Abstract
microRNAs (miRNAs) are the most abundant class of small RNAs in mammals. They play an important role in regulation of gene expression by inducing mRNA cleavage or translational inhibition. Each miRNA targets an average of 100–200 genes by binding, preferentially, to their 3′ UTRs by means of partial sequence complementarity. Most miRNAs are localized within transcriptional units, termed host genes, and show similar expression behavior with respect to their corresponding host genes. Considering the impact of miRNA in the regulation of gene expression and their involvement in a growing number of human disorders, it is vital to develop sensitive computational approaches able to identify miRNA target genes. The HOCTAR database (db) is a publicly available resource collecting ranked list of predicted target genes for 290 intragenic miRNAs annotated in human. HOCTARdb is a unique resource that integrates miRNA target prediction genes and transcriptomic data to score putative miRNA targets looking at the expression behavior of their host genes. We demonstrated, by testing 135 known validated target genes (either at the translational or transcriptional level) for different miRNAs, that the miRNA target prediction lists present in HOCTARdb are highly reliable. Moreover, HOCTARdb associates biological roles to each miRNA-controlled transcriptional network by means of Gene Ontology analysis. This information is easily accessible through a user-friendly query page. The HOCTARdb is available at http://hoctar.tigem.it/. We believe that a detailed relationship between miRNAs and their target genes and a constant update of the information contained in HOCTARdb will provide an extremely valuable resource to assist the researcher in the discovery of miRNA target genes.
Abbreviations: TFEB, transcription factor EB; HTT, huntingtin; TMEM49, transmembrane protein 49; AKT1, v-akt murine thymoma viral oncogene homolog 1
Keywords: microRNA, Target prediction
1. Introduction
MicroRNAs (miRNAs) are potent modulators of gene expression (He and Hannon, 2004). They have been found to play critical roles in a growing number of biological functions and human diseases (Thai et al.; Dalmay and Edwards, 2006; Chang and Mendell, 2007; Zhang, 2008; Meola et al., 2009). Structurally, miRNAs are single-stranded non-coding RNAs of 19–25 nucleotides in length, generated from hairpin-shaped transcripts. miRNAs cause translational repression and mRNA degradation by binding, with an imperfect pairing, to the 3′ untranslated region (3′ UTR) of their target genes (Kim, 2005; Giraldez et al., 2006). Current estimates indicate that each miRNA may regulate, on average, the expression of 100–200 mRNAs (Ambros, 2004; Lim et al., 2005). Currently, there are 718 annotated miRNAs in human (Griffiths-Jones, 2004), localized in intragenic (60% of cases) or in intergenic (40%) regions. Intragenic miRNAs are contained within transcriptional units termed “host genes” (Kim et al., 2009), which are generally protein-coding. Recent work demonstrated that many intronic miRNAs and their host genes are co-transcribed from a common promoter (Rodriguez et al., 2004; Kim and Kim, 2007). The large impact of miRNAs in the regulation of biological processes has determined a strong interest in novel technologies for the detection/prediction of miRNA target genes (Thomas et al., 2010) to dissect their regulatory gene networks and understand their function. Current efforts are dedicated to both experimental (Baek et al., 2008; Selbach et al., 2008) and bioinformatic (Krek et al., 2005; Betel et al., 2008; Friedman et al., 2009; Gennarino et al., 2009) approaches to address miRNA target identification. However, giving the laborious nature of the experiments needed for target validation, and considering that most ad-hoc developed high-throughput techniques (e.g. p-Silac) are costly and only validate a few target genes (Lim et al., 2005), it is imperative to improve in silico approaches to identify miRNA target genes.
Recently we have developed a new and efficient approach to perform miRNA target prediction, the HOCTAR (Host Gene Oppositely Correlated Targets) procedure (Gennarino et al., 2009). This procedure is based on gene expression analysis and exploits the inverse correlation existing between the expression behavior of a host gene and the target genes of the corresponding intragenic miRNA. The analysis of dozens of validated miRNA targets showed that the HOCTAR procedure is generally applicable to all intragenic miRNAs and that its performance overruns that of first-generation prediction softwares, which are based on sequence analysis alone (Gennarino et al., 2009).
In this paper we describe the HOCTAR database (HOCTARdb), which collects the results of HOCTAR analyses for 290 human intragenic miRNAs. The HOCTAR database is publicly available to the scientific community at the web address http://hoctar.tigem.it.
2. Materials and methods
2.1. The HOCTAR procedure
The lists of putative target genes were compiled, using default parameters, by retrieving the latest releases of PicTar (release March 2007) (Krek et al., 2005), TargetScan (release 5.1) (Friedman et al., 2009) and miRanda (release September 2008) (Betel et al., 2008). In the case of TargetScan (Friedman et al., 2009), we considered the predicted conserved targets. For PicTar, we selected the targets which were found to be conserved in mammalian genomes (Krek et al., 2005). For each intragenic miRNA, we compiled a non-redundant list of predicted mRNA targets by pooling all corresponding miRanda, TargetScan, and PicTar predictions. Expression correlation analysis of miRNA host genes and putative targets was based on total of 217 microarray data sets (3583 microarray experiments) downloaded from GEO repository (http://www.ncbi.nlm.nih.gov/geo/) (Barrett et al., 2007). All experiments were performed on the same microarray platform, the HG-U133A GeneChip array (GPL96, Feb 19, 2002) and each dataset was normalized and pre-processed independently. For each probe targeting a miRNA host gene, the 3% most anti-correlated probes (using Pearson correlation coefficient) were selected in each data set and all the lists were combined and ranked according to the number of occurrences and to the average rank in the single lists. As input to the HOCTAR procedure we used all the probes corresponding to the selected miRNA host genes and represented in the HG-U133A array, as assessed through the analysis of the Affymetrix website (http://www.affymetrix.com/index.affx).
2.2. HOCTAR database
The dataset is stored into a relationship database on MySQL platform, version 5.0.44, whereas all the necessary scripts to store and query the database were written in Perl v5.8.8 (built for i686-linux) object-oriented scripting language. The web interface was written using PHP 4.4.7, it is freely accessible and supported by all common internet browsers. All data are stored on a Gentoo LINUX Server. HOCTARdb is freely available at http://hoctar.tigem.it/.
2.3. Gene ontology
Gene Ontology (GO) analyses were performed with the web tool DAVID at http://david.abcc.ncifcrf.gov/home.jsp using default parameters (Sherman et al., 2007). GO analyses were performed on the first 30th, 50th, and 100th percentile of HOCTAR ranked lists for all 290 miRNA analyzed. After performing the analysis, only BP, MF and CC categories with a P-value ≤ 0.001, FDR ≤ 5, and fold enrichment ≥ 2 in the analysis of the 50th percentile lists were retained. Redundant terms and non-informative terms (e.g., multigene family) were discarded. Results are collected in the HOCTARdb at http://hoctar.tigem.it.
3. Results
3.1. Construction and contents
The lists of human intragenic miRNAs and corresponding host genes were downloaded from miRBase (release 13.0) (Griffiths-Jones, 2006). We considered host genes for these analysis only those that a) had a sequence overlapping that of the precursor miRNA either in introns, exons, or UTRs, b) were transcribed from the same strand as the miRNA, and c) were represented in the HG-U133A Affymetrix platform, which mostly includes known genes. By using HOCTAR we analyzed 230 host genes and 377 Affymetrix probes corresponding to 290 human miRNAs. 265,136 miRNA target predictions are present in HOCTARdb corresponding to a non-redundant total of 9963 genes. All host gene-intragenic miRNA relationships were manually verified by using the UCSC Genome Browser (release 2006/March; http://genome.ucsc.edu/). Gene Ontology (GO) analyses of putative miRNA targets were performed following HOCTAR analyses to assign a tentative biological function to the miRNA gene networks analyzed. Gene Ontology results have been deposited in HOCTARdb as charts, in which the fold enrichment for each significantly over-represented category is reported.
3.2. HOCTARdb overview
A first set of HOCTAR results was released online when we first described the principles of the procedure (Gennarino et al., 2009). HOCTARdb contains a substantial improvement over these preliminary data in terms of number of miRNAs analyzed (n = 290), number of expression datasets considered for the meta-analysis (217 microarray datasets), and number of putative target genes analyzed for each miRNA. HOCTARdb is the first and unique database to use expression data for scoring putative miRNA targets by examining the expression behavior of their host genes: HOCTARdb includes and re-analyzes all miRNA target predictions generated by softwares such as miRanda, TargetScan and PicTar (Krek et al., 2005; Betel et al., 2008; Friedman et al., 2009). For these reasons, we believe that HOCTARdb is a valuable resource for the ‘wet lab’ researcher willing to investigate miRNA regulatory networks, in that it provides exclusive and experimentally-supported information for the identification of target genes of human miRNAs.
3.3. HOCTAR validation
The HOCTAR procedure combines the observations that the expression of a given intragenic miRNA is mimicked by the expression of its host gene, and that the expression of a given miRNA is anti-correlated to the expression of its target genes. Following these assumptions, the HOCTAR procedure examines the expression behavior of miRNA host genes and putative target genes in multiple publicly available expression microarray datasets, and ranks putative targets from the most to the least anti-regulated with respect to the host gene.
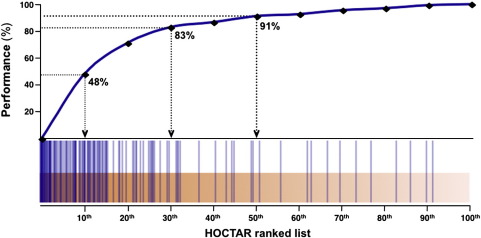
To test the efficiency of HOCTAR in predicting miRNA target genes, we analyzed 135 validated target genes for 28 different intragenic miRNAs selected from PubMed (http://www.ncbi.nlm.nih.gov/pubmed/) and DIANA TarBase (Papadopoulos et al., 2009) (see Supplementary Table S1 for a complete list). The results showed that, out of 135 validated targets, 125 (91%), 112 (83%) and 65 (48%) were included within the first 50th, 30th and 10th percentile of the HOCTAR ranked lists, respectively (Fig. 1). To further assess the performance of the HOCTAR procedure, we decided to test the two datasets of target genes validated by p-Silac procedure from Baek et al. and Selbach et al. (Baek et al., 2008; Selbach et al., 2008). We found that 95% (Selbach et al. gene subset) and 87% (Baek et al. gene subset) of the putative target genes of the intragenic miRNAs analyzed in the latter reports fall within the first 50th percentile of the HOCTAR ranked list (data not shown).
Fig. 1.
Testing the performance of HOCTAR. The diagram displays the ranking, in a percentile format, of 135 previously validated miRNA target genes (blue vertical lines). The distribution of the validated target genes is enriched at the top positions of the HOCTAR prediction ranked lists. In particular, 123 out of the 135 (91%), 112 out of the 135 (83%), and 65 out of the 135 (48%) validated targets fall within the first 50th, 30th and 10th percentile of the HOCTAR ranked lists, respectively.
Overall, this indicates that the genes present at the top of HOCTAR ranked list have a higher probability of representing bona fide targets of miRNAs (Fig. 1). Given the HOCTAR performance relative to the first 50th percentile of prediction ranked lists, we decided to fix this as a threshold to consider bona fide target genes. In HOCTARdb, the target genes that fall within the fixed threshold are highlighted (see Section 3.5). We observed that the average number of predicted target genes present in the first 50th percentile of HOCTAR (n = 357) is comparable with the number of targets predicted by miRanda (n = 1603), TargetScan (n = 721) and PicTar (n = 496).
3.4. Utility
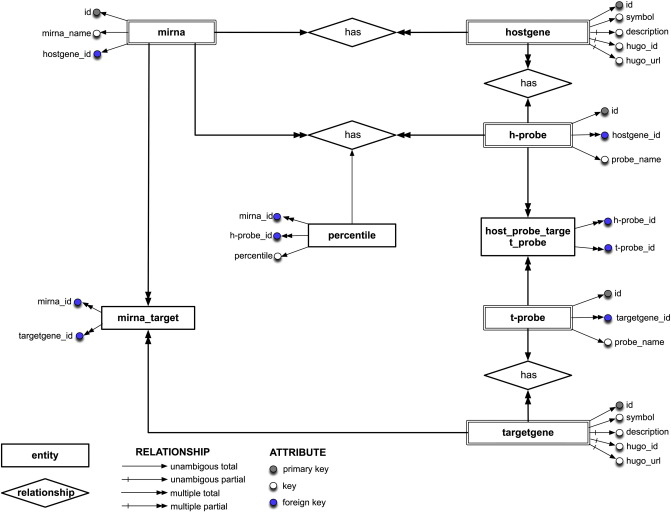
HOCTARdb is an interactive, user-friendly database for identifying and analyzing putative target genes for intragenic human miRNAs. The database is publicly available at http://hoctar.tigem.it. The Entity-Relation design connecting HOCTARdb 8 tables is illustrated in Fig. 2. The graph shows the attributes (arrow and circle) for each entity (tables) and the relationships (square and rhombus, respectively) among the entities. HOCTARdb can be queried by information regarding either miRNAs or target genes. The analyses of all examined miRNA regulatory networks include Gene Ontology results that tentatively assign them a specific function.
Fig. 2.
HOCTARdb relational framework. The Entity-Relationship diagram shows for each entities the corresponding attributes (arrow and circle) and the relationship between the entities (rhombus and square, respectively). Primary keys are indicated in gray, foreign keys in blue, and other keys in white. The ‘mirna’ entities represent the main body of the database.
3.5. User interface
HOCTARdb contents are accessible through a user-friendly retrieval system where simple and advanced search forms are available. The main page briefly summarizes HOCTAR aims, provides some results and allows the user to submit a search (Fig. 3). There are two main ways for interrogating HOCTARdb: (i) by selecting a miRNA using either an alphabetically sorted pull-down menu (e.g. hsa-miR-26a-2) in the “microRNA” query, or (ii) by selecting a gene through its gene symbol (HUGO Gene Name) in the “Target Gene Name” query to look for miRNAs that may possibly target it.
Fig. 3.
Main query page. The HOCTARdb homepage (see text for details).
3.5.1. “microRNA” query
Following the miRNA query, the user is redirected to a new page (Fig. 4) for selecting the host gene's Affymetrix probe of interest and retrieving the corresponding ranked list of target genes. Each prediction list is available for downloading as an excel (.xls) file. For each probe, the link “View Gene Ontology” provides access to the results of the GO analysis, represented as a chart (Fig. 5). Moreover, this page provides link to the miRBase (Griffiths-Jones, 2006) database, which contains further information on the genomic organization of miRNAs. Predicted target genes that fall within the first 50th percentile in the prediction ranked list are highlighted (bold characters). Finally, each predicted target gene is linked to the full list of its predicted targeting miRNAs (see below).
Fig. 4.
HOCTARdb intermediate query page. See description in the text.
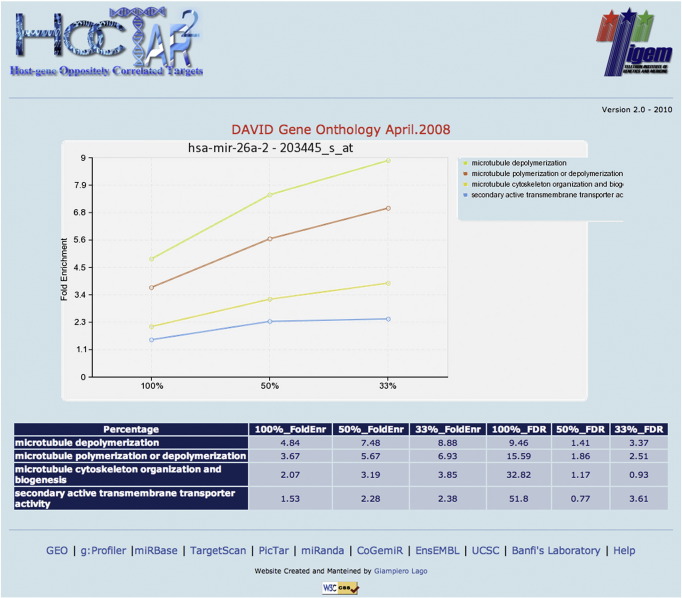
Fig. 5.
Gene Ontology analysis of HOCTARdb predictions. The GO chart, obtained through the DAVID server, shows the specific GO categories enriched in HOCTAR ranked prediction list.
3.5.2. “Target Gene Name” query
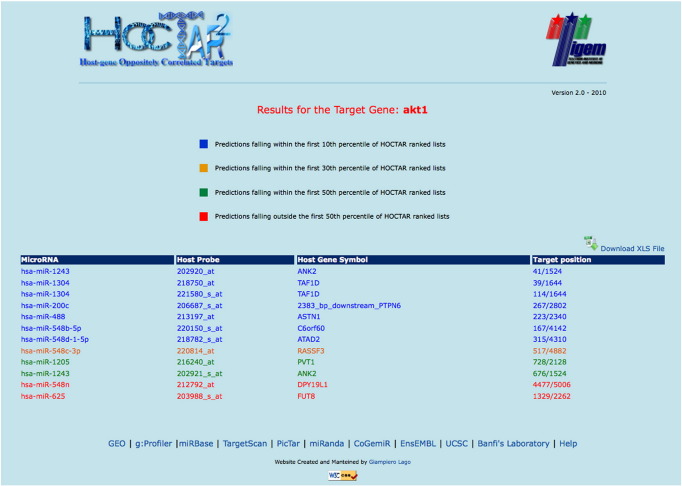
By using the “Target Gene Name” query the user can retrieve all intragenic miRNAs predicted to bind the selected target gene. Following the selection of the target gene of interest, the user is redirected to a page reporting the putative targeting miRNAs (Fig. 6). In this page, the green and the red colors indicate miRNAs whose predicted targeting fall, respectively, above or below the set threshold corresponding to the 50th percentile of the HOCTAR prediction ranked lists. Moreover, by clicking on each of these miRNAs the user is redirected to the corresponding ranked list of predicted targets miRNA (see above). Download is also available for the predicted miRNA binding lists.
Fig. 6.
HOCTARdb “Target Gene Name” selection. The miRNAs associated to prediction lists in which the selected target gene (AKT1 in the example) falls above or below the set threshold are indicated in green and red, respectively.
3.6. HOCTARdb integration with other databases
We integrated HOCTARdb with the miRBase database (Griffiths-Jones, 2006), which provides more detailed information about the genomic organization of analyzed miRNAs, including an evolutionary overview. Furthermore, each gene reported in HOCTARdb is linked to a gene card information page provided by the HUGO database (http://www.genenames.org/data/hgnc_data.php?hgnc_id:-). A link to the help section and several links to related online resources such as TargetScan, PicTar, miRanda, miRBase and g:Profiler (Kent et al., 2002; Griffiths-Jones, 2004; Krek et al., 2005; Reimand et al., 2007; Betel et al., 2008; Friedman et al., 2009; Hubbard et al., 2009) are provided in the main page.
4. Discussions and conclusions
miRNAs are small molecules that play an important role in gene regulation by controlling the expression of multiple target genes. miRNAs maintain different equilibrium of gene expression in different human tissues, and their dysregulation is implicated in various diseases (Thai et al.; Hebert and De Strooper, 2009; Latronico and Condorelli, 2009; Meola et al., 2009; Negrini et al., 2009; Xiao and Rajewsky, 2009). Based on multiple tests on known miRNA targets, we have demonstrated that the HOCTAR procedure is effective in identifying miRNA target genes.
HOCTAR integrates transcriptomic data and in silico prediction tools to perform miRNA target prediction. The main difference between HOCTARdb and already available resources (Krek et al., 2005; Betel et al., 2008; Friedman et al., 2009) is the procedure used to identify putative miRNA target genes. HOCTARdb includes all data generated by miRanda, TargetScan and PicTar and assigns to all identified putative targets a score based on their transcriptional behavior with respect to the miRNA host genes, which represent reliable proxies for the miRNAs themselves (Huang et al., 2007; Gennarino et al., 2009). Other strategies for miRNA target identification based on the evaluation of inverse expression relationships between miRNAs and mRNAs have been devised (Huang et al., 2007). However, an advantage of HOCTAR over the latter procedures is that it utilizes the same set of microarray experiments to simultaneously monitor the expression of both miRNAs and putative targets (Gennarino et al., 2009).
Obviously, the HOCTAR procedure cannot be used at present to identify miRNA targets exclusively regulated at the translational level. On the other hand, several observations confirmed that the transcriptional effects of miRNA action on the expression levels of their targets represent a rather widespread phenomenon (Lim et al., 2005; Baek et al., 2008; Selbach et al., 2008; Gennarino et al., 2009). Even if, recently it has been show that the impact of miRNAs on gene expression and destabilization of target mRNAs is the predominant reason for reduced protein output (Guo et al.).
Differently from other miRNA target prediction databases, HOCTARdb is able to provide different prediction target lists for identical miRNAs present in multiple copies and localized within different host genes. These lists differ in the ranking of the predictions, which are based on the expression data pertaining to the specific host gene. We believe that HOCTAR represents a very sensitive tool for miRNA target prediction (Fig. 1).
Recently, it has been reported that, in 30% of cases, intragenic miRNAs have their own promoters, which are independent from those of their host genes (Fujita et al., 2008; Ozsolak et al., 2008). Surprisingly, the HOCTAR procedure seems to be efficient also in the latter cases, such as for example, miR-21 and its “near host gene” TMEM49 which do not share the same promoter (Fujita et al., 2008). However, all miR-21 experimentally validated targets (see Supplementary Table S1) rank high in the HOCTAR prediction list (http://hoctar.tigem.it). This suggests that intragenic miRNAs and their corresponding host genes may be coregulated even if they have independent promoters although further studies are necessary to better understand the mechanisms regulating the expression of miRNAs.
Importantly, HOCTARdb assigns a tentative biological role to the examined miRNA regulatory networks by means of Gene Ontology analyses. By applying the HOCTAR tool we have recently identified TFEB, a master regulator of lysosomal-dependent degradation pathways, as a target gene for miR-128, which is involved in neurodegenerative diseases (Sardiello et al., 2009). We found that TFEB overexpression was able to prevent the accumulation of mutant huntingtin (HTT) in cell lines containing a transgenic, aggregate-prone HTT allele with 105 CAG repeats responsible for Huntington's disease (Sardiello et al., 2009). The discovery of miR-128 as a TFEB-targeting miRNA suggests a possible molecular pathway linking miR-128 overexpression to neurodegenerative diseases, highlighting the utility of the HOCTAR tool and database for the experimental researcher.
For these reasons, we decided to further implement the HOCTAR procedure and to deposit all HOCTAR information in a user-friendly database, the HOCTARdb. A constant update of HOCTARdb contents will help researchers in selecting miRNAs targeting their genes of interest for experimental work, and also will contribute to the dissection of the regulatory gene networks that involve miRNA control.
Link/URL:
-
1.
UCSC Genome Bioinformatics [http://genome.ucsc.edu/].
-
2.
NCBI PubMed [http://www.ncbi.nlm.nih.gov/pubmed/].
-
3.
HUGO Gene Name Card [http://www.genenames.org/data/hgnc_data.php?hgnc_id:-].
-
4.
NCBI Gene Expression Omnibus (GEO) [http://www.ncbi.nlm.nih.gov/geo/]
-
5.
Affimetrix NetAffx™ Analysis Center [http://www.affymetrix.com/analysis/index.affx]
The following are the supplementary materials related to this article.
Additional file 1 — List of experimentally supported miRNA-target pairs used for HOCTAR validation.
Acknowledgments
We are grateful to Mario Traditi and Gennaro Oliva for their technical support. This work was supported by the Italian Telethon Foundation.
References
- Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- Baek D., Villen J., Shin C., Camargo F.D., Gygi S.P., Bartel D.P. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett T., Troup D.B., Wilhite S.E., Ledoux P., Rudnev D., Evangelista C., Kim I.F., Soboleva A., Tomashevsky M., Edgar R. NCBI GEO: mining tens of millions of expression profiles—database and tools update. Nucleic Acids Res. 2007;35:D760–D765. doi: 10.1093/nar/gkl887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betel D., Wilson M., Gabow A., Marks D.S., Sander C. The microRNA.org resource: targets and expression. Nucleic Acids Res. 2008;36:D149–D153. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang T.C., Mendell J.T. microRNAs in vertebrate physiology and human disease. Annu. Rev. Genomics Hum. Genet. 2007;8:215–239. doi: 10.1146/annurev.genom.8.080706.092351. [DOI] [PubMed] [Google Scholar]
- Dalmay T., Edwards D.R. MicroRNAs and the hallmarks of cancer. Oncogene. 2006;25:6170–6175. doi: 10.1038/sj.onc.1209911. [DOI] [PubMed] [Google Scholar]
- Friedman R.C., Farh K.K., Burge C.B., Bartel D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita S., Ito T., Mizutani T., Minoguchi S., Yamamichi N., Sakurai K., Iba H. miR-21 gene expression triggered by AP-1 is sustained through a double-negative feedback mechanism. J. Mol. Biol. 2008;378:492–504. doi: 10.1016/j.jmb.2008.03.015. [DOI] [PubMed] [Google Scholar]
- Gennarino V.A., Sardiello M., Avellino R., Meola N., Maselli V., Anand S., Cutillo L., Ballabio A., Banfi S. MicroRNA target prediction by expression analysis of host genes. Genome Res. 2009;19:481–490. doi: 10.1101/gr.084129.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldez A.J., Mishima Y., Rihel J., Grocock R.J., Van Dongen S., Inoue K., Enright A.J., Schier A.F. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312:75–79. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S. The microRNA registry. Nucleic Acids Res. 2004;32:D109–D111. doi: 10.1093/nar/gkh023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S. miRBase: the microRNA sequence database. Meth. Mol. Biol. 2006;342:129–138. doi: 10.1385/1-59745-123-1:129. [DOI] [PubMed] [Google Scholar]
- Guo H., Ingolia N.T., Weissman J.S., Bartel D.P. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L., Hannon G.J. MicroRNAs: small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- Hebert S.S., De Strooper B. Alterations of the microRNA network cause neurodegenerative disease. Trends Neurosci. 2009;32:199–206. doi: 10.1016/j.tins.2008.12.003. [DOI] [PubMed] [Google Scholar]
- Huang J.C., Babak T., Corson T.W., Chua G., Khan S., Gallie B.L., Hughes T.R., Blencowe B.J., Frey B.J., Morris Q.D. Using expression profiling data to identify human microRNA targets. Nat. Meth. 2007;4:1045–1049. doi: 10.1038/nmeth1130. [DOI] [PubMed] [Google Scholar]
- Hubbard T.J., Aken B.L., Ayling S., Ballester B., Beal K., Bragin E., Brent S., Chen Y., Clapham P., Clarke L., Coates G., Fairley S., Fitzgerald S., Fernandez-Banet J., Gordon L., Graf S., Haider S., Hammond M., Holland R., Howe K., Jenkinson A., Johnson N., Kahari A., Keefe D., Keenan S., Kinsella R., Kokocinski F., Kulesha E., Lawson D., Longden I., Megy K., Meidl P., Overduin B., Parker A., Pritchard B., Rios D., Schuster M., Slater G., Smedley D., Spooner W., Spudich G., Trevanion S., Vilella A., Vogel J., White S., Wilder S., Zadissa A., Birney E., Cunningham F., Curwen V., Durbin R., Fernandez-Suarez X.M., Herrero J., Kasprzyk A., Proctor G., Smith J., Searle S., Flicek P. Ensembl 2009. Nucleic Acids Res. 2009;37:D690–D697. doi: 10.1093/nar/gkn828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim V.N. MicroRNA biogenesis: coordinated cropping and dicing. Nat. Rev. Mol. Cell Biol. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- Kim Y.K., Kim V.N. Processing of intronic microRNAs. EMBO J. 2007;26:775–783. doi: 10.1038/sj.emboj.7601512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim V.N., Han J., Siomi M.C. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- Krek A., Grun D., Poy M.N., Wolf R., Rosenberg L., Epstein E.J., MacMenamin P., da Piedade I., Gunsalus K.C., Stoffel M., Rajewsky N. Combinatorial microRNA target predictions. Nat. Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- Latronico M.V., Condorelli G. MicroRNAs and cardiac pathology. Nat. Rev. Cardiol. 2009;6:419–429. doi: 10.1038/nrcardio.2009.56. [DOI] [PubMed] [Google Scholar]
- Lim L.P., Lau N.C., Garrett-Engele P., Grimson A., Schelter J.M., Castle J., Bartel D.P., Linsley P.S., Johnson J.M. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- Meola N., Gennarino V.A., Banfi S. microRNAs and genetic diseases. Pathogenetics. 2009;2:7. doi: 10.1186/1755-8417-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negrini M., Nicoloso M.S., Calin G.A. MicroRNAs and cancer—new paradigms in molecular oncology. Curr. Opin. Cell Biol. 2009;21:470–479. doi: 10.1016/j.ceb.2009.03.002. [DOI] [PubMed] [Google Scholar]
- Ozsolak F., Poling L.L., Wang Z., Liu H., Liu X.S., Roeder R.G., Zhang X., Song J.S., Fisher D.E. Chromatin structure analyses identify miRNA promoters. Genes Dev. 2008;22:3172–3183. doi: 10.1101/gad.1706508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopoulos G.L., Reczko M., Simossis V.A., Sethupathy P., Hatzigeorgiou A.G. The database of experimentally supported targets: a functional update of TarBase. Nucleic Acids Res. 2009;37:D155–D158. doi: 10.1093/nar/gkn809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimand J., Kull M., Peterson H., Hansen J., Vilo J. g:Profiler—a web-based toolset for functional profiling of gene lists from large-scale experiments. Nucleic Acids Res. 2007;35:W193–W200. doi: 10.1093/nar/gkm226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez A., Griffiths-Jones S., Ashurst J.L., Bradley A. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004;14:1902–1910. doi: 10.1101/gr.2722704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardiello M., Palmieri M., di Ronza A., Medina D.L., Valenza M., Gennarino V.A., Di Malta C., Donaudy F., Embrione V., Polishchuk R.S., Banfi S., Parenti G., Cattaneo E., Ballabio A. A gene network regulating lysosomal biogenesis and function. Science. 2009;325:473–477. doi: 10.1126/science.1174447. [DOI] [PubMed] [Google Scholar]
- Selbach M., Schwanhausser B., Thierfelder N., Fang Z., Khanin R., Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- Sherman B.T., Huang da W., Tan Q., Guo Y., Bour S., Liu D., Stephens R., Baseler M.W., Lane H.C., Lempicki R.A. DAVID Knowledgebase: a gene-centered database integrating heterogeneous gene annotation resources to facilitate high-throughput gene functional analysis. BMC Bioinform. 2007;8:426. doi: 10.1186/1471-2105-8-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thai T.H., Christiansen P.A., Tsokos G.C. Is there a link between dysregulated miRNA expression and disease? Discov Med. 2010;10:184–194. [PubMed] [Google Scholar]
- Thomas M., Lieberman J., Lal A. Desperately seeking microRNA targets. Nat. Struct. Mol. Biol. 2010;17:1169–1174. doi: 10.1038/nsmb.1921. [DOI] [PubMed] [Google Scholar]
- Xiao C., Rajewsky K. MicroRNA control in the immune system: basic principles. Cell. 2009;136:26–36. doi: 10.1016/j.cell.2008.12.027. [DOI] [PubMed] [Google Scholar]
- Zhang C. MicroRNomics: a newly emerging approach for disease biology. Physiol. Genomics. 2008;33:139–147. doi: 10.1152/physiolgenomics.00034.2008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1 — List of experimentally supported miRNA-target pairs used for HOCTAR validation.