Abstract
Genetic studies have identified Drosophila Naked Cuticle (Nkd) as an antagonist of the canonical Wnt/β-catenin signaling pathway, but its mechanism of action remains obscure [Zeng, W., Wharton, K. A., Jr., Mack, J. A., Wang, K., Gadbaw, M., et al. (2000) Nature (London) 403, 789–795]. Here we have cloned a cDNA encoding a mammalian homolog of Drosophila Nkd, mNkd, and demonstrated that mNkd interacts directly with Dishevelled. Dishevelled is an intracellular mediator of both the canonical Wnt pathway and planar cell polarity (PCP) pathway. Activation of the c-Jun-N-terminal kinase has been implicated in the PCP pathway. We showed that mNkd acts in a cell-autonomous manner not only to inhibit the canonical Wnt pathway but also to stimulate c-Jun-N-terminal kinase activity. Expression of mNkd disrupted convergent extension in Xenopus, consistent with a role for mNkd in the PCP pathway. These data suggest that mNkd may act as a switch to direct Dishevelled activity toward the PCP pathway, and away from the canonical Wnt pathway.
Dishevelled, an intracellular modular protein, is required for the activation of several Wnt signaling pathways, including the canonical Wnt/β-catenin pathway and the planar cell polarity (PCP) pathway (1). These pathways are activated by a variety of Wnt ligands and receptors, and they signal downstream to at least two different cascades (1–3). One pathway, the canonical Wnt pathway, is mediated by GSK-3β and β-catenin and controls normal cell growth and cell-fate specification. Aberrant regulation of this pathway causes diverse developmental defects in organisms ranging from Drosophila to mammals. Notably, aberrant regulation of the pathway has also been found in a variety of cancer types, implicating the involvement of this pathway in tumorigenesis (4–7). Another Wnt signaling pathway, the PCP pathway, signals via small GTPases and activates c-Jun-N-terminal kinase (JNK) to control epithelial cell polarity in Drosophila (8, 9). Recently, the PCP pathway has also been shown to regulate cell polarity during convergent extension movements in developing Xenopus and zebrafish embryos (10–13).
Dishevelled is at the branchpoint of these pathways, and different conserved domains within Dishevelled are required for the specific activation of each pathway (10–12, 14–16). To understand how the Dishevelled protein works as a common component in multiple pathways, we screened for mouse Dishevelled (mDvl)-associated proteins in a yeast two-hybrid assay. Among those identified in this assay is a mouse homolog of Drosophila Naked Cuticle (Nkd). This finding is particularly interesting because loss-of-function mutation of nkd produces a defect that mimics the phenotype caused by excess Wingless, and nkd has recently been shown to encode an antagonist of Wingless signaling (17). However, the mechanism by which Nkd functions is unclear. The genetic data in flies suggest that Nkd could function non-cell-autonomously by regulating Wingless production and/or Wingless binding to its receptor, or it could function cell-autonomously by regulating the intracellular cascade of Wingless signaling (17). Here we show that the mammalian Nkd (mNkd)interacts directly with Dishevelled and functions as an intracellular, cell-autonomous regulator of multiple Dishevelled-mediated Wnt pathways.
Materials and Methods
Two-Hybrid Screen.
Yeast two-hybrid screening was carried out by using the complete coding region of mouse Dishevelled 3 (mDvl3) as a bait. A prey cDNA library made from 9.5- to 10.5-day-old (E9.5–E10.5) mouse embryo (18) was screened in the presence of 10 mM 3-amino-1,2,4-triazole (3-AT, Sigma).
Northern Blotting.
A mouse embryo RNA blot (CLONTECH) was hybridized with the radioactively labeled coding region of mNkd cDNA. A mouse multiple tissue RNA blot (CLONTECH) was hybridized with a radioactively labeled cDNA fragment of mNkd containing nucleotides 319–690, which corresponds to the Dishevelled-binding domain of mNkd isolated from the yeast two-hybrid screen.
Cloning of mNkd.
Full-length mNkd cDNA was cloned by a combination of mouse fetal cDNA library screening, reverse transcription–PCR (RT-PCR), and rapid amplification of cDNA ends (RACE) methodologies. A fetal mouse cDNA library (OriGene Technologies) was screened by PCR using oligo pairs: 5′-CCTCCAAGAAGCAGCTCAAGTT-3′ and 5′-TTGTGCTCTGCAGATCGGTATGG-3′. A Marathon-ready cDNA library from mouse lung (CLONTECH) was then used to obtain the 5′ end sequence of mNkd cDNA by 5′ RACE. The oligo sequence of mNkd cDNA used in the RACE was 5′-CCCGTCAGGAGCCACGGTGAGCTTCAC-3′.
Mammalian Cell Cultures and Luciferase Reporter Assay.
HEK293 and Cos7 cells were grown in DMEM supplemented with 10% (vol/vol) FBS. NIH 3T3 cells were grown in DMEM supplemented with 10% (vol/vol) calf serum. Transfections were performed by using Lipofectamine Plus (Life Technologies, Grand Island, NY) according to the manufacturer's protocols. Luciferase reporter assays were carried out in 12-well plates as described (19). The LEF-1 luciferase reporter activity of each sample was determined and normalized by using a dual-luciferase reporter assay system (Promega) according to the manufacturer's instructions.
Immunoblots and Immunoprecipitations.
Cell lysates were prepared with lysis buffer (150 mM NaCl/20 mM Tris⋅HCl, pH 7.5/1 mM EDTA/0.1% Triton X-100, supplemented with protease inhibitor mixture). Immunoblotting and immunoprecipitations were performed as described (19).
In Vitro Binding of mNkd with Dishevelled.
Glutathione S-transferase (GST) fusion proteins were expressed in Escherichia coli strain BL21 DE3 (plyS) and purified with glutathione beads (Amersham Pharmacia). Myc-mNkd protein was prepared by in vitro transcription and translation by using a TNT coupled reticulocyte lysate system (Promega) in the presence of [35S]-methionine. The 35S-labeled mNkd was precipitated for 3 h at 4°C with Myc antibody and protein A beads or by GST-fusion proteins immobilized on glutathione beads.
RT-PCR.
RNA for RT-PCR was prepared by using an RNeasy mini kit (Qiagen, Chatsworth, CA). RT reaction was performed by using a RETROscript kit (Ambion, Austin, TX). Quantitative PCR analyses were performed by using a LightCycler system from Roche Molecular Biochemicals.
Xenopus Experiments.
For secondary axis assay, in vitro transcribed mRNAs were injected into one ventral blastomere of four-cell-stage embryos, and secondary axes were scored at stage 30. For Keller explant assays, Xenopus embryos were injected into both dorsal blastomeres at the four-cell stage; embryos were coinjected with 100 pg of green fluorescent protein (GFP) mRNA as a tracer. Embryos were reared to stage 10.5, and Keller explants were removed as described (12) and cultured under coverglass until stage 20 in 1× Steinberg's medium. Only GFP-positive explants were scored.
JNK Assay.
The JNK assay was performed as described (15) with modifications. NIH 3T3 cells were transfected with a total of 2 μg of DNA containing a mixture of 0.4 μg of c-Jun with various amounts of mNkd or mDvl3 constructs. At 22 h after transfection, cells were lysed in SDS-sample buffer. Protein samples were separated by electrophoresis on a Tris-Glycine polyacrylamide gel (NOVEX, San Diego) and transferred onto a nitrocellulose membrane. The membrane was blotted with PhosphoPlus c-Jun (Ser-63) II antibody (New England Biolabs), which recognizes the phosphorylated serine at position 63 of c-Jun.
Results
Cloning of mNkd, a Mammalian Homolog of Drosophila Nkd.
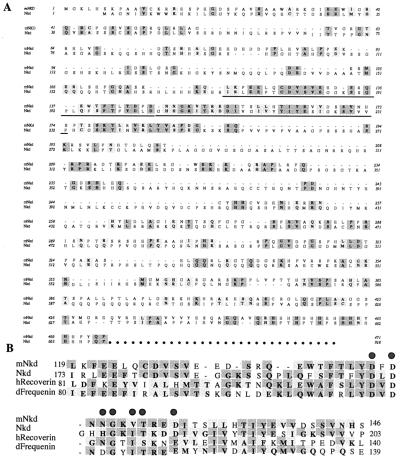
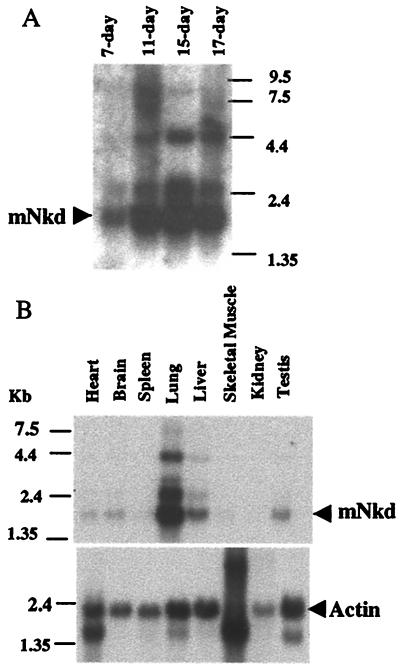
Using mDvl as bait in a yeast two-hybrid screen of a mouse embryonic cDNA library (18), we identified an interacting protein, mNkd, that is 30% identical to Drosophila Nkd (ref. 17; Fig. 1A). Both mNkd and Drosophila Nkd (17) contain a single EF-hand (common helix-loop-helix calcium-binding motif) calcium-binding motif which has the most similarity to the EF-hand found in the Recoverin family of calcium-binding proteins (Fig. 1B). The domain of mNkd that interacts with mDvl in the yeast two-hybrid experiments is located between amino acids 107 and 230 and includes the EF-hand (Fig. 1A). Northern blot analysis showed a major 1.7-kb mRNA corresponding to the size of the cloned mnkd cDNA. This mRNA was detected during all stages of embryonic development (Fig. 2A), which is consistent with the requirement of Drosophila Nkd for fly embryonic development. In adult mice, this mRNA was detected at high levels only in lung and liver and at low levels in heart, brain, and testis (Fig. 2B). We also observed mRNAs that are larger and less abundant than the 1.7-kb mRNA. It is likely that these mRNAs of larger sizes are other EF-hand-containing mRNAs that cross-hybridized with the probes. However, we cannot exclude the possibility that these larger mRNAs may be splice variants of the same gene.
Figure 1.
Sequence comparison of mNkd and the Drosophila Nkd. (A) Protein sequence alignment of mNkd and Drosophila Nkd. Deduced protein sequences of mNkd and Nkd (17) are compared with the Macvector clusterw program. Identical residues are highlighted in dark gray; conserved changes are highlighted in light gray. The EF-hand motifs and the surrounding amino acids in both proteins are underlined. (B) Alignment of the underlined amino acids of mNkd, Nkd (17), human Recoverin (41), and Drosophila Frequenin (42). Residues that are identical in more than two proteins are highlighted in dark gray; conserved changes are highlighted in light gray. Consensus residues in the EF-hand that bind calcium with high affinity are indicated by shaded dots above these residues (17, 20).
Figure 2.
mNkd expression. (A) Result of mouse-embryo Northern blotting. (B) Result of mouse multiple-tissue Northern blotting. Actin was used as an internal standard for RNA loading.
mNkd Interacts Directly with the Middle Region of Dishevelled.
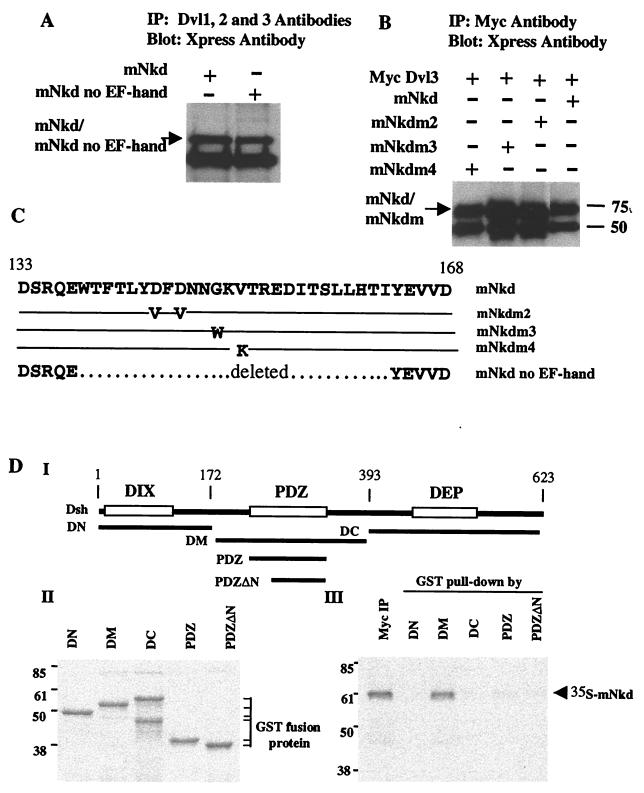
The interaction of mNkd with mDvl was recapitulated in mammalian cells by coimmunoprecipitation experiments. mNkd was transiently expressed in Cos7 or HEK293 cells; endogenous mDvl proteins were immunoprecipitated from total cell lysates. As shown in Fig. 3A, mNkd was detected in the Dishevelled immunocomplex by Western blotting. Because the EF-hand is included in the region of mNkd that is associated with mDvl in the yeast two-hybrid experiments and is highly conserved between the Drosophila Nkd and mNkd, the requirement of the EF-hand in the association with mDvl was investigated. Based on the crystal structure of the third EF-hand of the Recoverin protein (20), we made mutations that either changed the consensus residues in the calcium-binding loop or deleted the entire calcium-binding loop together with the surrounding amino acids (Fig. 3C). These mNkd mutants were expressed in HEK293 or Cos7 cells. Coimmunoprecipitation experiments revealed that none of these mutations significantly impaired the ability of these mNkd proteins to associate with mDvl (Fig. 3 A and B). These data showed that mNkd associates with mDvl in mammalian cells and that the intact EF-hand is not required for the association.
Figure 3.
mNkd interacts with Dishevelled. (A) Lysates were prepared from Cos7 cells expressing mNkd, mNkd no EF-hand, or GFP constructs. Total cell lysates were immunoprecipitated with monoclonal antibodies 1, 2, and 3 to Dishevelled (Santa Cruz Biotechnology); after SDS/PAGE and membrane transfer, the membrane was blotted with Xpress antibody (Invitrogen). (B) mNkd mutations in the EF-hand do not have a significant effect on the binding of mNkd to Dishevelled. Cell lysates prepared from HEK293 cells expressing Myc-tagged Dishevelled and Xpress-tagged mNkd proteins were immunoprecipitated with Myc antibody (Roche Molecular Biochemicals); after SDS/PAGE and membrane transfer, the membrane was blotted with Xpress antibody. (C) Summary of mutations in the EF-hand of mNkd. The mNkd mutant m2 contains amino acid changes D144V and D146V. The mNkd mutant m3 contains amino acid change G149W. The mNkd mutant m4 contains amino acid change V151K. –, identical amino acids as the wild type; … . . , deleted amino acids in the mNkd no EF-hand mutant. (D) mNkd binds to the conserved middle region of Dishevelled. (I) Schematic diagram of GST-fusion proteins containing different regions of Drosophila Dishevelled (Dsh). (III) Myc-tagged mNkd protein, labeled with 35S, was immunoprecipitated with Myc antibody or precipitated by GST-fusion proteins. The immunocomplexes were separated by SDS/PAGE and detected by autoradiography. (II) Equal amounts of GST-fusion proteins used in III were separated by SDS/PAGE and detected by Coomassie blue stain.
We also identified the domain on Dishevelled that associated with mNkd. Dishevelled is a highly conserved protein and contains three distinct domains (1). The N-terminal region has a DIX domain that is required for canonical Wnt signaling. The middle region of Dishevelled contains a PDZ domain that is known to bind GBP/Frat1 (21, 22) and CK1ɛ (23, 24), both positive regulators of the canonical Wnt pathway. The C-terminal region contains a DEP domain that is crucial for regulating the PCP pathway. Fragments corresponding to different regions of Drosophila Dishevelled (Dsh; Fig. 3 DI) were expressed in E. coli as GST-fusion proteins. Equal amounts of each fragment (Fig. 3 DII) were mixed with in vitro-translated mNkd in the binding buffer, precipitated with glutathione beads and separated by SDS/PAGE. mNkd associated with the DM fragment of Dsh that encompasses the PDZ domain with the adjoining N-terminal basic amino acid stretch. Notably, the PDZ domain alone is not sufficient for the association. Also, there is no association of mNkd with the DIX domain in the N-terminal region or the DEP domain in the C-terminal region of Dsh (Fig. 3 DIII). Thus mNkd is associated with a region within Dsh shared with GBP/Frat 1 and CK1ɛ.
mNkd Antagonizes Canonical Wnt Signaling.
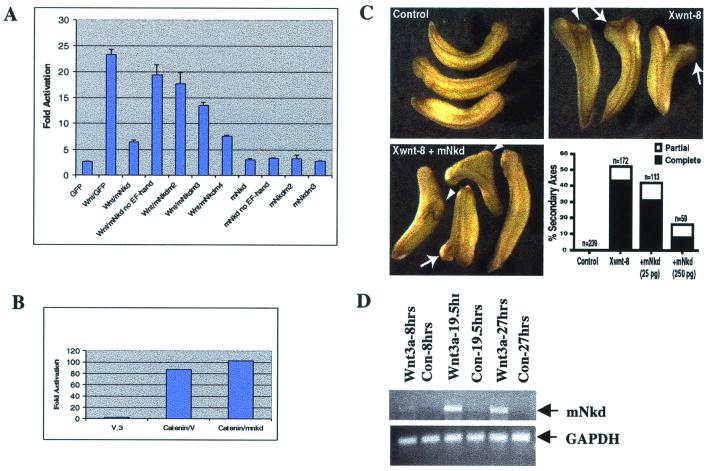
Because Dishevelled is a known positive regulator of the canonical Wnt pathway and mNkd is found here to be directly associated with Dishevelled, the role of mNkd in the canonical Wnt pathway was tested in mammalian cell culture by using a Wnt-1 ligand-responsive luciferase reporter assay (25). In multiple experiments, activation of the reporter by Wnt-1 was inhibited 75% by coexpression of mNkd in mammalian cells (Fig. 4A). Expression of wild-type mNkd, in the absence of Wnt-1, had no effect on the activity of the reporter. These data suggest that mNkd negatively regulates the canonical Wnt pathway and are consistent with the inhibitory effect that Drosophila Nkd has on Wingless signaling in genetic studies (17). Interestingly, the EF-hand mutants of mNkd showed an impaired ability to inhibit the canonical Wnt pathway (Fig. 4A), although these mutants were all capable of binding to Dishevelled (Fig. 3 A and B). These data suggest that the association of mNkd with Dishevelled alone is not sufficient to inhibit the canonical Wnt pathway, and that the intact EF-hand is required for the inhibitory function. Importantly, mNkd failed to inhibit the gene response elicited by overexpression of β-catenin, a result that placed mNkd upstream of β-catenin in the canonical Wnt pathway (Fig. 4B).
Figure 4.
mNkd inhibits the canonical Wnt pathway and mNkd-mRNA level is up-regulated by Wnt. (A) HEK293 cells were transfected with expression constructs that included 0.02 μg of LEF-1, 0.2 μg of luciferase reporter (25), 0.02 μg of pRL-TK (Promega), 0.1 μg of pCGWnt-1, and 0.2 μg of GFP, mNkd, or mNkd-mutant derivatives. LEF-1 luciferase reporter activities were determined. (B) mNkd did not inhibit the β-catenin-activated Lef-1 reporter. HEK293 cells were transfected with expression constructs that included 0.02 μg of LEF-1, 0.2 μg of luciferase reporter (25), 0.02 μg of pRL-TK (Promega), 0.1 μg of β-catenin, and 0.2 μg of vector or mNkd. (C) mNkd inhibited Xwnt-8-induced secondary axes formation in Xenopus. Control Xenopus embryos are shown at Upper Left. Embryos were injected ventrally with 5–10 pg of Xwnt-8 RNA and developed with secondary axes (Upper Right). Secondary axes were scored as complete (arrows) or partial (arrowheads) based on development of anterior structures such as eyes and cement glands. Coexpression of mNkd with Xwnt-8 decreased the frequency of secondary axes formation as well as the percentage of secondary axes that contained anterior structures (Lower Left and Lower Right). This effect was dose-dependent. (D) BALB/c LI mouse liver epithelial cells were treated with either Wnt-3a or Neo-conditioned medium (34) for indicated hours. PCR was carried out by using primer pairs 5′-TGTGAACCATTCCCCCACATCAA-3′ and 5′-AAATGGGGTGTCAAGGAGGTGGAA-3′. The PCR products were separated by agarose gel electrophoresis. GAPDH, glyceraldehyde-3-phosphate control.
We also used the induction of secondary axes by ectopic expression of Xwnt-8 in Xenopus embryo as an assay for the role of mNkd in the canonical Wnt pathway in vivo (26–28). Ventral blastomere injection of 5–10 pg of Xwnt-8 RNA induced secondary axes in over 50% of the embryos; most of these secondary axes contained anterior structures (Fig. 4C). Coinjection of 25 pg of mNkd RNA suppressed the effect of Xwnt-8 and resulted in fewer secondary axes (Fig. 4C). Coinjection of higher doses of mNkd (250 pg) resulted in even fewer secondary axes (Fig. 4C), only half of which contained anterior structures, indicating that mNkd inhibited the canonical Wnt pathway in vivo. The promoter of the Xenopus nodal-related-3 (Xnr-3) gene has been shown to be directly activated by the canonical Wnt/β-catenin signaling in the Xenopus embryo (29), and expression of Xwnt-8 RNA into Xenopus animal caps activated transcription from a coinjected Xnr-3-luciferase reporter plasmid in vivo (data not shown). Consistent with the results from mammalian cell culture (Fig. 4A) and the secondary axis assay (Fig. 4C), coinjection of mNkd RNA suppressed Xwnt-8 activation of the Xnr-3 promoter (data not shown). These data indicate that mNkd is an inhibitor of the canonical Wnt/β-catenin pathway both in vitro and in vivo. Together, our findings that mNkd interacted directly with Dishevelled and inhibited the canonical Wnt pathway upstream of β-catenin suggest that mNkd is an intracellular antagonist of the Wnt pathway.
mNkd mRNA Is Induced by Wnt Ligand in Mammalian Cells.
Recently, a number of genes have been identified that are induced by Wnt expression in mammalian cells (30–33). We also tested whether mNkd-mRNA levels changed when cells were treated with Wnt ligands. BALB/c LI mouse liver epithelial cells were treated with Wnt-3A-conditioned medium or control medium (34) for 8 h, 19.5 h, or 27 h, respectively. Levels of mNkd transcripts increased significantly in cells treated with Wnt-3a-conditioned medium for 19.5 h and 27 h compared with control treatments (Fig. 4D). Levels of the glyceraldehyde-3-phosphate dehydrogenase transcript remained constant in all conditions (Fig. 4D). The results of quantitative PCR recorded in the linear range using a LightCycler corroborated the PCR data (data not shown). Wnt-3a-conditioned medium also caused an increase in mRNA levels of mNkd in L cells (data not shown). The mRNA levels of mNkd also increased in BALB/c LI mouse liver epithelial cells treated with lithium chloride (data not shown), a known inhibitor of GSK-3 (35). Thus, the ability of mNkd to inhibit the intracellular signaling of the canonical Wnt pathway, in conjunction with the result that mNkd is itself a downstream target of the canonical Wnt signaling, suggests that mNkd is an intracellular cell-autonomous negative-feedback regulator of the canonical Wnt pathway.
mNkd Stimulates JNK Activity.
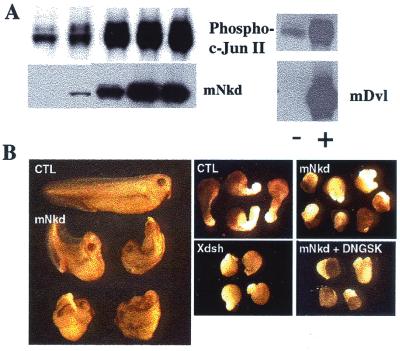
Genetic and biochemical studies have shown that Dishevelled controls cell polarity by acting as an upstream activator of the JNK pathway both in vivo and in vitro (15). Because mNkd is directly associated with Dishevelled, we tested whether mNkd participated in the JNK pathway. NIH 3T3 cells were transfected with expression constructs of mNkd and c-Jun, in which c-Jun served to monitor JNK activities. In this assay, expression of mNkd or mDvl alone induced a strong phosphorylation of c-Jun which was detected by blotting with an antibody specific for phosphoserine-63 (Fig. 5A). These data showed that mNkd had an effect similar to Dishevelled in activating the JNK pathway in mammalian cell culture assays.
Figure 5.
mNkd activates JNK in mammalian cell culture and alters convergent extension movements in Xenopus laevis. (A) The JNK assay was carried out as described (15). NIH 3T3 cells were transfected with plasmids expressing c-Jun and mNkd or c-Jun and mDvl3. Cell lysates were separated by SDS/PAGE and transferred onto a nitrocellulose membrane. The membrane was blotted with PhosphoPlus c-Jun (Ser-63) II antibody (New England Biolabs). The same membrane was then stripped and blotted with Xpress antibody (Invitrogen) to detect the amount of Xpress-tagged mNkd or blotted with Myc antibody to detect Dishevelled. (B) Dorsal expression of mNkd in developing Xenopus embryos inhibited the normal elongation and straightening of the anteroposterior axis (Left; control embryo at top, mNkd-injected embryos below). (Center Upper) Control open-face Keller explants of the dorsal marginal zone elongated and changed shape significantly. (Right Upper) Explants expressing mNkd failed to elongate or to change shape. (Center Lower) Overexpression of wild-type Xdsh mimicked overexpression of mNkd. (Right Lower) Downstream activation of the canonical Wnt pathway by coexpression of dominant-negative GSK-3β did not attenuate the inhibitory effect of mNkd on convergent extension. Expression of dominant-negative GSK-3β alone had no effect on convergent extension (data not shown).
mNkd Expression Inhibits Convergent Extension.
Activation of JNK seems to be an important step in the PCP pathway (8, 15), and it has been demonstrated recently that a vertebrate cognate of the Drosophila PCP pathway (8) controls convergent extension movements during vertebrate development (10–13). In both Xenopus and Drosophila, hyperactivation of this pathway disrupts PCP signaling without affecting the canonical Wnt pathway (12–15). Consistent with its ability to activate JNK in vitro (Fig. 5A), mNkd overexpression inhibited the normal elongation of Xenopus embryos (Fig. 5B, Left). The normal formation of anterior structures in these embryos indicates that the phenotype is not the result of ventralization, suggesting that mNkd inhibits convergent extension. To assess more directly the effects of mNkd on convergent extension, we examined open-face Keller explants of the dorsal mesoderm (12). Such explants made from control embryos elongated and changed shape significantly (Fig. 5B, Center Upper), whereas explants made from embryos expressing mNkd failed to elongate (Fig. 5B, Right Upper). These effects are similar to those elicited by overexpression of other wild-type components of the planar cell polarity cascade, such as Xdsh (Fig. 5B, Center Lower; ref. 12) and Xfz-8 (13), indicating a role of mNkd in controlling the PCP pathway.
Because mNkd is an inhibitor of the canonical Wnt pathway (Fig. 4A), it was important to test whether the effects of mNkd on convergent extension were simply a consequence of the effects of mNkd on the canonical Wnt pathway. Expression of dominant-negative GSK-3β, a strong activator of the canonical Wnt pathway in Xenopus (36, 37), did not attenuate the inhibitory effects of mNkd on convergent extension (Fig. 5B, Right Lower). Taken together, these data suggest that mNkd inhibits convergent extension by overstimulating the PCP-signaling cascade, and that this effect is independent of its inhibitory role on the canonical Wnt pathway.
Discussion
Previous genetic studies identified Drosophila Nkd as an important negative regulator of Wnt signaling. However, the mechanism by which Nkd effects this regulation has not been elucidated. In this study, we showed that mNkd, a mammalian homolog of Nkd, also negatively regulates canonical Wnt signaling. Furthermore, we have demonstrated that mNkd associates with Dishevelled directly and functions as an intracellular cell-autonomous Wnt-inducible negative regulator of the canonical Wnt pathway at a step upstream of β-catenin. Our data also revealed the importance of the intact EF-hand calcium-binding domain in regulating Wnt signaling, a result that is particularly intriguing in light of other studies that reported stimulation of intracellular calcium release by Wnt ligands (38–40). In addition, we have shown here that mNkd not only suppresses the canonical Wnt pathway but also activates JNK, a component of the PCP pathway. Furthermore, mNkd expression inhibits convergent extension in Xenopus in a manner similar to overexpression of other PCP pathway components.
Because mNkd binds to Dishevelled and Dishevelled is a known regulator of both the canonical Wnt pathway and the PCP pathway (1), the data described here suggest the possibility that mNkd could act as a switch that directs Dishevelled toward the JNK/PCP pathway and away from the canonical Wnt/β-catenin pathway. Finally, as an intracellular negative regulator of the canonical Wnt pathway, mNkd shares properties with the tumor suppressors Axin and APC.
Acknowledgments
We are grateful to Drs. R. Grosschedl, D. Sussman, S. Hollenberg, and R. Takada for reagents. We thank S. Harrison, V. Chan, M. Haertel-Wiesmann, R. Halenbeck, D. Rohrer, and M. Rohan for helpful discussions. We also thank M. Del Rosario and Dr. E. Casey for technique support and Betty Cheung for proofreading of the manuscript. This work was funded partially by unrestricted awards from the Howard Hughes Medical Institute and Bristol-Myers Squibb (to L.T.W.). Work in the laboratory of R.M.H. is supported by National Institute of Health grants. J.B.W. is supported by a postdoctoral research fellowship from the American Cancer Society.
Abbreviations
- PCP
planar cell polarity
- JNK
c-Jun-N-terminal kinase
- Nkd
Drosophila Naked Cuticle
- RT-PCR
reverse transcription–PCR
- GST
glutathione S-transferase
- EF-hand
common helix–loop–helix calcium binding motif
- mNkd
mammalian homolog of Nkd
- Dsh
Drosophila Disheveled
- En
embryo n days old
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF343352).
References
- 1.Boutros M, Mlodzik M. Mech Dev. 1999;83:27–37. doi: 10.1016/s0925-4773(99)00046-5. [DOI] [PubMed] [Google Scholar]
- 2.McEwen D G, Peifer M. Curr Biol. 2000;10:R562–R564. doi: 10.1016/s0960-9822(00)00611-4. [DOI] [PubMed] [Google Scholar]
- 3.Peifer M, Polakis P. Science. 2000;287:1606–1609. doi: 10.1126/science.287.5458.1606. [DOI] [PubMed] [Google Scholar]
- 4.Cadigan K, Nusse R. Genes Dev. 1997;11:3286–3305. doi: 10.1101/gad.11.24.3286. [DOI] [PubMed] [Google Scholar]
- 5.Wodarz A, Nusse R. Annu Rev Cell Dev Biol. 1998;14:59–88. doi: 10.1146/annurev.cellbio.14.1.59. [DOI] [PubMed] [Google Scholar]
- 6.Smalley M, Dale T C. Cancer Metastasis Rev. 1999;18:215–230. doi: 10.1023/a:1006369223282. [DOI] [PubMed] [Google Scholar]
- 7.Polakis P. Genes Dev. 2000;14:1837–1851. [PubMed] [Google Scholar]
- 8.Shulman J M, Perrimon N, Axelrod J D. Trends Genet. 1998;14:452–458. doi: 10.1016/s0168-9525(98)01584-4. [DOI] [PubMed] [Google Scholar]
- 9.Mlodzik M. EMBO J. 1999;18:6873–6879. doi: 10.1093/emboj/18.24.6873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heisenberg C-P, Tada M, Rauch G J, Saude L, Concha M L, Geisler R, Stemple D L, Smith J C, Wilson S W. Nature (London) 2000;405:76–81. doi: 10.1038/35011068. [DOI] [PubMed] [Google Scholar]
- 11.Tada M, Smith J C. Development (Cambridge, UK) 2000;127:2227–2238. doi: 10.1242/dev.127.10.2227. [DOI] [PubMed] [Google Scholar]
- 12.Wallingford J B, Rowning B A, Vogeli K M, Rothbacher U, Fraser S E, Harland R M. Nature (London) 2000;405:81–85. doi: 10.1038/35011077. [DOI] [PubMed] [Google Scholar]
- 13.Wallingford J B, Vogeli K M, Harland R M. Int J Dev Biol. 2001;45:225–227. [PubMed] [Google Scholar]
- 14.Axelrod J D, Miller J R, Shulman J M, Moon R T, Perrimon N. Genes Dev. 1998;12:2610–2622. doi: 10.1101/gad.12.16.2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boutros M, Paricio N, Strutt D I, Mlodzik M. Cell. 1998;94:109–118. doi: 10.1016/s0092-8674(00)81226-x. [DOI] [PubMed] [Google Scholar]
- 16.Li L, Yuan H, Xie W, Mao J, Caruso A M, McMahon A, Sussman D J, Wu D. J Biol Chem. 1999;274:129–134. doi: 10.1074/jbc.274.1.129. [DOI] [PubMed] [Google Scholar]
- 17.Zeng W, Wharton K A, Jr, Mack J A, Wang K, Gadbaw M, Suyama K, Klein P S, Scott M P. Nature (London) 2000;403:789–795. doi: 10.1038/35001615. [DOI] [PubMed] [Google Scholar]
- 18.Hollenberg S M, Sternglanz R, Cheng P F, Weintraub H. Mol Cell Biol. 1995;15:3813–3822. doi: 10.1128/mcb.15.7.3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sakanaka C, Weiss J B, Williams L T. Proc Natl Acad Sci USA. 1998;95:3020–3023. doi: 10.1073/pnas.95.6.3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Flaherty K M, Zozulya S, Stryer L, McKay D B. Cell. 1993;75:709–716. doi: 10.1016/0092-8674(93)90491-8. [DOI] [PubMed] [Google Scholar]
- 21.Yost C, Farr G H, III, Pierce S B, Ferkey D M, Chen M M, Kimelman D. Cell. 1998;93:1031–1041. doi: 10.1016/s0092-8674(00)81208-8. [DOI] [PubMed] [Google Scholar]
- 22.Li L, Yuan H, Weaver C D, Mao J, Farr G H, III, Sussman D J, Jonkers J, Kimelman D, Wu D. EMBO J. 1999;18:4233–4240. doi: 10.1093/emboj/18.15.4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peters J M, McKay R M, McKay J P, Graff J M. Nature (London) 1999;401:345–350. doi: 10.1038/43830. [DOI] [PubMed] [Google Scholar]
- 24.Sakanaka C, Leong P, Xu L, Harrison S D, Williams L T. Proc Natl Acad Sci USA. 1999;96:12548–12552. doi: 10.1073/pnas.96.22.12548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hsu S C, Galceran J, Grosschedl R. Mol Cell Biol. 1998;18:4807–4818. doi: 10.1128/mcb.18.8.4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McMahon A P, Moon R T. Cell. 1989;58:1075–1084. doi: 10.1016/0092-8674(89)90506-0. [DOI] [PubMed] [Google Scholar]
- 27.Smith W C, Harland R M. Cell. 1991;67:753–765. doi: 10.1016/0092-8674(91)90070-f. [DOI] [PubMed] [Google Scholar]
- 28.Sokol S, Christian J L, Moon R T, Melton D A. Cell. 1991;67:741–752. doi: 10.1016/0092-8674(91)90069-b. [DOI] [PubMed] [Google Scholar]
- 29.McKendry R, Hsu S C, Harland R M, Grosschedl R. Dev Biol. 1997;192:420–431. doi: 10.1006/dbio.1997.8797. [DOI] [PubMed] [Google Scholar]
- 30.Pennica D, Swanson T A, Welsh J W, Roy M A, Lawrence D A, Lee J, Brush J, Taneyhill L A, Deuel B, Lew M, et al. Proc Natl Acad Sci USA. 1998;95:14717–14722. doi: 10.1073/pnas.95.25.14717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Howe L R, Subbaramaiah K, Chung W J, Dannenberg A J, Brown A M. Cancer Res. 1999;59:1572–1577. [PubMed] [Google Scholar]
- 32.Haertel-Wiesmann M, Liang Y, Fantl W J, Williams L T. J Biol Chem. 2000;275:32046–32051. doi: 10.1074/jbc.M000074200. [DOI] [PubMed] [Google Scholar]
- 33.Spiegelman V S, Slaga T J, Pagano M, Minamoto T, Ronai Z, Fuchs S Y. Mol Cell. 2000;5:877–882. doi: 10.1016/s1097-2765(00)80327-5. [DOI] [PubMed] [Google Scholar]
- 34.Shibamoto S, Higano K, Takada R, Ito F, Takeichi M, Takada S. Genes Cells. 1998;3:659–670. doi: 10.1046/j.1365-2443.1998.00221.x. [DOI] [PubMed] [Google Scholar]
- 35.Klein P S, Melton D A. Proc Natl Acad Sci USA. 1996;93:8455–8459. doi: 10.1073/pnas.93.16.8455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pierce S B, Kimelman D. Development (Cambridge, UK) 1995;121:755–765. doi: 10.1242/dev.121.3.755. [DOI] [PubMed] [Google Scholar]
- 37.He X, Saint-Jeannet J P, Woodgett J R, Varmus H E, Dawid I B. Nature (London) 1995;374:617–622. doi: 10.1038/374617a0. [DOI] [PubMed] [Google Scholar]
- 38.Slusarski D C, Corces V G, Moon R T. Nature (London) 1997;390:410–413. doi: 10.1038/37138. [DOI] [PubMed] [Google Scholar]
- 39.Slusarski D C, Yang-Snyder J, Busa W B, Moon R T. Dev Biol. 1997;182:114–120. doi: 10.1006/dbio.1996.8463. [DOI] [PubMed] [Google Scholar]
- 40.Sheldahl L C, Park M, Malbon C C, Moon R T. Curr Biol. 1999;9:695–698. doi: 10.1016/s0960-9822(99)80310-8. [DOI] [PubMed] [Google Scholar]
- 41.Murakami A, Yajima T, Inana G. Biochem Biophys Res Commun. 1992;187:234–244. doi: 10.1016/s0006-291x(05)81483-4. [DOI] [PubMed] [Google Scholar]
- 42.Pongs O, Lindemeier J, Zhu X R, Theil T, Engelkamp D, Krah-Jentgens I, Lambrecht H G, Koch K W, Schwemer J, Rivosecchi R, et al. Neuron. 1993;11:15–28. doi: 10.1016/0896-6273(93)90267-u. [DOI] [PubMed] [Google Scholar]