γ lobe (Fig. 3, 18, 26, 34). OAMB expression was also consistent with our model, being high in the α′/β′ lobes, and the heel/γ lobe, but low in the α/β lobes (Fig. 3, 21, 29, 37). Taken together, the expression patterns of these markers suggested the following tripartite organization of mushroom body projections: (1) medial peduncle fibers project to the α and β lobe; (2) lateral peduncle fibers project to the heel and γ lobe; and (3) central peduncle fibers project to the α′ and β′ lobes. Considering the large number of mushroom body neurons, exceptions to these proposed types are not unlikely. Still, it is striking that of the diverse markers we examined here, all had expression patterns consistent with our proposed configuration. This suggests that many mushroom body neurons have one of three projection types.
All of the markers except FASII had some general neuropil expression in the brain. A number of the markers examined here also showed expression in the central complex. FASII, LEO, DRK, DCO, RII, and OAMB were all expressed in the ellipsoid body. FASII, DCO, RII, DAMB, and DRK were lightly expressed in the fan-shaped body.
ADULT MUSHROOM BODY MARKERS LABEL LARVAL MUSHROOM BODIES
The expression of the markers was also analyzed in paraffin sections of wandering third instar larvae. The preferential labeling of mushroom bodies was striking for all, as exemplified by anti-FASII, anti-RII, anti-DCO, and anti-DRK staining in Figures 5 and 6. Furthermore, the subcellular distribution patterns were conserved, where DCO, LEO, OAMB, and RII were detected in both the dendritic calyx and the axonal lobes, and DAMB, DRK, FASII, and RUT were confined to axons. Anti-FASII staining of frontal brain sections nicely revealed the morphology of the larval mushroom bodies (Fig. 5). The overall organization of the larval mushroom bodies was the same in the larva as in the adult; dorsoposterior cells project to form a calyx and a peduncle that branches into a heel, medially projecting lobes, and dorsally projecting lobes. There appeared to be two dorsally projecting lobes, one large and medial, the other smaller and curved laterally (Fig. 5f,g). By relative size and position, the large one may be α, and the smaller one α′. Likewise, there are several medially projecting lobes that may correspond to β, β′, and γ. There is also a fiber bundle projecting anteriorly from the heel, which appears round in frontal sections, as seen in Figure 5h. The identity of the larval lobes could not be analyzed by marker expression patterns as they were not stained differentially. This developmental change, from apparent uniform to restricted expression in the lobes, has also been reported for dnc (Nighorn et al. 1991).
Figure 5.
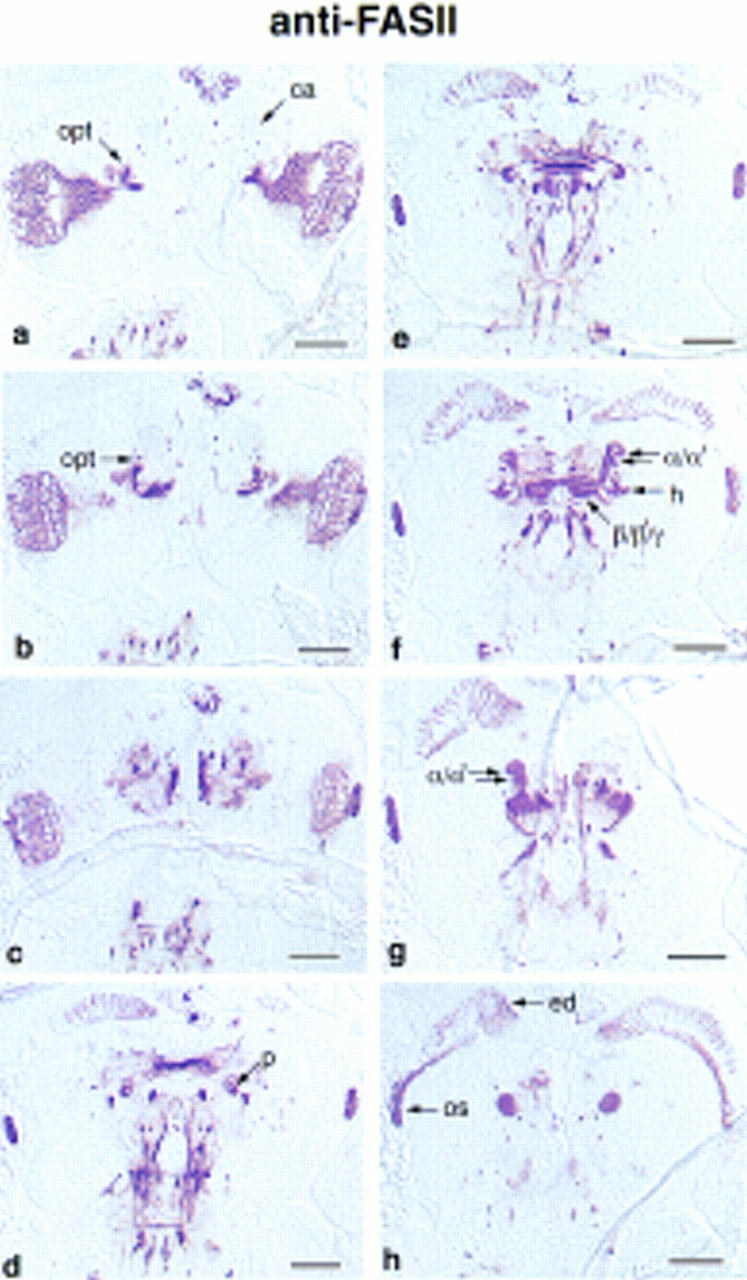
Frontal sections through the brain of a wandering third instar larva stained with anti-FASII. Sections advance from posterior to anterior. (a) Calyces. The developing lamina, medulla, and optic tract (opt) are heavily stained. Presumptive ring gland neurons are visible between the brain hemispheres. The stained longitudinal connectives and lateral commissures of the vental nerve cord are visible at the bottom of the photograph. (c) Optic tract appears to be reaching the calyces; (d) hollow peduncles; (f) heel and lobes; (g) lobes; only the γ lobe is visible on the right side. (h) Section through the fiber bundle that extends anteriorally from the heel. The eye disc (ed) is extending fibers into the brain through the optic stalk (os). Bar, 50 μm. Abbreviations are as in Fig. 3.
Figure 6.
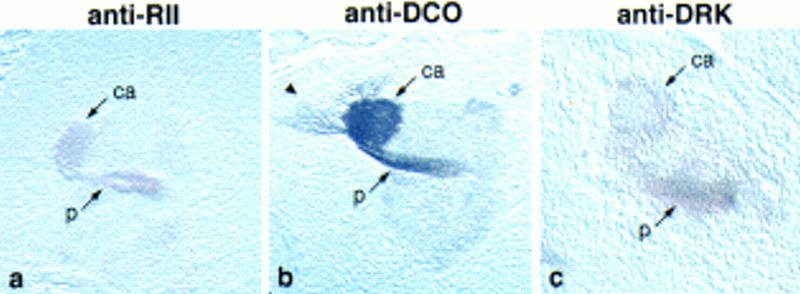
Sagittal sections through single brain hemispheres of wandering third instar larvae showing the calyx and peduncle stained with (a) anti-RII, (b) anti-DCO, and (c) anti-DRK. Fibers from the Kenyon cells are labeled with anti-DCO, as indicated by the arrowhead.
One curious feature of the larval mushroom bodies was the reduced marker expression in the core of the peduncle and lobes, as demonstrated with FASII in Figure 5. Even LEO and RUT, which are distributed throughout the mushroom bodies in adults, do not stain this core. A similar staining pattern has also been described for some P[Gal4]/UAS–lacZ reporter lines (Tettamanti et al. 1997). General protein stains, such as Bodian and hematoxylin/eosin, show reduced staining in the core, suggesting that it may have a different composition (Demerec 1994; J. Crittenden et al., unpubl.). Indeed, electron microscopy shows that the center of the larval peduncle consists of very thin fibers, which are maintained during the retraction of mushroom body axons that occurs in pupariation (Technau and Heisenberg 1982). One possibility is that these are pathfinding neurons that have specialized gene expression. They do not correspond completely to the fibers growing out during embryogenesis, however, as several of our markers are expressed in embryonic mushroom bodies (described below).
Kenyon cells derive from four neuroblasts that divide continuously throughout development (Ito and Hotta 1992). These neuroblasts lie at the dorsoposterior surface of the embryonic and larval brain, and produce columns of neuronal and glial progeny (Ito et al. 1997). Columns of Kenyon cells stained with anti-LEO are visible descending from large neuroblasts in Figure 7. Only three mushroom body neuroblasts were visible in any one section, reflecting their consistently nonplanar arrangement. None of the markers were detected in the neuroblast cell bodies of wandering third instar larvae, indicating that their expression may be differentiation dependent.
Figure 7.
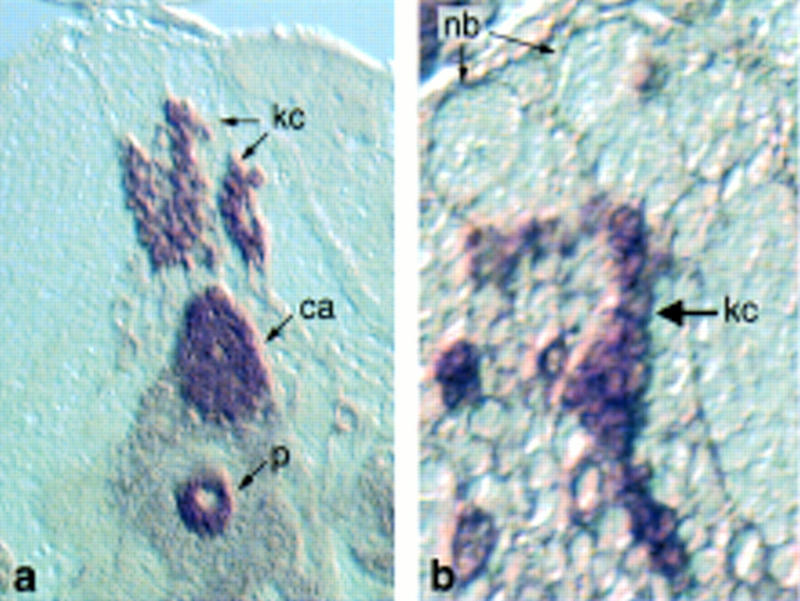
Frontal section of the mushroom body from a wandering third instar larva stained for LEO. (a) Columns of LEO-positive Kenyon cells projecting to the calyx, (b) posterior section showing columns of Kenyon cells delaminating from presumptive mushroom body neuroblasts (nb).
EMBRYONIC MUSHROOM BODIES
Little is known about how the mushroom body structure initially forms during embryogenesis, but the late expression of our mushroom body markers suggests that substantial development occurs after the beginning of stage 17. By this time, the fundamental body plan has been established, and the animal has secreted a tough, hydrophobic larval cuticle. The impenetrability of this cuticle to antibodies confounds the examination of development that occurs in the hours just before hatching. To overcome this technical problem, we dissected the central nervous system from embryos, performed immunocytochemistry with potential mushroom body markers, and sectioned the brain to examine mushroom body development and gene expression.
Plastic sections of brains stained with anti-FASII revealed that the mushroom body structure is recognizable by 21 hr (25°C) of development, just before hatching (Fig. 8). This is consistent with the observation that first instar larvae already have 300 fibers in the peduncle (Technau and Heisenberg 1982). The peduncle and α lobes are visible in a sagittal section stained with anti-FASII (Fig. 8a), and an oblique frontal section reveals the β/γ lobes (Fig. 8b). Additional FASII expression was observed in the cervical connectives, as described in the larval brain. Embryonic mushroom body expression was also observed with the markers DCO, RII, and LEO, as shown from a sagittal perspective in Figure 9. Antibodies to DCO decorated the Kenyon cells, the peduncle, and the α/α′ lobes (Fig. 9a). The calyx, just under the Kenyon cell bodies, was revealed by anti-LEO, as were the anterior portion of the β/γ lobes (Fig. 9b). The intense neuropil labeling by anti-RII made the mushroom body neurites difficult to distinguish, but expression in the Kenyon cells was clearly visible (Fig. 9c). All four markers displayed the same subcellular localization as in adults and larvae; DCO, RII, and LEO were found throughout the cell, whereas FASII was restricted to axons. Also corresponding to the adult profile was the high-contrast staining of anti-FASII, and general neuropil expression of DCO, RII, and LEO. In summary, the mushroom body calyx, peduncle, and dorsally and medially projecting lobes are all formed by late stages of embryogenesis.
Figure 8.
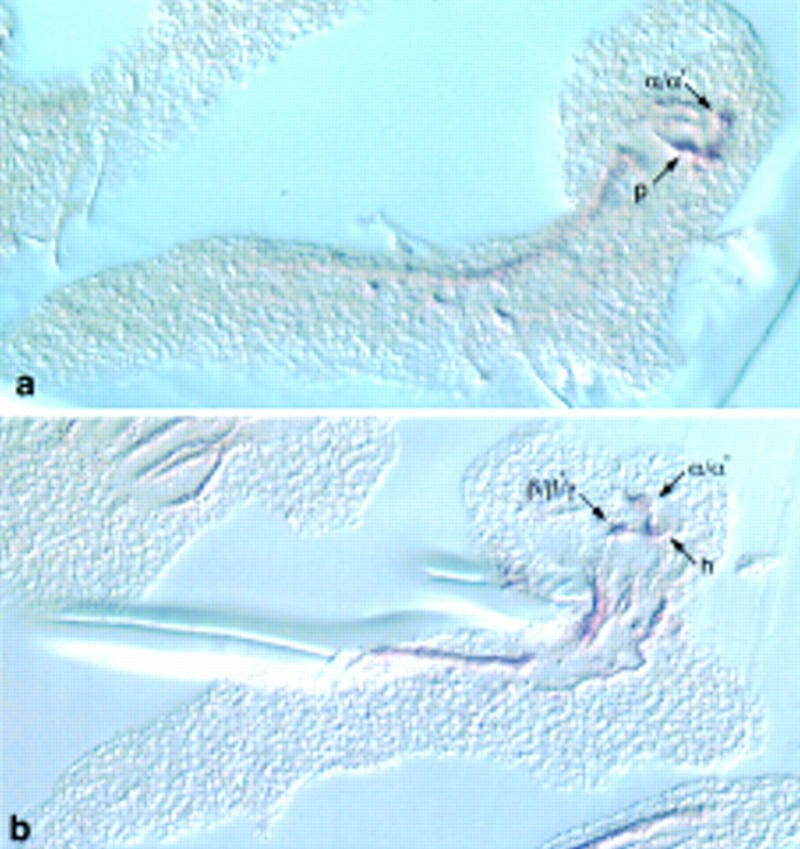
Sections of embryonic central nervous system stained to detect anti-FASII. (a) Sagittal section of FASII-positive peduncle and dorsally projecting lobes (α/α′); (b) oblique frontal section of dorsally and medially projecting lobes (β/β′).
Figure 9.
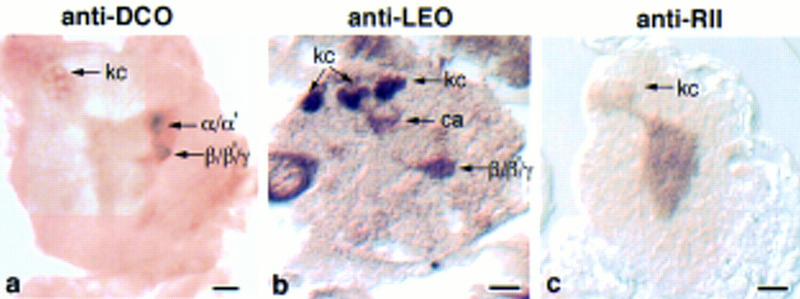
Sagittal sections of embryonic brains stained for markers. (a) Anti-DCO labels the Kenyon cells (kc) and dorsally projecting lobes. (b) anti-LEO detects the calyx and anterior lobes, anti-MEF2 labels the Kenyon cell nuclei. (c) Anti-RII labels the Kenyon cells. Bar, 10 μm.
MUSHROOM BODY NEUROPIL IS NO MORE DENSE THAN OTHER BRAIN NEUROPIL
We addressed the issue of whether the observed marker expression patterns were attributable to differential fiber density. To test this, we used two methods, staining with anti-horseradish peroxidase (HRP) and hematoxylin/eosin. Anti-HRP was used as a neuropil marker because it detects the β subunit of a Na+/K+ ATPase (Sun and Salvaterra 1995), a protein expected to be expressed on all neuronal membranes. Anti-HRP did not decorate preferentially any one of the mushroom body lobes, suggesting that they are not dramatically different in fiber density (Fig. 10). The only area of the mushroom bodies that showed preferential HRP staining was a short stretch of the peduncle, just anterior to the calyx (Fig. 10b). Hematoxylin/eosin staining of the brain similarly gave no indication that any of the mushroom body lobes are particularly compact, relative to each other, or to other brain areas (data not shown). Therefore, the staining patterns of our markers are a reflection of actual protein concentration and not fiber density.
Figure 10.
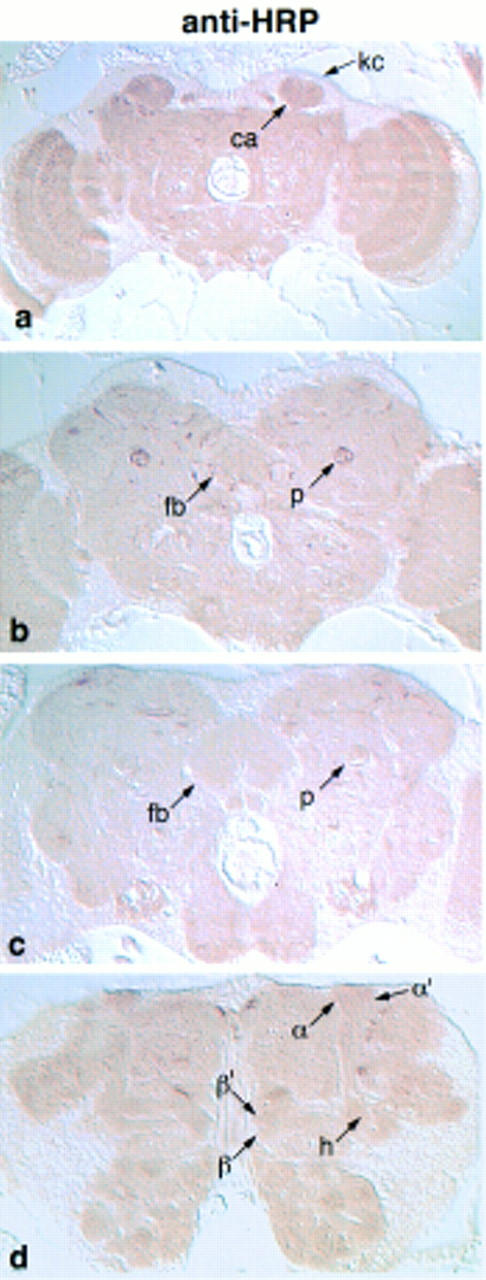
Frontal paraffin sections through an adult brain challenged with anti-HRP. Sections advance from posterior to anterior. (b) Staining of the peduncle at the posterior edge of the fan-shaped body. This is the only area of the mushroom body that showed preferential anti-HRP labeling.
Discussion
MUSHROOM BODY ARCHITECTURE
Several morphological findings have emerged from our analysis of mushroom body sections stained for antigenic markers. Our markers labeled two dorsally projecting mushroom body lobes and three medially projecting lobes. Six of the markers were expressed differentially expressed between the lobes, in patterns that were constant from animal to animal; therefore, we were able to identify consistently each lobe from multiple perspectives. For example, by examining both frontal and sagittal sections, we were able to determine that one of the dorsally projecting lobes, α′, wraps mediolaterally around the other, α. Likewise, by differential staining, the markers allowed us to resolve the interleaving of the medially projecting lobes, β, β′, and γ. β′ lies along the dorsal surface of β, and together they fill the neuropil space between the ellipsoid body and the γ lobe (Fig. 4g). β and β′ have a similar shape when viewed in horizontal sections, as if they are duplicate structures. The γ lobe lies anterior to the β and β′ lobes, and enwraps the anteriodorsal face of β′ (Fig. 4g–i). The marker expression patterns were also used to determine how the peduncle fibers are organized into lobes. In frontal sections, at the level of the fan-shaped body, the peduncle is visibly divided into three concentric fiber tracts, termed the medial, central, and lateral peduncles. We reasoned that a tract was continuous with the lobes that exhibited equivalent marker staining. The deduced organization was again consistent among markers, and indicated that the medial peduncle projects to the α and β lobes, the central peduncle to the α′ and β′ lobes, and the lateral peduncle to the heel and γ lobe. From frontal and horizontal sections (data not shown), we observed that the organization of the peduncle into three tracts occurs just posterior to the fan-shaped body. The peduncle originates, however, from the fasciculation of four axon tracts that derive from four Kenyon cell clusters (Ito et al. 1997). Interestingly, each of the Kenyon cell clusters contributes to all five lobes and the heel (Ito et al. 1997). Therefore, between the calyx and the fan-shaped body, the four peduncular tracts reorganize into three, such that fibers derived from distinct cell clusters come together and project to the same lobe. A similar case exists for the honeybee, where projections from separate Kenyon cell clusters fasciculate to form an individual strata in the peduncle (Mobbs 1982).
Information regarding the axon morphology of individual mushroom body cells was derived from directly comparing the expression patterns of each marker. Because a single mushroom body cell can extend an axon that branches into a dorsally projecting lobe and a medially projecting lobe (Mobbs 1982; Yang et al. 1995), a marker expressed in that cell will stain each of the two lobes (assuming it is distributed homogeneously). If the marker is expressed preferentially in the group of cells with the same projection pattern (i.e., α and β), that marker will stain preferentially the corresponding lobes. Many of our markers stained preferentially specific pairs of lobes. Furthermore, there was a correlation between which lobes were stained, such that equal staining intensities were observed in the following sets: (1) α and β lobes, (2) α′ and β′ lobes, and (3) the γ lobe and heel. Different markers were expressed in different sets of lobes, indicating that the staining levels were not simply a reflection of fiber density. Consistent with this, the density of neuropil in the lobes appeared equivalent with the control neuronal marker anti-HRP. Remarkably, all of our markers defined the same three sets of lobes, suggesting that the majority of mushroom body cells may be classified into one of three projection types. Of course, the classification of neurons according to their lobe projections does not prove that they are similar in other aspects. In the honeybee, Kenyon cells of differing dendritic morphologies send projections into both the α and β lobes (Mobbs 1982). The honeybee mushroom bodies are further divided into seven strata per lobe, according to the Kenyon cell type and calycal region that they comprise (Mobbs 1982). We anticipate further divisions of the Drosophila mushroom bodies as well, as the organization proposed here reflects only one, albeit fundamental, parameter of cell classification.
To determine the proportion of Kenyon cells that extend axons according to our proposed architecture, it would be of benefit to analyze more markers. One superb resource for this are the many P[Gal4] mushroom body enhancer trap lines that have been isolated in recent years (Yang et al. 1995; Ito et al. 1997). We have examined the published expression pattern of one P[Gal4]/UAS–lacZ line, 201Y (Yang et al. 1995). This line shows β-galactosidase staining in the heel, α, β, and γ lobes. There does not appear to be staining in the α′ or β′ lobe. This pattern is consistent with our proposed collaterals. Furthermore, Yang et al. (1995) observed age-dependent loss of staining in the medial peduncle, α lobe and β lobe–one collateral group. Staining in the other bundle, lateral peduncle fibers projecting to the heel and γ lobe, was maintained. The loss of lacZ expression in the α lobe but not in the γ lobe suggests that there may not be collateral fibers between these lobes. Consistent with this is the expression pattern driven by the P[Gal4] line H24, which is restricted to the γ lobe as described in this issue (M. Heisenberg, pers. comm.). Reporter gene lines are particularly useful to confirm our proposed collateral organization because products such as β-galactosidase are unlikely to be differentially transported into different branches of one axon.
It is interesting to note the similarity between the α/β and α′/β′ bundles; they have a similar shape and location, such that they contact some of the same surrounding neuropil. The α and α′ lobes extend dorsoposteriorly, while the β and β′ lobes both abut the anterior face of the ellipsoid body. The heel/γ lobe bundle is distinct, as it may not have a dorsal collateral, does not contact the ellipsoid body, and has a more tubular shape. The structural similarity of the α/β and α′/β′ groups suggests that they may have arisen by duplication. Because each of the four mushroom body neuroblasts contribute to each of the lobe groups, the duplication of the α/β group may have preceded the multiplication of mushroom body neuroblasts. Interestingly, only some of the lobe groups are found in the honeybee, which have collateral α and β lobes, but no indication of a α′/β′ grouping nor of a γ lobe (Mobbs 1982). Taken together, one might propose a model where a common ancestor of Drosophila and honeyee had a single mushroom body neuroblast that produced only the α/β fiber bundle. This organization was retained in the honeybee. In contrast, further changes occurred in Drosophila, so that the neuroblast gave rise to the three distinct lobe groups. Finally, multiplication of this neuroblast produced the four mushroom body neuroblasts we see today.
MUSHROOM BODY MARKERS IN TIME
We have followed the markers throughout development. All of the markers in Figure 3 were expressed in larval mushroom bodies. FASII, DCO, RII, and LEO were expressed in embryonic mushroom bodies as well. The basic structure of larval and embryonic mushroom bodies corresponded to that of adults, with a calyx, a heel, and medially and dorsally projecting lobes. The brain somata comprised a large portion of the larval and embryonic brains, so that the mushroom body neuropil appeared compacted in the center. Of the neuropil, however, a large proportion comprised the mushroom body structure. Perhaps this reflects the burrowing life-style of the larvae, where olfactory cues may predominate. Indeed, mushroom bodies are important for olfactory conditioning in larvae, as demonstrated by defects in mbm (Heisenberg et al. 1985). Does larval learning require the same signal transduction mechanisms as those mediating adult olfactory conditioning? This is likely to be true of the cAMP pathway, as mutants for the mushroom body gene dnc, which encodes a cAMP phosphodiesterase, are deficient in larval olfactory conditioning (Aceves-Pina and Quinn 1979; Tully et al. 1994). DCO, LEO, and RUT may also be involved in larval learning, as they are expressed in the larval mushroom bodies and are required for normal adult olfactory conditioning. It will be interesting to determine whether the physiological functions of the other mushroom body markers are conserved between larvae and adults as well.
MUSHROOM BODY STRUCTURE AND FUNCTION
The ultimate goal of defining the anatomical subdivisions of the mushroom bodies is to understand the underlying functional divisions. The identity of the genes presented here point to which signal transduction mechanisms may be at work in different lobe sets. For example, the preferential expression of the dopamine receptor DAMB in the α′ and β′ lobes implicates these lobes as the sites of dopaminergic input within the mushroom bodies. Moreover, it may be possible to map directly the function of learning and memory genes to specific lobes by spatially restricted rescue experiments; mutants for DCO, rut, and leo have impaired olfactory conditioning, therefore by testing mutants that express wild-type transgenes in different sets of lobes, one may determine which lobes are involved in learning. Similar experiments might even identify specific cell groups that receive olfactory inputs preferentially, as is the case with the cells that project to the lip of the honeybee calyx (Mobbs 1982). Another approach for determining the functional correlates of mushroom body subdivisions are lobe-specific ablation experiments (Mizunami et al. 1993). Ablation of lobes by genetic means would be especially desirable to establish populations of mutant animals. Toxins that induce cell death or inhibit synaptic transmission could be used to interfere with the function of a specific lobe set by way of the Gal4/UAS system (Kunes and Steller 1991; Bellen et al. 1992; Moffat et al. 1992; Brand and Perrimon 1993; Sweeney et al. 1995). It may also be feasible to ablate lobes with genetic lesions as disrupting the formation of the mushroom bodies is not a lethal event (de Belle and Heisenberg 1994). Immunohistochemical screens for mutants defective in an individual lobe may be expediated with the use of the markers presented here. Dissecting the physiology of the mushroom body substructure will enlighten our comprehension of complex behavior.
Acknowledgments
We thank K.-H. Wu for his discovery that FASII is expressed in mushroom bodies. We also thank S. Ahmed and B. Schoeder for technical assistance, and S. Meuser for critical reading of the manuscript. This work was supported by a predoctoral National Institute of Mental Health grant to J.R.C., and an National Institutes of Health grant to R.L.D. R.L.D. is the recipient of the R.P. Doherty-Welch Chair in Science at the Baylor College of Medicine.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
References
- Aceves-Pina EO, Quinn WG. Learning in normal and mutant Drosophila larvae. Science. 1979;206:93–95. doi: 10.1126/science.206.4414.93. [DOI] [PubMed] [Google Scholar]
- de Belle JS, Heisenberg M. Associative learning in Drosophila abolished by chemical ablation of mushroom bodies. Science. 1994;263:692–695. doi: 10.1126/science.8303280. [DOI] [PubMed] [Google Scholar]
- Bellen HJ, D’Evelyn D, Harvey M, Elledge SJ. Isolation of temperature-sensitive diphtheria toxins in yeast and their effects on Drosophila cells. Development. 1992;114:787–796. doi: 10.1242/dev.114.3.787. [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Demerec M. Biology of Drosophila. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- Erber J. Response characteristics and after effects of multimodal neurons in the mushroom body area of the honey bee. Physiol Entomol. 1978;3:77–89. [Google Scholar]
- Erber J, Masuhr T, Menzel R. Localization of short-term memory in the brain of the bee, Apis mellifera. Physiol Entomol. 1980;5:343–358. [Google Scholar]
- Gronenberg W. Anatomical and physiological properties of feedback neurons of the mushroom bodies in the bee brain. Exp Biol. 1987;46:115–125. [PubMed] [Google Scholar]
- Hammer M, Menzel R. Learning and memory in the honeybee. J Neurosci. 1995;15:1617–1630. doi: 10.1523/JNEUROSCI.15-03-01617.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han P-L, Levin LR, Reed RR, Davis RL. Preferential expression of the Drosophila rutabaga gene in mushroom bodies, neural centers for learning in insects. Neuron. 1992;9:619–627. doi: 10.1016/0896-6273(92)90026-a. [DOI] [PubMed] [Google Scholar]
- Han K-A, Millar NS, Grotewiel MS, Davis RL. DAMB a novel dopamine receptor expressed specifically in Drosophila mushroom bodies. Neuron. 1996;16:1127–1135. doi: 10.1016/s0896-6273(00)80139-7. [DOI] [PubMed] [Google Scholar]
- Han, K.-A., N.S. Millar, and R.L. Davis. 1998. A novel octapamine receptor with preferential expression in Drosophila mushroom bodies. J. Neurosci. (in press). [DOI] [PMC free article] [PubMed]
- Heisenberg M. Mutants of brain structure and function: What is the significance of the mushroom bodies for behaviour? In: Siddiqi O, Babu P, Hall LM, Hall JC, editors. Development and biology of Drosophila. New York, NY: Plenum; 1980. pp. 337–390. [DOI] [PubMed] [Google Scholar]
- Heisenberg M, Borst A, Wagner S, Byers D. Drosophila mushroom body mutants are deficient in olfactory learning. J Neurogenet. 1985;2:1–30. doi: 10.3109/01677068509100140. [DOI] [PubMed] [Google Scholar]
- Ito K, Hotta Y. Proliferation pattern of postembryonic neuroblasts in the brain of Drosophila melanogaster. Dev Biol. 1992;149:134–148. doi: 10.1016/0012-1606(92)90270-q. [DOI] [PubMed] [Google Scholar]
- Ito K, Awano W, Suzuki K, Hiromi Y, Yamamoto D. The Drosophila mushroom body is a quadruple structure of clonal units each of which contains a virtually identical set of neurones and glial cells. Development. 1997;124:761–771. doi: 10.1242/dev.124.4.761. [DOI] [PubMed] [Google Scholar]
- Kunes S, Steller H. Ablation of Drosophila photoreceptor cells by conditional expression of a toxin gene. Genes & Dev. 1991;5:970–983. doi: 10.1101/gad.5.6.970. [DOI] [PubMed] [Google Scholar]
- MacLeod K, Laurent G. Distinct mechanisms for synchronization and temporal patterning of odor-encoding neural assemblies. Science. 1996;274:976–979. doi: 10.1126/science.274.5289.976. [DOI] [PubMed] [Google Scholar]
- Mizunami M, Weibrecht JM, Strausfeld NJ. A new role for the insect mushroom bodies: Place memory and motor control. In: Beer RD, Ritzmann R, McKenna T, editors. Biological neural networks in invertebrate neuroethology and robotics. Cambridge, MA: Academic Press; 1993. pp. 199–225. [Google Scholar]
- Mobbs PG. The brain of the honeybee Apis mellifera. I. The connections and spatial organization of the mushroom bodies. Philos Trans R Soc Lond Ser B. 1982;298:309–354. [Google Scholar]
- Moffat KG, Gould JH, Smith HK, O’Kane CJ. Inducible cell ablation in Drosophila by cold-sensitive ricin A chain. Development. 1992;114:681–687. doi: 10.1242/dev.114.3.681. [DOI] [PubMed] [Google Scholar]
- Muller U. Neuronal cAMP-dependent protein kinase type II is concentrated in mushroom bodies of Drosophila melanogaster and the honeybee Apis mellifera. J Neurobiol. 1997;33:33–44. doi: 10.1002/(sici)1097-4695(199707)33:1<33::aid-neu4>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- Nighorn A, Healy MJ, Davis RL. The cyclic AMP phosphodiesterase encoded by the Drosophila dunce gene is concentrated in the mushroom body neuropil. Neuron. 1991;6:455–467. doi: 10.1016/0896-6273(91)90253-v. [DOI] [PubMed] [Google Scholar]
- Rybak J, Menzel R. Anatomy of the mushroom bodies in the honey bee brain: The neuronal connections of the alpha-lobe. J Comp Neurol. 1993;334:444–465. doi: 10.1002/cne.903340309. [DOI] [PubMed] [Google Scholar]
- Skoulakis EMC, Davis RL. Olfactory learning deficits in mutants for leonardo, a Drosophila gene encoding a 14-3-3 protein. Neuron. 1996;17:931–944. doi: 10.1016/s0896-6273(00)80224-x. [DOI] [PubMed] [Google Scholar]
- Skoulakis EMC, Kalderon D, Davis RL. Preferential expression in mushroom bodies of the catalytic subunit of protein kinase A and its role in learning and memory. Neuron. 1993;11:197–208. doi: 10.1016/0896-6273(93)90178-t. [DOI] [PubMed] [Google Scholar]
- Strausfeld NJ. Atlas of an insect brain. Heidelberg, Germany: Springer Verlag; 1976. [Google Scholar]
- Sun B, Salvaterra PM. Two Drosophila nervous system antigens, Nervana 1 and 2 and homologous to the beta subunit of Na+, K+-ATPase. Proc Natl Acad Sci. 1995;92:5396–5400. doi: 10.1073/pnas.92.12.5396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney ST, Broadie K, Keane J, Niemann H, O’Kane CJ. Targeted expression of tetanus toxin light chain in Drosophila specifically eliminates synaptic transmission and causes behavioral defects. Neuron. 1995;14:341–351. doi: 10.1016/0896-6273(95)90290-2. [DOI] [PubMed] [Google Scholar]
- Technau G, Heisenberg M. Neural reorganization during metamorphosis of the corpora pedunculata in Drosophila melanogaster. Nature. 1982;295:405–407. doi: 10.1038/295405a0. [DOI] [PubMed] [Google Scholar]
- Tettamanti M, Armstrong JD, Endo K, Yang MY, Furukubo-Tokunaga K, Kaiser K, Reichert H. Early development of the Drosophila mushroom bodies, brain centres for associative learning and memory. Dev Genes Evol. 1997;207:242–252. doi: 10.1007/s004270050112. [DOI] [PubMed] [Google Scholar]
- Tully T, Cambiazo V, Kruse L. Memory through metamorphosis in normal and mutant Drosophila. J Neurosci. 1994;14:68–74. doi: 10.1523/JNEUROSCI.14-01-00068.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang MY, Armstrong JD, Vilinsky I, Strausfeld NJ, Kaiser K. Subdivision of the Drosophila mushroom bodies by enhancer-trap expression patterns. Neuron. 1995;15:45–54. doi: 10.1016/0896-6273(95)90063-2. [DOI] [PubMed] [Google Scholar]
- Zito K, Fetter RD, Goodman CS, Isacoff EY. Synaptic clustering of Fasciclin II and Shaker: Essential targeting sequences and role of Dlg. Neuron. 1997;19:1007–1016. doi: 10.1016/s0896-6273(00)80393-1. [DOI] [PubMed] [Google Scholar]


