Abstract
Most attempts to localize physical correlates of memory in the central nervous system (CNS) rely on ablation techniques. This approach has the limitation of defining just one of an unknown number of structures necessary for memory formation. We have used the Drosophila rutabaga type I Ca2+/CaM-dependent adenylyl cyclase (AC) gene to determine in which CNS region AC expression is sufficient for memory formation. Using pan-neural and restricted CNS expression with the GAL4 binary transcription activation system, we have rescued the memory defect of the rutabaga mutant in a fast robust spatial learning paradigm. The ventral ganglion, antennal lobes, and median bundle are likely the CNS structures sufficient for rutabaga AC- dependent spatial learning.
Many attempts have been made to locate the physical correlates of a memory by mapping necessary brain structures in a given learning/memory paradigm in both vertebrates and invertebrates (for review, see Davis 1996; Heisenberg 1998; Milner et al. 1998). Recently, transgenic approaches have implicated NMDA receptor- and calcium/calmodulin-dependent protein (CaM) kinase-dependent brain structures necessary for different learning tasks (Tsien et al. 1996; Joiner and Griffith 1999). In insects, much attention has centered on the mushroom bodies, a paired neuropil structure in the central brain. This is because animals with disrupted mushroom bodies fail in an associative olfactory conditioning paradigm (but not other paradigms) and several genes implicated in this task are preferentially expressed there (Heisenberg et al. 1985; de Belle and Heisenberg 1994; Hammer and Menzel 1995; Connolly et al. 1996; Davis 1996; Heisenberg 1998; Wolf et al. 1998). All studies using ablation have the limitation of identifying one or several necessary structures and cannot determine whether the structures are the unique site of memory formation or are but one of several cooperating structures. Here we complement ablation strategies by searching for structures sufficient for a given learning/memory task. We use transgenes encoding a Ca2+/CaM-dependent adenylyl cyclase (AC) in the learning/memory mutant rutabaga to determine where in the central nervous system (CNS) the enzyme needs to be supplied for rescue of behavioral plasticity.
The regulation of cAMP levels is important for learning and memory in both vertebrates and invertebrates. ACs are responsible for the regulated synthesis of cAMP and were first implicated in a nonassociative learning task in the Aplysia marine slug (Brunelli et al. 1976; Klein and Kandel 1980; Schacher et al. 1993). One AC, type I, is a proposed molecular signal integrator responding synergistically to an increase in intracellular Ca2+ and stimulation by heterotrimeric G proteins. It is the coincidence of these two signals that permits neuronal synaptic plasticity underlying associative learning/memory (Lechner and Byrne 1998). Therefore, an animal lacking the type I AC would be expected to lack normal synaptic plasticity and fail in tasks such as memory formation. If one could target in this animal the expression of type I AC to defined neuronal populations, then these cells would be the only ones showing normal synaptic plasticity.
A mutational analysis of mouse and Drosophila showed that a type I AC is necessary for both associative and nonassociative learning tasks. Mice mutant for the type I AC show a memory defect in the Morris water maze, which measures spatial learning and memory (Wu et al. 1995). The Drosophila type 1 AC mutation (rutabaga) has been well studied in several nonassociative and associative learning tasks. Nonassociative tasks that are aberrant in rutabaga mutant flies include sensitization and habituation of the proboscis extension reflex, giant fiber-induced escape response, odor-induced jump habituation, and landing habituation (Duerr and Quinn 1982; Rees and Spatz 1989; Engel and Wu 1996; T. Tully and S. Koss, unpubl.). Associative learning paradigms in which rutabaga mutants fail include visual, olfactory, and spatial learning, as well as courtship suppression (Folkers 1982; Aceves-Pina et al. 1983; Gailey et al. 1984; Han et al. 1992; Wustmann et al. 1996). Several physiological and structural defects have also been described at the larval neuromuscular junction and adult ventral ganglion in rutabaga mutants. However, the adult brain is largely unaffected (Balling et al. 1987; Corfas and Dudai 1990, 1991; Zhong and Wu 1991; Zhong 1995; Zhong and Pena 1995; Barth et al. 1997; Guo et al. 1997). The conserved role of ACs in memory formation made the rutabaga type I AC our choice for defining a minimally sufficient brain region for memory formation.
An operant spatial learning paradigm was designed in which individual flies are trained to avoid one side of a narrow chamber by associating it with a temporal heat punishment. Several Drosophila olfactory learning mutants, including rutabaga, are defective in this learning paradigm (Wustmann et al. 1996; Wustmann and Heisenberg 1997). The neuronal basis of this learning is, however, poorly understood. From its turns and forward motion the fly presumably derives a sense of the space in the chamber and its position within it. Where in the fly this space is represented and how a position in it is associated with danger or safety is not known. We use GAL4 enhancer trap lines with selective expression of the GAL4 transcription factor in subsets of CNS neurons to drive the expression of a wild-type rutabaga cDNA. The behavioral rescue, achieved through regional expression, indicates that performance in the learning/memory task requires only a small subset of the CNS.
Materials and Methods
EXPERIMENTAL SETUP
The chamber (Fig. 1A) is a box (length, 40 mm; height, 2 mm; breadth, 5 mm) with floor and ceiling consisting of peltier elements that control the temperature in the chamber (built in the Genetics workshops, University of Wuerzburg). Clear plastic sides allow the transmission and detection of infrared light from an LED 100 mm away, and a light gate array (bar code reader). Thus, when a fly walks along the length of the chamber, it casts a shadow on the light gate array and this signal is sent to a computer. The fly's position is monitored at a frequency of 10 Hz and when a fly is in the predetermined “punished” half of the chamber, the entire chamber is heated to a maximum of 37°C under control of a thermistor in the chamber (custom software by R. Wolf). When the fly moves to the “unpunished” half, the chamber returns to 24°C. To test learning and memory, flies are subjected to a series of training/test phases. During a 30-sec pretest the fly can explore the chamber while the temperature stays at 24°C. The following training lasts 4 min, binned in 1-min blocks for analysis. Memory of the dangerous side is tested in the next 3 min (also binned) in which the heat is permanently turned off. This corresponds to minutes 5 to 7 in Figure 2. A single training session is a half-experiment, and the side the flies are trained to avoid is alternated between them. Therefore, half of the flies are trained to avoid one side of the chamber and half to avoid the other. This removes spontaneous side preferences in the chamber. A performance index (PI) is determined for different training/test phases as a measure of heat avoidance (during training) and memory (after training). It is calculated as the time spent in the unpunished half of the chamber minus the time spent in the punished half of the chamber, divided by the total time. The PI can have values ranging from −1 to 1, with flies that are perfect heat avoiders having a value of 1. A value of zero indicates no side preference. Pretest side preference (30 sec) was measured for individual flies. The mean pretest side preference was usually <0.05 and this was subtracted from the PIs in Figure 2 and Table 1.
Figure 1.
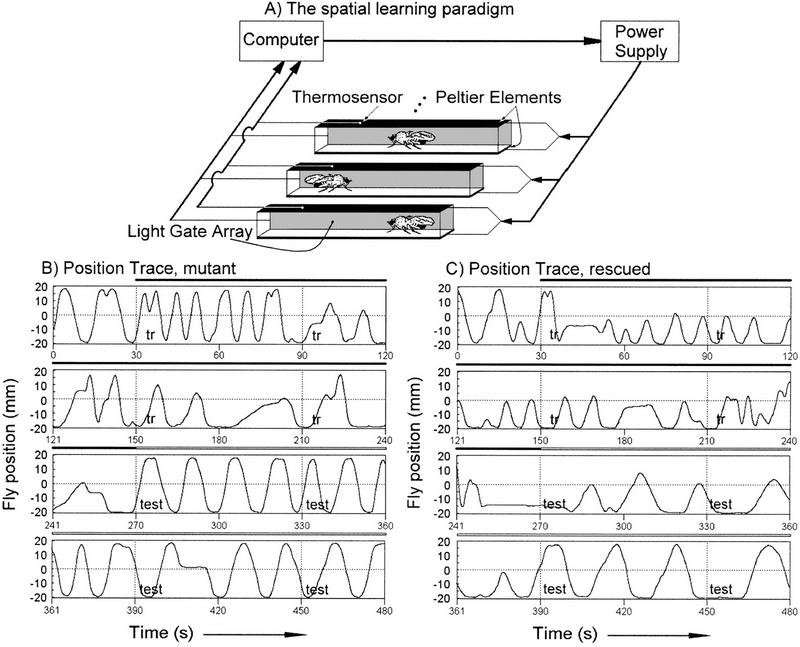
Spatial learning paradigm in the heat box. (A) A schematic diagram of the apparatus with 3 of the 15 parallel chambers shown. A computer receives position information for individual flies from a light gate array and this is used to calculate PIs and control heat punishment by the Peltier elements. (B, C) Raw data from a single mutant (B) and normal (C) fly are presented. The chamber length is displayed on the y-axis of the time traces, and time is the x-axis. The experiment consists of a 30-sec pretest, followed by 4 min of training in which the fly is punished if it is on the “wrong” side of the chamber, and finally a 3.5-min memory test in which the heat is permanently turned off. Performance data during training and testing are binned in 1-min intervals for analysis.
Figure 2.
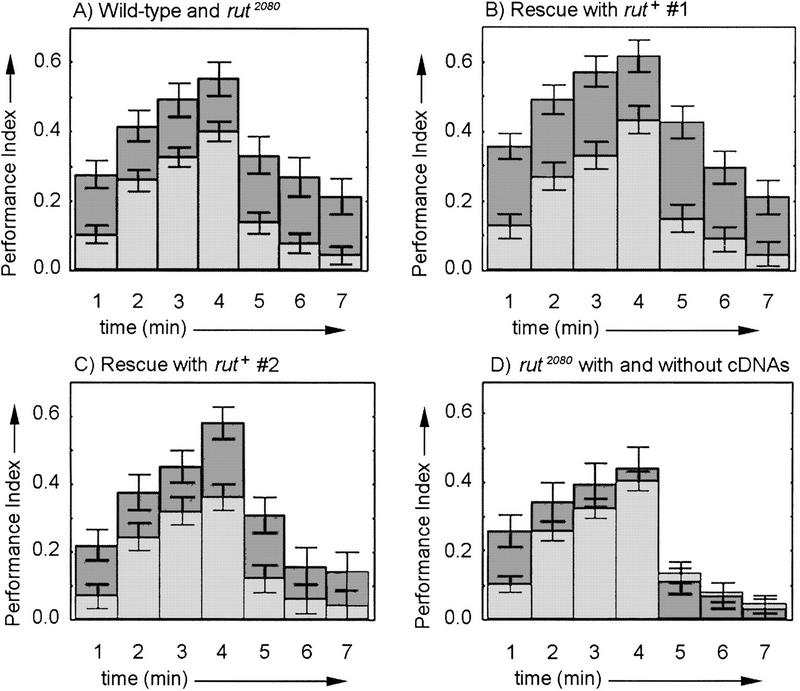
Neuronal expression of a wild-type cDNA rescues the rutabaga learning and memory phenotype. Columns show PIs for consecutive 1-min periods of the training and test phases. (A) Wild-type flies (dark gray bars) have significantly higher avoidance scores than rutabaga mutants (light gray bars) during training and test (n = 510; F = 16.30; P = 0.000062). Wild-type is w CS10 with either of the two UASGAL4–rut+ cDNA transgenes, and rutabaga mutant data are from rut2080 with either of the two UASGAL4–rut+ cDNA transgenes. (B) rutabaga mutant flies with neuronal expression (GAL4–elav) of wild-type UASGAL4–rut+ cDNA #1 (dark bars) have significantly higher avoidance and learning/memory scores than rutabaga mutant flies with UASGAL4–rut+ cDNA #1 and no GAL4 driver (gray bars, n = 420; F = 22.02; P = 0.000004). (C) A second UASGAL4–rut+cDNA (#2, dark bars) with neuronal expression also shows significantly higher avoidance in training and test phases thanrutabaga mutant flies with the UASGAL4–rut+ cDNA but no GAL4 driver (gray bars, n = 301; F = 9.38; P = 0.0024). (D) The UASGAL4–rut+ cDNAs without driver (dark bars) do not increase avoidance scores compared with the parental rut2080 mutant flies (gray bars, n = 478; P > 0.05). No significant difference is observed between wild-type and rescued rut2080 mutants (UASGAL4–rut + #1,n = 324; P > 0.05; for UASGAL4–rut + #2, n = 268; P > 0.05 nor between rut2080 mutants with or without GAL4–elav but no UASGAL4–rut+ cDNA (data not shown, n = 481;P > 0.05). Columns are mean PIs and error bars are s.e.m..
Table 1.
The rutabaga mutant memory defect is rescued with GAL4–elav and four GAL4 enhancer trap lines
| GAL4 line
|
3-Min PI
|
3-Min PI
|
||||
|---|---|---|---|---|---|---|
|
rut+ cDNA #1
|
|
P value
|
rut+ cDNA #2
|
|
P value
|
|
| elav | 0.312 ± 0.041 | n = 197 | 0.00001 | 0.204 ± 0.044 | n = 141 | 0.02 |
| c115 | 0.244 ± 0.076 | n = 56 | 0.05 | 0.331 ± 0.087 | n = 46 | 0.006 |
| c271 | 0.219 ± 0.084 | n = 50 | 0.054 (0.048) | 0.261 ± 0.104 | n = 32 | 0.035 |
| c772 | 0.257 ± 0.075 | n = 74 | 0.005 | 0.354 ± 0.090 | n = 46 | 0.002 |
| c522 | 0.293 ± 0.077 | n = 46 | 0.005 | 0.162 ± 0.050 | n = 72 | 0.28 (0.11) |
| 30y | 0.106 ± 0.089 | n = 41 | N.S. | −0.071 ± 0.103 | n = 28 | N.S. |
| 72y | 0.054 ± 0.078 | n = 36 | N.S. | 0.056 ± 0.053 | n = 30 | N.S. |
| 103y | 0.153 ± 0.076 | n = 36 | N.S. | 0.091 ± 0.071 | n = 32 | N.S. |
| 201y | 0.132 ± 0.089 | n = 35 | N.S. | −0.071 ± 0.096 | n = 32 | N.S. |
| 238y | 0.167 ± 0.081 | n = 38 | N.S. | 0.197 ± 0.095 | n = 30 | N.S. |
| 1407 | 0.076 ± 0.072 | n = 36 | N.S. | 0.174 ± 0.071 | n = 30 | N.S. |
| c232 | 0.155 ± 0.063 | n = 57 | N.S. | 0.139 ± 0.072 | n = 30 | N.S. |
| c492b | 0.127 ± 0.074 | n = 38 | N.S. | 0.107 ± 0.090 | n = 30 | N.S. |
| c739 | 0.141 ± 0.082 | n = 48 | N.S. | 0.213 ± 0.075 | n = 46 | N.S. |
| 121y | 0.189 ± 0.071 | n = 49 | N.S. | N.D. | ||
| 188y | 0.055 ± 0.061 | n = 47 | N.S. | N.D. | ||
| 189y | 0.064 ± 0.050 | n = 46 | N.S. | N.D. | ||
| H24 | 0.180 ± 0.073 | n = 48 | N.S. | N.D. | ||
| No driver | 0.096 ± 0.029 | n = 223 | 0.075 ± 0.034 | n = 160 | ||
| rut2080 | 0.034 ± 0.051 | n = 95 | N.S. | 0.034 ± 0.051 | n = 95 | N.S. |
Mutant rut2080 flies were tested for rescue of the 3-min memory defect using 17 GAL4 enhancer trap lines plus GAL4-elav with two UASGAL4 rut+ cDNAs. GAL4–elav, c115, c522, and c772 showed significantly higher memory scores with UASGAL4 rut+ ⧣1 compared to rut2080 mutant flies both with (no driver) and without (rut2080) the UASGAL4 rut+ but no GAL4 driver. c271 showed significantly higher memory scores with UASGAL4 rut+ ⧣1 compared only to the original rut2080 mutant. GAL4–elav, c115, c271, and c772 rescued with UASGAL4 rut+ ⧣2 compared to rutabaga mutant flies both with and without the UASGAL4 rut+ but no GAL4 driver. c522 with UASGAL4 rut+ ⧣2 was not significantly different from either mutant control. Mean values and s.e.m. are shown; (n) number of flies; (N.S.) not significantly different (P > 0.05) from the no driver control.
Thermosensitivity is recorded adjusting the temperature in the front versus back half of the chamber independently and also independent of the flies' actions. It is stepped from 24°C on both sides to 24°C/30°C, 24°C/37°C, and 24°C/45°C for 1-min intervals, sequentially alternating the side with the higher temperature. A single run from low to high temperature difference is again a half-experiment and the side with higher temperature is alternated between half experiments. A fly's time spent on a given side is measured and heat avoidance indices are calculated as above (PI).
GENETIC MANIPULATIONS
The rut2080; ry double mutant (in a Canton-S genetic background) and the white-eyed Canton-S line wCS10 were used as mutant and wild-type strains in all experiments. Two independent lines were generated by standard techniques in which the wild-type rut+ cDNA under control of the UASGAL4 promoter was inserted onto the second (#2) and third (#1) chromosomes of the w CS10 strain. The UASGAL4cDNAs were crossed into the rut2080 background using a cantonized FM7a; SM1 or, respectively, FM7a; TM2 line (which were themselves generated by chromosome replacement using the w CS10 line). It should be noted that only male flies were used in the behavioral tests and were F1 progeny of a genetic cross (e.g., a rut2080; UASGAL4–rut+ female crossed to a GAL4 enhancer trap line male). Several of the GAL4 enhancer trap lines used to rescue the rutabaga phenotype were crossed for more than six generations to the w CS10 line to effectively replace the genetic background to w CS10. Other GAL4 lines were not subjected to outcrossing but did not affect learning negatively in control experiments (e.g., c522).
The wild-type rutabaga cDNA was subcloned from the Bluescript vector (Levin et al. 1992) with KpnI and XbaI into the pUAST vector (Brand and Perrimon 1993). This step removes 294 nucleotides of the cloned 3′ untranslated sequence but leaves the putative stop codon unchanged.
STATISTICS
A repeated measures ANOVA (Sachs 1992) in the Statistica 4.5 software program was used to determine any differences in performance indices between genotypes with the two rut+ cDNA transgene rescue lines, wild type, and rutabaga mutants during both training and test phases (Fig. 2). To test the 3-min memory scores and heat avoidance, an ANOVA was performed on single genotype pairs, potentially rescued line versus rutabaga mutant with either rescue cDNA but no GAL4 driver or the parental rut2080 (Table 1), and wild-type Canton-S versus rut2080 (heat avoidance; Fig. 3). GAL4 lines with PIs for 3-min memory significantly different from the rutabaga mutant were determined using the P < 0.05 criterion. Differences between w CS10 wild-type and rutabaga mutant lines with either of the two rut+ cDNAs were examined with a repeated measures ANOVA and no significant differences were found (n = 127 for w cs10 with either rut+ cDNA and n = 382 for rut2080 with either rut+ cDNA, F = 0.611, P = 0.61). Therefore, data were pooled for these lines.
Figure 3.
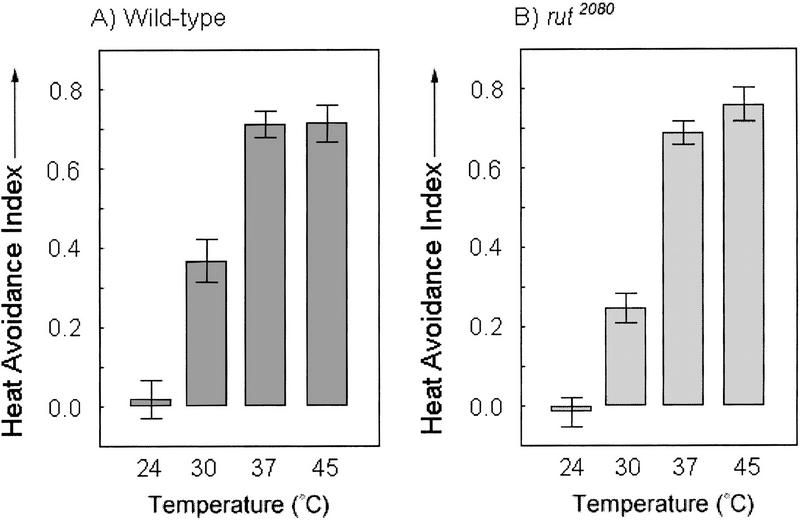
Thermosensitivity of wild-type (A) and rut2080 mutant (B) flies. The experimental setup is similar to that of Fig. 1A. In this assay half of the chamber is heated to the indicated temperature independent of the fly's position. The other half is kept at a reference temperature of 24°C. Heated side is alternated each minute (see Materials and Methods). A heat-avoidance index was calculated as in the performance index of the learning experiment. There are no significant differences between wild-type (n = 104) and rutabaga mutant (n = 78) flies at any of the temperatures assayed. The columns are the mean PIs and error bars are s.e.m..
IMMUNOHISTOCHEMISTRY
GAL4 enhancer trap lines of interest were examined for expression patterns using the UASGAL4–Tau reporter (Ito et al. 1997). Approximately 20 flies were fixed (Carnoy) for mass histology as described (Heisenberg and Boehl 1979). Seven-micrometer sections were blocked for 3 hr with normal horse serum (1:50) in PBS (pH 7.4) plus 0.1% Triton X-100 (PBT) and challenged with anti-Tau antiserum (1:1000) in PBT overnight at 4°C. A series of washes and challenge with biotinylated horse anti-mouse serum (1:100) in PBT for 1 hr at room temperature followed. Signal was detected following Vectastain ABC elite kit instruction with a modified substrate preparation (0.67 mg/ml 3,3′diaminobenzidine dihydrochloride and 0.03% H2O2 in PBT).
Results and Discussion
The spatial learning paradigm, in which the position of a fly can be continuously monitored, is shown in Figure 1A. Individual flies' responses to the training protocol were examined and reveal several qualitative changes in behavior. The spontaneous behavior of flies in this chamber is to walk from one end to the other (Fig. 1B,C). This behavior is evident in the pretest period (the first 30 sec of the position traces in Fig. 1B,C) and in control experiments in which flies are allowed to run without punishment in the chamber for several minutes (not shown). There are no obvious differences between rutabaga mutant (Fig. 1B), wild-type (not shown), and rescued (see below, Fig. 1C) flies in this behavior. Walking speed first increases during training, perhaps as a reaction to the heat, and later slows to below spontaneous levels (period marked with “tr” and a black bar). Also evident during training is a marked preference for the chamber side not associated with punishment. There are differences between rutabaga mutant and wild-type flies during this phase. The rutabaga mutant flies have a longer high speed walking phase compared with wild type and are slower to avoid the side associated with punishment. The last phase in this experiment is a test without heat punishment (the period labeled with “test” and a hollow bar). Memory of training is evident in the continued avoidance of the chamber side associated with punishment, even after a fly re-explores that side. This avoidance eventually disappears and flies return to the pretraining behavior of walking from chamber end to end. The difference between rutabaga mutant and wild-type flies in this phase is the length of time before the pretraining behavior returns.
The performance deficits observed in rutabaga mutant flies are rescued with neuronal expression of a wild-type rutabaga cDNA. Both rutabaga mutant alleles (rut1 and rut2080) tested in the spatial learning paradigm are defective (Wustmann et al. 1996; Fig. 2). Indeed, both the training and memory test avoidance performances are lower in rutabaga mutant flies. This, together with the thermosensitivity assay results (see below) imply that these flies are defective in the learning process itself. Two independent insertions of an UASGAL4–rut+ cDNA were tested for their ability to rescue this defect when combined with the elav neuronal GAL4 driver (Luo et al. 1994; Yao and White 1994; Ito et al. 1998). The UASGAL4–rut+ cDNAs are labeled rut+ cDNA #1 (B) and rut+ cDNA #2 (C) throughout the text. The training period in Figure 2, A–D, is the first 4 min and is followed by 3 min of testing. Figure 2A shows that the white-eyed wild-type strain w CS10, with either rut+ cDNA insert #1 or #2 (dark gray bars), have significantly higher avoidance scores during training and test phases than the rutabaga mutants with either rut+ cDNA but no GAL4 driver (light gray bars, see figure legend for detailed genotypes). There were no significant differences between the w CS10 wild-type flies with rut+ cDNA #1 or with rut+ cDNA #2. Therefore, the wild-type data were pooled in Figure 2A for easier comparison to the rutabaga mutant. Figure 2B also shows significantly higher avoidance scores during training and testing for flies carrying the rut+ cDNA #1 and the GAL4–elav driver (dark gray bars) than for the rutabaga mutant with the rut+ cDNA #1 but no GAL4 driver (light gray bars). The second rut+ cDNA in the rutabaga mutant background with the GAL4–elav driver (Fig. 2C, dark gray bars) also shows significantly higher training/test phase avoidance scores compared to the rutabaga mutant control (light gray bars). Finally, Figure 2D shows that in the rutabaga mutant with either rut+ cDNA #1 or #2, but no GAL4 driver (dark gray bars, pooled data), learning/memory scores are not significantly different from the parental rutabaga mutant strain (light gray bars). In addition, no significant difference was observed between the w CS10 wild-type UASGAL4–rut+ cDNAs and either rescue line (Fig. 2A–C, dark gray bars). Also, no differences were detected comparing the rutabaga mutant with and without the GAL4–elav driver (data not shown). These results demonstrate the full rescue of the rutabaga mutant learning/memory phenotype in the spatial learning paradigm. In addition, because rutabaga mutant flies are rescued with expression in all or most neurons, ectopic expression is not deleterious to the fly for this behavior. Moreover, the rescue provides an essential tool to determine rutabaga's minimally sufficient spatial expression in the brain for rescue of performance in the spatial learning paradigm. Finally, together with the identification of the molecular defects in several rutabaga alleles (Levin et al. 1992) the rescue clearly identifies the rutabaga gene as a Drosophila type I AC.
In a first attempt to localize the minimal brain region sufficient for rutabaga memory formation, arbitrarily chosen GAL4 enhancer trap lines were used to express the rut+ cDNAs in subsets of neurons. The mean 3-min memory was tested for rescue in 17 lines (Table 1). GAL4 enhancer trap lines 30y, 72y, 103y, 201y, 238y, 1407, c115, c232, c271, c492b, c739, and c772 with either of the two rut+ cDNAs were tested. Lines c115, c271, and c772 showed full rescue of the rutabaga mutant memory defect with both rut+ cDNAs compared with the GAL4–elav rescue flies and wild type w CS10. With line c271 rescue was slightly (but not significantly) lower than with lines c115 and c772. With the rut+ cDNA #1 rescue in this line was significantly different from only the rutabaga mutant but not the “no driver” control. The other GAL4 enhancer trap lines did not show significantly higher memory scores with either rut+ cDNA. Five other GAL4 lines (121y, 188y, 189y, H24, and c522) were initially tested with only rut+ cDNA #1. Line c522 had the second highest rescue score of all GAL4 enhancer trap lines with rut+ cDNA #1, but surprisingly, the performance scores with rut+ cDNA #2 failed to reach wild-type levels. It is possible that a threshold for rut+ expression is not reached with this combination of GAL4 driver and UASGAL4–rut+. Thus, four of 17 GAL4 enhancer trap lines rescued the rutabaga 3-min memory. Three of the four showed rescue with both rut+ cDNA inserts, whereas line c522 only rescued with rut+ cDNA #1.
Several conclusions can be drawn from these rescue experiments. First, the rescue of the learning/memory phenotype with four GAL4 enhancer trap lines corroborates the GAL4–elav driven rescue, arguing against, for example, unknown genetic interactions in the GAL4–elav rescue flies. Second, only one of 13 GAL4 enhancer trap lines rescues with only one of the two rut+ cDNAs. This consistency gives weight to the specificity of the ability of GAL4 enhancer trap lines to rescue and argues against genetic background effects. Finally, if the rescue in the four lines is indeed due to the specific expression of the rut+ cDNA in the neurons labeled in these strains, this suggests that the cAMP-dependent synaptic plasticity is required in only a small subset of neurons for this behavioral plasticity to work properly (see below).
We tested the thermal sensitivity of wild-type and rutabaga mutant flies to address the possibility that the rutabaga learning defect was simply due to a defect in heat perception. In the experiment of Figure 3, flies are exposed to a reference temperature (24°C) and a higher test temperature in a chamber with split peltier elements so that only half of the chamber is heated to a preset temperature independent of the fly's behavior. The side of the reference temperature changes, and the test temperature increases in a step-like manner (30°C, 37°C, and 45°C) every minute, alternating sides in each half-experiment. A fly's avoidance of the higher temperatures is an indication of its thermosensitivity. Wild-type and rutabaga mutant flies are similar in sensitivity to temperature differences over the range tested. The thermosensitivity profile of wild-type and rutabaga flies is drastically different from flies lacking antennae, which have no thermopreference at 30°C, yet show normal performance indices in a learning experiment in the spatial learning paradigm (Sayeed and Benzer 1996; T. Zars, unpubl.). It is clear from these results that the rutabaga mutation does not affect temperature perception per se, but likely affects the association of spatial position with temperature punishment.
The expression patterns of the GAL4 enhancer trap lines were examined to identify the CNS structures sufficient for memory formation in this paradigm. Frontal brain sections of the four GAL4 enhancer trap lines that rescued the rutabaga phenotype and three that did not are shown in Figure 4. Sagittal brain sections of the four rescue lines are shown in Figure 5. The ventral ganglion was also examined with serial sections (Fig. 5N, and data not shown). At a gross level, the rescuing GAL4 lines show expression in six common structures. These are the antennal lobes, mushroom bodies, median bundle, ellipsoid body, fan-shaped body, and ventral ganglion. Closer examination shows that different substructures are often labeled. We use the nomenclature of Hanesch et al. (1989) and Crittendon et al. (1998) to describe the central complex and mushroom body substructures. The horizontal layers in the fan-shaped body (I–VI) and ellipsoid body (I–III) are diagrammed in Figure 5H. We propose that the ellipsoid body can be divided into three vertical layers (A–C; hence nine domains are distinguished in the ring by the intersections of horizontal and vertical layers) based on differences in GAL4 enhancer trap expression patterns. This is an elaboration of anterior and posterior rings previously proposed (Hanesch et al. 1989). The layers in the fan-shaped body and ellipsoid body, as well as the three lobe systems in the mushroom body (α/β, α′/β′,γ), allow a rough mapping of expression patterns within these structures.
Figure 4.
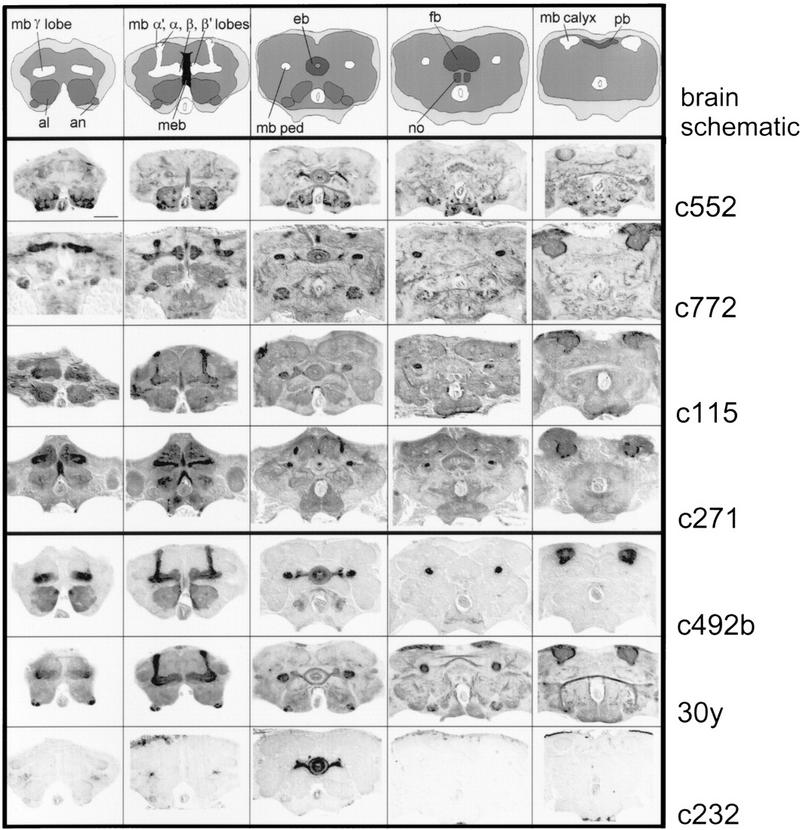
Adult expression patterns of rescue and nonrescue GAL4 enhancer trap lines. Serial frontal paraffin sections of fixed fly heads were examined for GAL4 expression using UASGAL4–Tau as a reporter (Ito et al. 1997). The first four GAL4 enhancer trap lines rescue the rutabaga learning phenotype and the final three do not. The brain schematic highlights the antennal lobes (al) and antennal nerve (an) in a medium gray; the mushroom body (mb) lobes (α,α′, β,β′,γ), peduncle (ped), and calyx in white; the ellipsoid body (eb), fan-shaped body (fb), noduli (no), and protocerebral bridge (pb) of the central complex in dark gray; and the median bundle in black (meb). The brain neuropil is light gray and is surrounded by the cell body rind in off-white. The labeled structures in the rescue GAL4 enhancer trap lines include the mushroom bodies, ellipsoid body, fan-shaped body, antennal lobes, and median bundle. The sections are from anterior (left) to posterior (right), dorsal is up. Bar, 50 μm.
Figure 5.
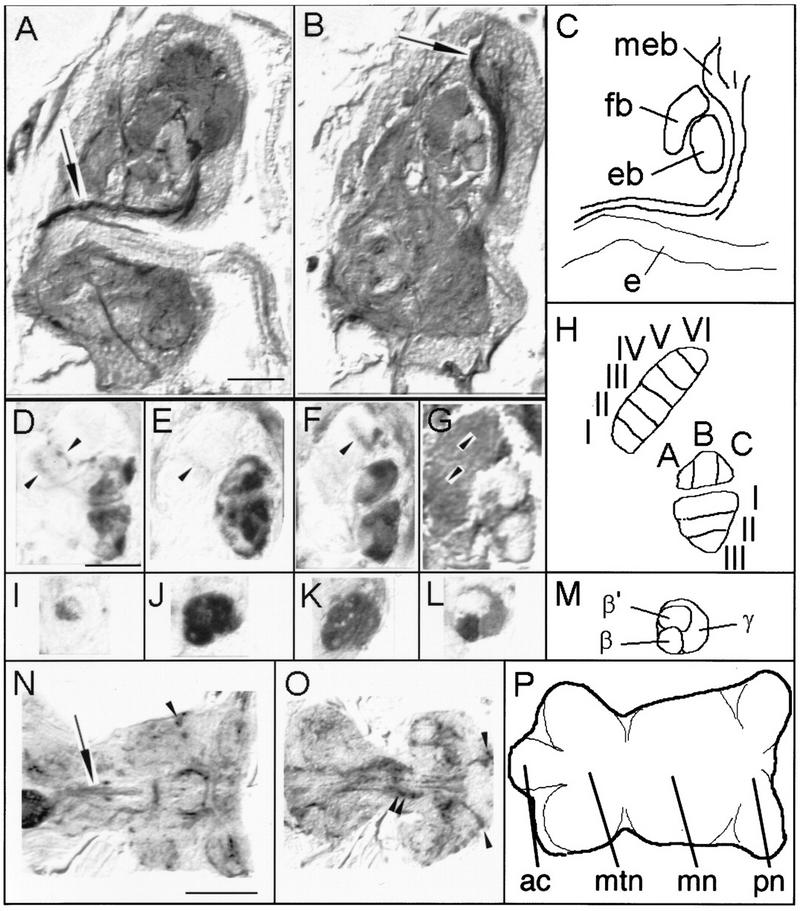
The four rescue lines have different expression patterns in the mushroom bodies, fan-shaped body, and ellipsoid body as well as expression in the ventral ganglion. (A–C) Serial sagittal paraffin sections of fixed fly heads were examined for GAL4 expression using UASGAL4–Tau as a reporter. Two slightly oblique consecutive sagittal sections of rescue line c271 show the projection of the median bundle (arrows) from the cervical connective to the superior medial protocerebrum (diagram in C). (D–G) Medial sagittal sections through the fan-shaped body and ellipsoid body label different layers in the different GAL4 enhancer trap rescue lines (arrowheads indicate immunopositive layers in the fan-shaped body). (H) The fan-shaped body can be divided into six horizontal layers. In the ellipsoid body three horizontal and three vertical layers can be distinguished dividing the ring into nine “domains”. The horizontal layers are drawn in only the ventral region and the vertical layers in the dorsal. (I–M) Sagittal sections of the medially projecting mushroom body lobes indicate difference in expression between lines. Dorsal is up; anterior is to the right in A–M. Images D and I are line c522, E and J are line c772, F and K are line c115, and G and L are line c271. (N–P) Two horizontal sections of the ventral ganglion of rescue line c772. Immunopositive structures include the median dorsal abdominal tract (arrow in N) and the ventral median tract of ventral cervical fasciculus (O), part of the ventral ellipse (double arrowheads in O), as well as various cell bodies (arrowheads in N and O). Abbreviations are as in Fig. 4; (e) esophagus; (ac) abdominal center; (mtn) metathoracic neuromere; (mn) mesothoracic neuromere; (pn) prothoracic neuromere. (A,B) Bar, 50 μm; (D–L) bar, 25 μm; (N–P) bar, 100 μm.
Line c522 strongly labels a subset of antennal lobe glomeruli, as well as fibers in the antennomechanosensory center and the subesophageal ganglion. Weak labeling is observed in the α′ and β′ lobes of the mushroom bodies and the corresponding peduncular and calycal regions (Figs. 4 and 5I). Further stained are subsets of ellipsoid body ring neurons (Figs. 4 and 5D, possibly R1 and R4 neurons). These strongly innervate domains BI and CIII and spare domains AI and AII. A central portion of the median bundle (Fig. 4), two layers of the fan-shaped body (possibly II and III, Figs. 4 and 5D) from two different medial fan-shaped body large field neurons, and other neuron tracts are highlighted.
Line c772 weakly labels a subset of the antennal lobe glomeruli. More strongly labeled are fibers in the antennomechanosensory tract and subsets of Kenyon cells in all three mushroom body substructures (Figs. 4 and 5J). The ellipsoid body is labeled in most domains, also possibly from R1 and R4 type ring neurons. They spare domain AII and are restricted to the periphery in BIII and possibly AII (Figs. 4 and 5E). A middle layer (possibly III) of the fan-shaped body is faintly labeled from a lateral type large field tangential neuron (Figs. 4 and 5E). Finally, the central part of the median bundle is strongly stained (Fig. 4).
In line c115, most of the antennal lobe glomeruli as well as most or all parts of the mushroom bodies are detailed. The central part of the median bundle, many of the ellipsoid body ring neurons (sparing AII), and a distal layer (possibly V) of the fan-shaped body are also stained (Figs. 4 and 5 F,K). Finally, line c271 displays a subset of antennal lobe glomeruli and the α,β, and γ (but not α′β′) mushroom body lobes. In the ellipsoid body, exclusively small parts of domains AI and AII are innervated from a novel “ring” neuron that does not have arborizations in the lateral triangles. Labeled fibers project through the ellipsoid body canal. Also, most or all of the median bundle (Figs. 5A,B), and two layers of the fan-shaped body (possibly II and V) are innervated from a median large field fan-shaped body fiber (Figs. 4 and 5G,L).
The ventral ganglions of the four rescuing GAL4 lines were also examined. In all rescuing GAL4 lines at least some expression was found in the leg neurons, the dorsal and ventral paths of the cervical fasciculi, the ventral ellipse, several commissures, as well as other structures (Fig. 5; data not shown). Some of the immunopositive neuropil in the ventral ganglion arose from cell bodies in the ventral ganglion rind. Interestingly, the dorsal and ventral projection pathways from the cervical fasciculi include neurons of the median bundle (Power 1948). The ventral ellipse is an intriguingly organized ventral ganglion structure. Its function is, however, unknown.
Three GAL4 enhancer trap lines represent the 13 expression patterns that do not rescue the rutabaga learning phenotype (Fig. 4). Some patterns are more restricted than those in the rescue lines but others have very similar structures labeled. In line c232, for instance, only a subset of the ellipsoid body ring neurons are marked. This is the most selective pattern in our collection. Line c492b shows subsets of Kenyon cells in all three mushroom body lobes, a subset of the antennal lobe glomeruli, a portion of the median bundle (lateral), and some ring neurons of the ellipsoid body. No expression is detected in the fan-shaped body. Finally, line 30y displays expression in a subset of the antennal lobe glomeruli, all mushroom body lobe systems, some ring neurons of the ellipsoid body, large field neurons of the fan-shaped body, and lateral fibers of the median bundle (not seen in these sections). Expression patterns for lines c772, c271, c492b, 30y, c232, and GAL4–elav have been described in some detail before (Luo et al. 1994; Yao and White 1994; O'Dell et al. 1995; Yang et al. 1995; Tettamanti et al. 1997; Armstrong et al. 1998; Ito et al. 1998). Lines c522 and c115 (as well as others) have been displayed on the Flytrap website (http://www.molgen.gla.ac.uk/flytrap/). The difference in overall gray levels in rescue versus nonrescue GAL4 lines is due to inadvertent differences in signal maturation and the images used here were picked because of their well-preserved cytoarchitecture. Control experiments in which serial sections of all GAL4 enhancer trap lines were treated identically on the same slide showed that c115 had the highest qualitative level of overall expression in the protocerebrum and c232 had the least. The other rescue and nonrescue lines had similar levels of expression in the protocerebrum (data not shown). A correlation between the number of brain cells expressing GAL4 in the different lines and their ability to rescue is not evident. Lines c522, c772, c115, c271, c492b, and 30y have detectable signal in the range of 60 cell bodies per hemisphere (not counting Kenyon cell bodies, which were in the range of hundreds per hemisphere). This is in contrast to c232 that has ∼20 stained cells per hemisphere and no detectable expression in Kenyon cells. This can be compared to the estimated total of 200,000 neurons in the Drosophila central brain (M. Heisenberg, unpubl.) of which ∼70,000 are in a single optic lobe (see Heisenberg and Wolf 1984). The fraction of immunopositive ventral ganglion cells were estimated to be <20% (data not shown). We have, therefore, successfully rescued the rutabaga learning/memory phenotype with expression in ∼1% of the central brain neurons (600/60,000) and a small fraction of ventral ganglion cells.
Can we determine the structure minimally sufficient for rutabaga-dependent learning/memory formation in this paradigm? The first conservative conclusion from these experiments is that in the rescue GAL4 enhancer trap lines, the sufficient set of cells (in the maximal range of a few percent) is expressing the sufficient level of transgenic rutabaga protein to allow rutabaga mutant flies to learn in the spatial paradigm. The second conclusion is that the sufficient cells for learning in this paradigm are neurons because the GAL4–elav line drives expression specifically in neurons (Luo et al. 1994; Yao and White 1994; Ito et al. 1998). Then, each of the rescue lines defines a set of neurons that are sufficient for learning and memory in the spatial learning paradigm. Assuming the UASGAL4–Tau expression represents the expression of the UASGAL4–rut+ cDNA, we can now identify the common labeled brain structures in the rescue lines as the sufficient set. The most conspicuous regions of the central brain, the mushroom bodies, ellipsoid body, and fan-shaped body are all labeled to some extent. However, different lobe systems of the mushroom bodies are labeled in different lines [e.g., c522 (α′β′) vs. c271 (α,β,γ)]. Also, it has been found that the mushroom bodies are not necessary for learning in this paradigm (Wolf et al. 1998). The GAL4 enhancer trap lines c522, c772, and c115 have strong expression in several types of ring neurons of the ellipsoid body. c271, however, has weak expression in the ellipsoid body and this is in a novel ring neuron that does not have projections in the lateral triangles and seems not to be labeled in the rescue lines c522 and c115. Hence, the four rescue lines appear to have no ellipsoid body neurons stained in common. The fan-shaped body has the most diverse innervation pattern between lines, with some expressing in medially or laterally projecting large field neurons or small field neurons. No common staining pattern in the fan-shaped body emerges from the four lines. The differences in ellipsoid body, fan-shaped body, and mushroom body expression between the four rescue lines at the neuronal level strongly argue against these structures functioning in rutabaga-dependent spatial learning. Only if these structures function in a redundant fashion (e.g., rutabaga expression in any part of the ellipsoid body or fan-shaped body being sufficient) could these neuropil structures still be involved in rutabaga-dependent spatial learning.
This leaves the antennal lobes and median bundle in the central brain and the ventral ganglion. The antennal lobe is labeled in all four rescue lines and, several stained regions of the antennal lobe may be similar between them. The ventral ganglion expression may also have some common structures such as the dorsal and ventral projection pathways from the cervical fasciculi and the ventral ellipse. Finally, expression in the median bundle is similar between the four lines, with expression in at least the medial portion of the bundle and projections to the superior medial protocerebrum.
The median bundle is a good candidate for an integration site from two sensory inputs. The first should be a thermosensory input, possibly from the antennae as has been detected in flies and other insects (Altner and Loftus 1985; Sayeed and Benzer 1996). This could explain the common antennal lobe expression in rescue lines (although the antennal lobe has not been shown in any insect species to be the projection site of antennal thermosensitive neurons). The second is a postulated path integrator (Wehner 1992, 1996). Although the training chamber is relatively simple compared to the desert and forest environments that ants, bees, and spiders must be able to navigate, there is still no light (or other external cue) in this chamber by which Drosophila can orient (Wustmann and Heisenberg 1997). No nervous center has been proposed as yet to house the path integrator. However, the ventral ganglion is a candidate site and, interestingly, it is connected to the superior medial protocerebrum through the median bundle in Drosophila and large flies (Power 1948; Strausfeld 1976). Also, in animals in which necessary structures for a given behavior have been mapped, synaptic modifications generally occur in the sensory input pathway (Milner et al. 1998). This is also consistent with the putative role of the antennal lobe, ventral ganglion, and median bundle. It will be quite exciting to determine whether expression in any single structure can rescue the rutabaga-dependent learning in this paradigm.
Is the wild-type Rutabaga protein expressed in any of the structures labeled in the rescue lines? The wild-type Rutabaga protein has elevated expression in the mushroom bodies, as well as expression in the ellipsoid body, antennal lobes, optic lobes, ventral ganglion, and a general low level staining in the protocerebrum (Han et al. 1992; Crittenden et al. 1998). There is also expression in a subset of neurons in the median bundle (T. Zars, unpubl. data). This expression is strongly reduced (but not absent) in a rutabaga P-element allele with the same insertion site as the one used in this study (Han et al. 1992). Therefore, in wild-type flies the candidate structures for minimally sufficient rutabaga-dependent spatial learning have endogenous Rutabaga protein. Whether the exact same neurons labeled in the median bundle, ventral ganglion, and antennal lobes of the rescue lines contain the Rutabaga type I AC is unknown and may be addressed with double labeling experiments.
In conclusion we have, for the first time, rescued the Drosophila rutabaga learning mutation using a wild-type cDNA encoding a type I Ca2+/CaM-dependent AC, confirming the identity of the Drosophila rutabaga gene. We have done so using the GAL4 transcription activation system (Brand and Perrimon 1993) that can be used to turn on gene expression in a spatially restricted fashion. We have localized the cells sufficient for rutabaga learning to neurons and likely localized the CNS regions sufficient for learning in this spatial learning paradigm to the antennal lobe, ventral ganglion, and the median bundle.
It should be remembered that there are several limitations of this study. First, the spatially restricted rescue of rutabaga mutants can only address rutabaga-dependent synaptic plasticity. There could be other plastic synapses in these flies that are unaffected by the mutation and rescue. Second, the role of the rutabaga type I AC in CNS synaptic plasticity is inferred from the larval neuromuscular junction (Zhong and Wu 1991). Third, only the UASGAL4–Tau reporter was available to address spatial expression, as all rescue experiments were done in a strong hypomorphic, but not null, rutabaga mutant background (Han et al. 1992). Fourth, precise single neuron anatomical analysis is not yet available nor is temporal control of spatially restricted expression. Finally, ablation-type studies that would give information about necessary brain structures have not yet been exhaustively performed in this paradigm. Once all these technical advances are available and the uncertainties are removed, the anatomy of this memory task will be experimentally accessible in unprecedented detail. Knowledge of the necessary and sufficient brain structures for memory formation may shed new light on the circuit properties of brains in general.
Acknowledgments
We thank D. Armstrong, K. Kaiser, P. Kolodziej, and T. Raabe for kindly providing GAL4 enhancer trap lines used in this study. B. Mühlbauer, K. Öchsner, and H. Kadeschabek gave technical assistance throughout this study. The Mitarbeitergruppe: M. Reif, B. Brembs, R. Ernst, A. Keller, G. Putz, and S. Kramer, contributed to many stimulating conversations. This study was supported in part by the Deutsche Forschungsgemeinschaft to M.H. and a National Institutes of Health grant, the Mathers Foundation, and R.P. Doherty-Welch Chair in Science to R.L.D. T.Z. was supported by a postdoctoral-fellowship from the Alexander von Humbold Foundation.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
References
- Aceves-Pina EO, Booker R, Duerr JS, Livingstone MS, Quinn WG, Smith RF, Sziber PP, Tempel BL, Tully TP. Learning and memory in Drosophila, studied with mutants. Cold Spring Harb Symp Quant Biol. 1983;48:831–840. doi: 10.1101/sqb.1983.048.01.086. [DOI] [PubMed] [Google Scholar]
- Altner H, Loftus R. Ultrastructure and function of insect thermo- and hygroreceptors. Ann Rev Entomol. 1985;30:273–295. [Google Scholar]
- Armstrong JD, de Belle JS, Wang Z, Kaiser K. Metamorphosis of the mushroom bodies; Large-scale rearrangements of the neural substrates for associative learning and memory in Drosophila. Learn & Mem. 1998;5:102–114. [PMC free article] [PubMed] [Google Scholar]
- Balling A, Technau GM, Heisenberg M. Are the structural changes in adult Drosophila mushroom bodies memory traces? Studies on biochemical learning mutants. J Neurogenet. 1987;4:65–73. [PubMed] [Google Scholar]
- Barth M, Hirsch HVB, Meinertzhagen IA, Heisenberg M. Experience-dependent developmental plasticity in the optic lobe of Drosophila melanogaster. J Neurosci. 1997;17:1493–1504. doi: 10.1523/JNEUROSCI.17-04-01493.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Brunelli M, Castellucci V, Kandel ER. Synaptic facilitation and behavioral sensitization in Aplysia: Possible role of serotonin and cyclic AMP. Science. 1976;194:1178–1181. doi: 10.1126/science.186870. [DOI] [PubMed] [Google Scholar]
- Connolly JB, Roberts IJ, Armstrong JD, Kaiser K, Forte M, Tully T, O'Kane CJ. Associative learning disrupted by impaired Gs signaling in Drosophila mushroom bodies. Science. 1996;274:2104–2107. doi: 10.1126/science.274.5295.2104. [DOI] [PubMed] [Google Scholar]
- Corfas G, Dudai Y. Adaptation and fatigue of a mechanosensory neuron in wild-type Drosophila and in memory mutants. J Neurosci. 1990;10:491–499. doi: 10.1523/JNEUROSCI.10-02-00491.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Morphology of a sensory neuron in Drosophila is abnormal in memory mutants and changes during aging. Proc Natl Acad Sci. 1991;88:7252–7256. doi: 10.1073/pnas.88.16.7252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crittenden JR, Skoulakis EMC, Han K-A, Kalderon D, Davis RL. Tripartite mushroom body architecture revealed by antigenic markers. Learn & Mem. 1998;5:38–51. [PMC free article] [PubMed] [Google Scholar]
- Davis RL. Physiology and biochemistry of Drosophila learning mutants. Physiol Rev. 1996;76:299–317. doi: 10.1152/physrev.1996.76.2.299. [DOI] [PubMed] [Google Scholar]
- de Belle JS, Heisenberg M. Associative odor learning in Drosophila abolished by chemical ablation of mushroom bodies. Science. 1994;263:692–695. doi: 10.1126/science.8303280. [DOI] [PubMed] [Google Scholar]
- Duerr JS, Quinn WG. Three Drosophila mutations that block associative learning also affect habituation and sensitization. Proc Natl Acad Sci. 1982;79:3646–3650. doi: 10.1073/pnas.79.11.3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel JE, Wu CF. Altered habituation of an identified escape circuit in Drosophila memory mutants. J Neurosci. 1996;16:3486–3499. doi: 10.1523/JNEUROSCI.16-10-03486.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folkers E. Visual learning and memory of Drosophila melanogaster wild-type C-S and the mutants dunce, amnesiac, turnip, and rutabaga. J Insect Physiol. 1982;28:535–539. [Google Scholar]
- Gailey DA, Jackson FR, Siegel RW. Conditioning mutations in Drosophila melanogaster affect an experience-dependent behavioral modification in courting males. Genetics. 1984;106:613–623. doi: 10.1093/genetics/106.4.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H-F, Hannan F, Bernards A, Zhong Y. Requirement of Drosophila NF1 for activation of adenylyl cyclase by PACAP38-like neuropeptides. Science. 1997;276:795–798. doi: 10.1126/science.276.5313.795. [DOI] [PubMed] [Google Scholar]
- Hammer M, Menzel R. Learning and memory in the honeybee. J Neurosci. 1995;15:1617–1630. doi: 10.1523/JNEUROSCI.15-03-01617.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han PL, Levin LR, Reed RR, Davis RL. Preferntial expression of the Drosophila rutabaga gene in mushroom bodies, neural centers for learning in insects. Neuron. 1992;9:619–627. doi: 10.1016/0896-6273(92)90026-a. [DOI] [PubMed] [Google Scholar]
- Hanesch U, Fischbach K-F, Heisenberg M. Neuronal architecture of the central complex in Drosophila melanogaster. Cell Tissue Res. 1989;257:343–366. [Google Scholar]
- Heisenberg M. What do the mushroom bodies do for the insect brain? An introduction. Learn & Mem. 1998;5:1–10. [PMC free article] [PubMed] [Google Scholar]
- Heisenberg M, Boehl K. Isolation of anatomical brain mutants of Drosophila melanogaster by histological means. Z Naturforsch C. 1979;34:143–147. [Google Scholar]
- Heisenberg M, Wolf R. Vision in Drosophila; Genetics of microbehavior. In: Braitenberg V, Barlow HB, Bullock TH, Florey E, Gruesser O-J, Peters A, editors. Studies of brain function. Vol. 12. Berlin, Germany: Springer-Verlag; 1984. [Google Scholar]
- Heisenberg M, Borst A, Wagner S, Byers D. Drosophila mushroom body mutants are deficient in olfactory learning. J Neurogenet. 1985;2:1–30. doi: 10.3109/01677068509100140. [DOI] [PubMed] [Google Scholar]
- Ito K, Sass H, Urban J, Hofbauer A, Schneuwly S. GAL4-responsive UASGAL4–tau as a tool for studying the anatomy and development of the Drosophila central nervous system. Cell Tissue Res. 1997;290:1–10. doi: 10.1007/s004410050901. [DOI] [PubMed] [Google Scholar]
- Ito K, Suzuki K, Estes P, Ramaswami M, Yamamoto D, Strausfeld NJ. The organization of extinsic neurons and their implications in the functional roles of the mushroom bodies in Drosophila melanogaster Meigen. Learn & Mem. 1998;5:52–77. [PMC free article] [PubMed] [Google Scholar]
- Joiner M-lA, Griffith LC. Mapping of the anatomical circuit of CaM kinase-dependent courtship conditioning in Drosophila. Learn & Mem. 1999;6:177–192. [PMC free article] [PubMed] [Google Scholar]
- Klein M, Kandel ER. Mechanism of calcium current modulation underlying presynaptic facilitation and behavioral sensitization in Aplysia. Proc Natl Acad Sci. 1980;77:6912–6916. doi: 10.1073/pnas.77.11.6912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner HA, Byrne JH. New perspectives on classical conditioning: A synthesis of Hebbian and non-Hebbian mechanisms. Neuron. 1998;20:355–358. doi: 10.1016/s0896-6273(00)80977-0. [DOI] [PubMed] [Google Scholar]
- Levin LR, Han PL, Hwang PM, Feinstein PG, Davis RL, Reed RR. The Drosophila learning and memory gene rutabaga encodes a Ca2+/Calmodulin-responsive adenylyl cyclase. Cell. 1992;68:479–489. doi: 10.1016/0092-8674(92)90185-f. [DOI] [PubMed] [Google Scholar]
- Luo L, Liao YJ, Jan LY, Jan YN. Distinct morphogenetic functions of similar small GTPases: Drosophila Drac1 is involved in axonal outgrowth and myoblast fusion. Genes & Dev. 1994;8:1787–1802. doi: 10.1101/gad.8.15.1787. [DOI] [PubMed] [Google Scholar]
- Milner B, Squire LR, Kandel ER. Cognitive neuroscience and the study of memory. Neuron. 1998;20:445–468. doi: 10.1016/s0896-6273(00)80987-3. [DOI] [PubMed] [Google Scholar]
- O'Dell KM, Armstrong JD, Yang MY, Kaiser K. Functional dissection of the Drosophila mushroom bodies by selective feminization of genetically defined subcompartments. Neuron. 1995;15:55–61. doi: 10.1016/0896-6273(95)90064-0. [DOI] [PubMed] [Google Scholar]
- Power ME. The thoracico-abdominal nervous system of an adult insect, Drosophila melanogaster. J Comp Neurol. 1948;88:347–409. doi: 10.1002/cne.900880303. [DOI] [PubMed] [Google Scholar]
- Rees CT, Spatz HC. Habituation of the landing response of Drosophila wild-type and mutants defective in olfactory learning. J Neurogenet. 1989;5:105–118. doi: 10.3109/01677068909066201. [DOI] [PubMed] [Google Scholar]
- Sachs L. Angewandte Statistik. Berlin, Germany: Springer-Verlag; 1992. [Google Scholar]
- Sayeed O, Benzer S. Behavioral genetics of thermosensation and hygrosensation in Drosophila. Proc Natl Acad Sci. 1996;93:6079–6084. doi: 10.1073/pnas.93.12.6079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schacher S, Kandel ER, Montarolo P. cAMP and arachidonic acid simulate long-term structural and functional changes produced by neurotransmitters in Aplysia sensory neurons. Neuron. 1993;10:1079–1088. doi: 10.1016/0896-6273(93)90056-w. [DOI] [PubMed] [Google Scholar]
- Strausfeld NJ. Atlas of an insect brain. Berlin, Germany: Springer-Verlag; 1976. [Google Scholar]
- Tettamanti M, Armstrong JD, Endo K, Yang MY, Furukubo-Tokunaga K, Kaiser K, Reichert H. Early development of the Drosophila mushroom bodies, brain centres for associative learning and memory. Dev Genes Evol. 1997;207:242–252. doi: 10.1007/s004270050112. [DOI] [PubMed] [Google Scholar]
- Tsien JZ, Huerta PT, Tonegawa S. The essential role of hippocampal CA1 NMDA receptor-dependent synaptic plasticity in spatial memory. Cell. 1996;87:1327–1338. doi: 10.1016/s0092-8674(00)81827-9. [DOI] [PubMed] [Google Scholar]
- Wehner R. Arthropods. In: Papi F, editor. Animal homing. London, UK: Chapman and Hall; 1992. pp. 45–144. [Google Scholar]
- ————— Middle-scale navigation: The insect case. J Exp Biol. 1996;199:125–127. doi: 10.1242/jeb.199.1.125. [DOI] [PubMed] [Google Scholar]
- Wolf R, Wittig T, Liu L, Wustmann G, Eyding D, Heisenberg M. Drosophila mushroom bodies are dispensible for visual, tactile, and motor learning. Learn & Mem. 1998;5:166–178. [PMC free article] [PubMed] [Google Scholar]
- Wu ZL, Thomas SA, Villacres EC, Xia Z, Simmons ML, Chavkin C, Palmiter RD, Storm DR. Altered behavior and long-term potentiation in type I adenylyl cyclase mutant mice. Proc Natl Acad Sci. 1995;92:220–224. doi: 10.1073/pnas.92.1.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wustmann G, Heisenberg M. Behavioral manipulation of retrieval in a spatial memory task for Drosophila melanogaster. Learn & Mem. 1997;4:328–336. doi: 10.1101/lm.4.4.328. [DOI] [PubMed] [Google Scholar]
- Wustmann G, Rein K, Wolf R, Heisenberg M. A new paradigm for operant conditioning of Drosophila melanogaster. J Comp Physiol [A] 1996;179:429–436. doi: 10.1007/BF00194996. [DOI] [PubMed] [Google Scholar]
- Yang MY, Armstrong JD, Vilinsky I, Strausfeld NJ, Kaiser K. Subdivision of the Drosophila mushroom bodies by enhancer-trap expression patterns. Neuron. 1995;15:45–54. doi: 10.1016/0896-6273(95)90063-2. [DOI] [PubMed] [Google Scholar]
- Yao KM, White K. Neural specificity of elav expression: Defining a Drosophila promoter for directing expression to the nervous system. J Neurochem. 1994;63:41–51. doi: 10.1046/j.1471-4159.1994.63010041.x. [DOI] [PubMed] [Google Scholar]
- Zhong Y. Mediation of PACAP-like neuropeptide transmission by coactivation of Ras/Raf and cAMP signal transduction pathways in Drosophila. Nature. 1995;375:588–592. doi: 10.1038/375588a0. [DOI] [PubMed] [Google Scholar]
- Zhong Y, Pena LA. A novel synaptic transmission mediated by a PACAP-like neuropeptide in Drosophila. Neuron. 1995;14:527–536. doi: 10.1016/0896-6273(95)90309-7. [DOI] [PubMed] [Google Scholar]
- Zhong Y, Wu CF. Altered synaptic plasticity in Drosophila memory mutants with a defective cyclic AMP cascade. Science. 1991;251:198–201. doi: 10.1126/science.1670967. [DOI] [PubMed] [Google Scholar]