Abstract
Multisubunit tethering complexes of the CATCHR (Complexes Associated with Tethering Containing Helical Rods) family are proposed to mediate the initial contact between an intracellular trafficking vesicle and its membrane target. To begin elucidating the molecular architecture of one well-studied example, the COG (conserved oligomeric Golgi) complex, we reconstituted its essential subunits (Cog1, Cog2, Cog3, and Cog4) and used single-particle electron microscopy to reveal a y-shaped structure with three flexible, highly extended legs. Labeling experiments established that the N-termini of all four subunits interact along the proximal segment of one leg, whereas three of the four C-termini are located at the tips of the legs. Our results suggest that the central region of the Cog1-4 complex, as well as the distal regions of at least two legs, all participate in interactions with other components of the intracellular trafficking machinery.
Keywords: Intracellular trafficking, COG complex, multisubunit tethering complex, electron microscopy, CATCHR complex
Introduction
The intracellular sorting via vesicular transport of proteins and other molecules requires at least four discrete operations: vesicle budding, tethering, docking, and fusion. Of these, vesicle tethering is perhaps the least well understood. Protein factors thought to mediate tethering include eight multi-subunit tethering complexes (MTCs), which function in various transport pathways and differ in molecular weight (250–800 kDa) and number of subunits (3–10)1–4. The conserved oligomeric Golgi (COG) complex is an MTC that comprises eight subunits (Cog 1 to 8) and has a combined molecular weight of 590–750 kDa, depending on species5. The COG complex was first isolated from bovine brain on the basis of its ability to stimulate Golgi transport in vitro6,7. It was identified independently in the yeast Saccharomyces cerevisiae8, and was found to include proteins (Cog2/Sec35 and Cog3/Sec34) already recognized as essential for vesicle trafficking in the early secretory pathway9–11. The discovery of interactions between the COG subunits and other Golgi trafficking proteins, including Rab GTPases9,10,12–14, SNAREs9,10,12,13,15,16, and COPI coat components12,13, led to the hypothesis that the COG complex acts primarily in retrograde transport to and within the Golgi apparatus, helping to maintain the proper cisternal distribution of Golgi enzymes. Consistent with this idea, depleting Cog3 in HeLa cells blocked retrograde traffic, but not anterograde traffic, within the Golgi apparatus17.
In yeast, three of the eight COG complex subunits (Cog2, 3, and 4) are encoded by essential genes11,18, while a deficiency in Cog1 gives rise to severe growth defects8,19. By contrast, null mutations in the genes encoding the remaining four subunits (Cog5, 6, 7, and 8) result in much milder defects8. Thus, it is possible to categorize the COG subunits into two classes, one containing essential (or “nearly essential”) subunits (Cog1, 2, 3, and 4), the other containing non-essential subunits (Cog5, 6, 7, and 8). Yeast two-hybrid and biochemical approaches have been used to map subunit connectivity within the mammalian and yeast COG complexes20,21. Notably, the essential subunits form a discrete sub-network within both the mammalian and yeast maps. Thus, it appears that Cog1, 2, 3, and 4 may constitute a functional unit, acting in an essential process for which the remaining subunits are not required.
The absence of structural information has been a major impediment in formulating specific models for MTC function. Of the eight known MTCs, four (the COG, Dsl1, exocyst, and GARP complexes) are members of the CATCHR family1–4. A model of the Dsl1 complex, based on overlapping crystal structures, was recently reported22,23. Two of the three subunits of the Dsl1 complex display homology to partial structures of Cog224 and Cog425 and four of the eight subunits composing the exocyst complex26–28. It remains unclear, however, whether the CATCHR complexes adopt similar quaternary structures, particularly in light of their significantly different subunit composition. To begin to investigate this question, we have reconstituted a core COG complex consisting of the four essential S. cerevisiae subunits, and have characterized its overall structure and subunit organization by negative stain EM and image analysis.
Results
Cog1, Cog2, Cog3, and Cog4 support wild-type growth in yeast
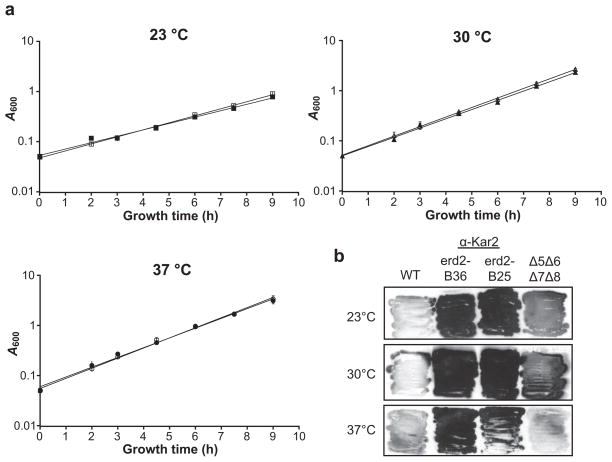
As noted above, deleting individual yeast COG genes produces widely varying growth phenotypes, ranging from wild-type growth (Δcog5, Δcog6, Δcog7, or Δcog8) to severe growth defects (Δcog1) or lethality (Δcog2, Δcog3, or Δcog4). Given these results, we sought to determine whether the Cog1, Cog2, Cog3, and Cog4 subunits, acting together, could provide wild-type COG function. Haploid strains carrying KanMX cassettes in place of the COG5, COG6, COG7, or COG8 genes were obtained from the Saccharomyces Genome Deletion Project. In each of these strains, we replaced the KanMX cassette with a unique marker (see Methods for details). After several rounds of mating and sporulation, we generated a haploid strain lacking all four genes (Δcog5 Δcog6 Δcog7 Δcog8), as confirmed by Southern blotting (Supplementary Fig. 1). Strikingly, the quadruple knockout strain exhibited no significant growth defect, relative to the wild-type parent strain, at 23°C, 30°C, or 37°C (Fig. 1a). A modest defect in retrograde trafficking was observed at 23°C and 30°C, but not at 37°C, as judged by elevated secretion of the ER-localized protein Kar2 (Fig. 1b). Single and double COG subunit knockouts were, in all cases, similar to wild-type (data not shown). Overall, the results show that Cog1, Cog2, Cog3, and Cog4 are jointly capable of providing essential COG complex function, and we therefore refer to these four COG subunits as the ‘core complex’.
Figure 1.
Analysis of the quadruple knockout strain for growth and trafficking defects. (a) Δcog5 Δcog6 Δcog7 Δcog8 cells displayed no growth defect or temperature sensitivity. Each growth curve represents either the haploid wild-type (open symbols) or Δcog5 Δcog6 Δcog7 Δcog8 (filled symbols) strain. Error bars represent the standard deviation (s.d.) for three independent cultures. (b) Haploid strains were grown on solid GNA medium at the indicated temperatures and exposed to nitrocellulose membranes; membranes were immunoblotted for secreted Kar2. erd2-B36 and erd2-B25 strains45 contain mutations in the HDEL receptor, responsible for the retrieval of ER-localized proteins, and therefore display high levels of inappropriate Kar2 secretion.
Reconstitution and EM analysis of the Cog1-4 core complex
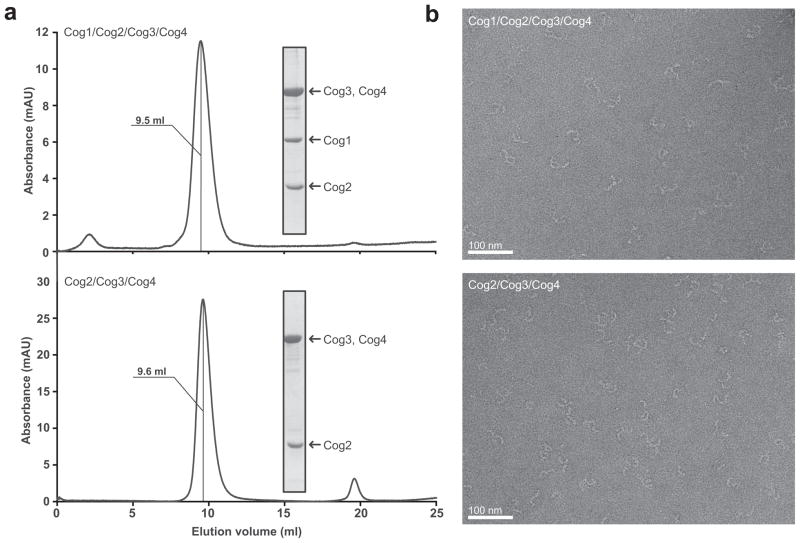
We initially attempted to purify each of the essential (or, in the case of Cog1, “nearly essential”) yeast COG subunits in Escherichia coli. Of the four proteins, only Cog2 was monodisperse and soluble, as observed previously9,24. Cog1 could be overexpressed and purified, but low yields and poor solubility suggested that it might not be properly folded. Cog3 and Cog4 were mostly insoluble, and only very small quantities could be recovered from E. coli lysates. Since previous studies showed that Cog2 interacts with Cog1, Cog3, and Cog420, pairs of subunits, each including Cog2, were co-expressed. All three of these binary complexes (Cog2/Cog1, Cog2/Cog3, and Cog2/Cog4) could be successfully purified. In each case, the solubility of the second subunit (Cog1, Cog3, or Cog4) was significantly improved by the presence of Cog2. Extending this concept, we used the pQLink multiple-insert plasmid system29 to co-express larger sets of subunits. Two of the resulting complexes, Cog2/Cog3/Cog4 (denoted Cog2-4) and Cog1/Cog2/Cog3/Cog4 (denoted Cog1-4), were much more soluble than any of the binary complexes. In each complex, Cog2 carried an N-terminal 7×His tag. The purified Cog2-4 and Cog1-4 complexes were monodisperse as judged by gel filtration (Fig. 2a). Interestingly, Cog2-4 and Cog1-4 eluted at similar volumes, but earlier than expected for globular proteins of the same molecular weights, suggesting that the hydrodynamic radii of the complexes are similar and relatively large.
Figure 2.
Purification and electron microscopy of the COG subcomplexes. (a) Gel filtration of purified Cog2-4 and Cog1-4 subcomplexes. Purified samples were analyzed on a Superdex 200 10/300 GL column; elution volume is indicated for each complex. Insets: Coomassie-stained gels for each purified complex. Note that Cog3 and Cog4 co-migrate as a single band. (b) EM analysis of purified Cog2-4 and Cog1-4. Representative raw images of negatively stained particles are shown. The scale bars represent 100 nm.
To further investigate their structural characteristics, we prepared negatively stained specimens of the purified Cog1-4 and Cog2-4 complexes. EM images showed that the shape of both complexes resembles a “y” with three flexible “legs” (Fig. 2b). We selected approximately 10,000 particles of each complex and classified them into 200 classes (Fig. 3a; Supplementary Figs. 2 and 3). The class averages for the Cog2-4 complex reveal two curved legs. Some averages also contain an additional, less well-defined leg. By contrast, class averages of the Cog1-4 complex are somewhat more uniform. Many Cog1-4 averages display three well-defined legs, designated A, B, and C (Fig. 3b), connected at a central junction; other averages appear incomplete, presumably due to poor staining or the “averaging out” of flexible elements (Supplementary Fig. 3). In well-defined averages, each leg has a distinctive curvature and unique features that together allow its unambiguous identification. Especially notable is a kink at the distal end of leg C, followed by a further extension we designate a “foot”. The foot structure is not observed in any of the Cog2-4 class averages, suggesting that this feature is most likely contributed by Cog1 (see below). Legs A and B are indistinguishable in length (16 nm), while leg C is longer (23 nm; Fig. 3b, c). The “joint” linking the three legs appears to be flexible (Fig. 3a, Supplementary Fig. 3), although we cannot rule out the possibility that the various conformations are artificially induced by the adsorption of the complexes to the carbon support film. The width of the legs (approximately 3 nm; Fig. 3d) is consistent with the diameter of the helix bundles estimated from the reported structures (or partial structures) of COG, Dsl1, and exocyst subunits (e.g., Exo70; Fig. 3e).
Figure 3.
Projection structures of the Cog2-4 and Cog1-4 (“core”) complexes. (a) Selected class averages from purified Cog1-4 and Cog2-4 complexes. The number in the bottom right-hand corner of each panel indicates the number of particles in the class average. The side length of each panel is approximately 45 nm. (b) Cartoon representation of Cog1-4 denoting key structural elements. (c) Length measurements of Cog1-4 leg structures. Bar graph represents length measurements from raw particles using ImageJ (± s.d.; n=3). Lengths are also represented in b. (d) The thicknesses of the legs of the complex in class averages were measured and compared using ImageJ at the points indicated on the cartoon. Notable is the increased thickness of leg A (position 1) relative to the others and the presence of a swelling on leg B (position 3) (± s.d.; n=12). (e) The x-ray structure of the CATCHR-family subunit Exo7026, provided for comparison as described in the text.
Organization of the subunits in the Cog1-4 complex
Having established the overall structure of the core Cog1-4 complex, we next used GFP labeling to investigate the organization of individual subunits within the complex. Each subunit was tagged with GFP at its N- or C-terminus and co-expressed with the remaining three subunits in order to reconstitute eight different Cog1-4 complexes, each carrying a single GFP tag. In the two complexes in which Cog2 was GFP-tagged, the 7×His tag used for purification was moved to the N-terminus of Cog1. The eight GFP-tagged complexes could be purified by means of the same procedure used for the original complex, although their solubility and stability were somewhat variable. Nonetheless, all four subunits were recovered in each of the eight purifications, allowing each of the complexes to be analyzed by negative stain EM (Fig. 4a). Approximately 6,000–10,000 particles of each complex were selected and classified into 200 classes. For seven of the eight samples a single GFP tag, appearing as a globular density approximately 4 nm in diameter, was clearly visible in multiple class averages (Fig. 4a). For each of these samples, the position of the GFP tag relative to the overall structure was consistent across all class averages resolving the GFP tag. This result strongly implies that, as suggested by the analysis of Coomassie-stained gels, the four subunits of the Cog1-4 complex are present in 1:1:1:1 stoichiometry (Fig. 4a; Supplementary Fig. 4). We could not resolve the GFP tag in the sample containing C-terminally tagged Cog4. As an alternative approach, we co-expressed Cog1-3 with a C-terminally truncated Cog4 comprising residues 1–553 and labeled with GFP at its N-terminus (Fig. 4a).
Figure 4.
Model for subunit organization in the Cog1-4 core complex. (a) Localization of GFP tags from GFP-labeled Cog1-4 complexes. Four representative class averages are shown for each tagged complex. The tag position is indicated by an arrowhead in one of the four averages. The Cog4 C-terminus was localized by using a C-terminally truncated version of Cog4 (residues 1–553) carrying an N-terminal GFP tag. The inferred C-terminus of Cog4 is indicated by a white arrowhead. The number in the bottom right-hand corner of each panel indicates the number of particles in the class average. The side length of each panel is approximately 45 nm. (b) Model for subunit localization in the Cog1-4 complex; arrowheads indicate C-termini.
The GFP labeling results allowed us to unequivocally localize each of the four Cog subunits and to generate a model of the organization of these proteins within the Cog1-4 complex (Fig. 4b). In this model, the C-terminus of Cog3 lies at the distal end of leg A. Based on the sample in which the C-terminus of Cog4 was deleted, we conclude that Cog4 forms leg B, with its C-terminus likely positioned at the outer end. The C-terminus of Cog1 maps to the distal tip of the foot at the end of leg C, while the C-terminus of Cog2 lies near the ”ankle” of leg C. The N-termini of all four subunits, on the other hand, map to the proximal portion of leg C (Fig. 4b). Their orientations demand that all four subunits overlap along this 10-nm segment of leg C. Moreover, because this leg segment is not noticeably thicker than other leg segments (Fig. 3d), we reason that the ‘overlap region’ cannot simply represent the side-by-side association of four subunits with helical bundle folds (see Discussion below).
Cog1 connects the Cog1-4 and Cog5-8 networks
Cog5-8 do not appear to be required for normal growth under laboratory conditions, a property they share with a range of well-conserved components of the yeast secretory pathway. This is generally explained by the plasticity of recycling trafficking routes in yeast, and indeed defects in these subunits are more obviously deleterious in other eukaryotes. For instance, a mutation in human Cog7 causes a lethal congenital disorder of glycosylation30. There is also good evidence that the C-terminal regions of human Cog1 and Cog8 interact, thereby connecting (or helping to connect) Cog1-4 to Cog5-821,31,32. The severing of this connection, by means of short C-terminal truncations of either Cog1 or Cog8, is apparently responsible for two additional congenital disorders of glycosylation31,33. To test whether an interaction between yeast Cog1 and Cog8 might, in a similar way, serve as a bridge between the essential and non-essential subunits, we incubated Cog8 with Cog1-4 carrying a marker GFP tag at the Cog4 N-terminus. Cog8 bound tightly to the Cog1-4 complex and the resulting Cog1/2/3/4/8 complex could be isolated by gel filtration (Supplementary Fig. 5). This complex was analyzed by EM as detailed above. The averages obtained from the classification of ~6,500 particles revealed that Cog8 indeed binds to the C-terminus of Cog1 (Fig. 5a). Consistent with this conclusion, close examination of Cog1-4 class averages in which Cog1 carries a C-terminal GFP tag suggests that, in the absence of Cog8, the C-terminus of Cog1 is disordered (Fig. 5a).
Figure 5.
Functional architecture of the Cog1-4 core complex. (a) Selected class averages of the Cog1/2/3/4/8 complex are shown (side length = 50 nm); the Cog4 N-terminus carries a GFP tag. Cog8 forms a curved extension from the tip of the Cog1 foot structure and ends in a globular structure. Also shown is a class average from the Cog1-GFP/2/3/4 complex, in which a pronounced gap between the tip of the Cog1 foot and the C-terminal GFP tag is evident. The number in the bottom right-hand corner of each panel indicates the number of particles in the class average. (b) Cartoon representation of the Cog1-4 core complex colored by subunit, with expanded views highlighting functionally important regions. The crystal structure of human Cog4 (C-terminal residues 537–785) is shown, together with a magnified image of the salt bridge interaction between Arg 729 and Glu 764 that positions and/or stabilizes the C-terminal domain25. The N-terminal subunit interaction region is also highlighted, together with the approximate inferred interaction sites for human Syntaxin 5 and Sly116,37. Finally, the NMR structure of yeast Cog2 (C-terminal residues 109–262)24 is shown, indicating its approximate position within the core complex.
A previous study of binary interactions within the yeast COG complex reported that Cog1 occupies a central position, interacting directly with all of the other COG subunits except for Cog720. As we have demonstrated here, the N-terminal region of Cog1 mediates its interactions with Cog2, Cog3, and Cog4, while the C-terminal region of Cog1 interacts with Cog8. The structure of Cog8, as visualized by EM, seems to differ from that of the other subunits. Similar to one of the Dsl1 complex subunits (Tip20)23, Cog8 has a sharp hook, while the large globular distal region appears to be unprecedented among the tethering subunits of known structure. We have been unable to reconstitute larger complexes, containing Cog5 or Cog6 in addition to Cog1/2/3/4/8, so although these subunits may also interact with the C-terminal region of Cog1 we lack direct proof.
Discussion
The COG and exocyst complexes are among the most intensively-studied MTCs, and partial structures of individual subunits demonstrate that they share homology on the tertiary structural level22–28. Relatively little progress has, however, been made in characterizing these complexes in their assembled states. Previous quick-freeze/deep-etch EM analyses of the COG and exocyst complexes revealed evidence for multiple structural domains6,34. Brief glutaraldehyde fixation in these studies, however, drastically altered the structural features of both complexes, leaving open the question of which samples the unfixed samples or the fixed samples – were more native-like in their properties. For COG, the fixed samples collapsed to form two globular lobes, leading to the prediction that one lobe contains the subunits essential in yeast (Cog1-4) while the other lobe contains the remaining subunits (Cog5-8)6. While this division into functional domains has stood the test of time, the model of COG as composed of globular lobes is at odds with the results reported here. Instead, the essential (and presumably the remaining) COG subunits form long, thin legs in a decidedly spindly structure. This long-legged structure seems well-suited to a role in vesicle tethering at a distance. We note, however, that clathrin triskelia, which bear superficial resemblance to the Cog1-4 core complex, play an entirely different role in vesicle trafficking, serving as the protomers from which vesicle coats are assembled. Put differently, function does not necessarily follow form.
The four subunit N-termini play a fundamental role in complex assembly by bundling together along the proximal portion of one leg (Fig. 5b). Consistent with this finding, deletion of 97 N-terminal residues from Cog2 is lethal24. While our EM data could not further define the molecular details of the intersubunit interactions in the region in which all four subunits interdigitate, one attractive possibility is that each N-terminus contributes one (or more) α-helices to an extended helix bundle along this length of leg C. The diameter of such a bundle, were it to contain four helices, would be roughly the width of the leg as observed by EM. Furthermore, as first noted by Whyte and Munro1,8, the N-terminal regions of COG subunits (and other CATCHR-family subunits) often contain regions predicted to form amphipathic α-helices; for yeast Cog2, the N-terminal region has been shown experimentally to adopt an ensemble of highly α-helical conformations24. We therefore suggest that, in the Cog1-4 complex, the α-helical propensity of each subunit is further reinforced by quaternary interactions with the other subunits to form a bundle of four (or more) helices. Intriguingly, the core SNARE complex is a four-helix bundle of similar length35. In the SNARE case, however, all four helices are parallel, whereas in our speculative model, the Cog1 and Cog2 helices would be antiparallel to the Cog3 and Cog4 helices. Finally, we note that a more general involvement of N-terminal regions in forming the quaternary interactions that stabilize CATCHR-family tethering complexes would explain why almost none of the known structures of isolated subunits include this region.
The distal ends of at least two of the three legs play important roles in COG complex function. The distal end of leg C, representing the C-terminus of Cog1, is important because it serves as a bridge to the non-essential subunits, Cog5-8. The distal end of leg B is assigned to the C-terminal portion of Cog4 based primarily on images of the core complex containing a C-terminal Cog4 truncation in Cog4 (Fig. 3a). Also consistent with this assignment is the observation that the distal end of leg B is roughly congruent with the crystal structure of a 27-kDa C-terminal fragment of human Cog425 (Fig. 5b). The crystal structure includes the site of an Arg 729 to Trp missense mutation that contributes to a congenital disorder of glycosylation36 and that results in severe glycosylation defects in HeLa cells without affecting the assembly or stability of the COG complex25 . Moreover, yeast harboring a C-terminally truncated version of Cog4 are inviable25. These and other functional results25 strongly suggest that the distal end of leg B contains a binding site for an as-yet-unidentified trafficking factor. In the future, it will be interesting to investigate whether the C-terminus of Cog3, forming the distal tip of leg A, is likewise implicated in interactions with other components of the trafficking machinery. Finally, we note that human Cog2 is considerably larger than its yeast counterpart. Based on our model (Fig. 5b), we propose that the human Cog1-4 complex may be “H”-shaped, with the fourth leg derived from Cog2. As the human Cog1 subunit is also larger, leg C may also be longer.
Other functionally important interaction sites map nearer the central junction of the Cog1-4 core complex. The N-terminal 150 residues of human Cog4 has been reported to harbor binding sites for the SNARE protein Syntaxin 5 and the Sec1/Munc18-family protein Sly116,37. In the yeast COG core complex, this region of Cog4 resides along the proximal portion of leg C where all four subunits intertwine (Fig. 5b). Overall, we conclude that the Cog1-4 complex likely engages other components of the trafficking machinery (including Cog5-8) using widely-distributed sites located at the tips of at least two legs and within the central core.
The structural organization of the COG complex presented here invites comparison with the Dsl1 complex, whose structure was reported recently22,23. At the tertiary structure level, there are significant similarities between some COG and Dsl1 complex subunits22–25. At the quaternary structure level, the interactions that stabilize each complex involve regions at or near the termini of the individual subunits. All four of the essential COG subunits interact by means of their N-terminal segments; likewise, two of the three Dsl1 complex subunits (Tip20 and Dsl1) interact via the anti-parallel pairing of N-terminal helices23. A second similarity between the COG and Dsl1 complexes is that each of them contains a single subunit (Cog1 and Dsl1, respectively) that functions as a bridge by interacting at (or near) each end with different subunits. Finally, like the COG complex, the Dsl1 complex contains spatially distant sites important for interaction with other components of the trafficking machinery22,23. Nonetheless, despite these similarities, the overall architecture of the Dsl1 complex and the COG core complex are quite different. Further study will be needed to determine the extent to which these complexes, as well as the exocyst and GARP complexes, operate according to common mechanistic principles.
Conclusion
Our results establish that four of the eight COG subunits, Cog 1 to 4, form a stable core complex sufficient to support wild-type growth and reasonably efficient retrograde trafficking in yeast. The four N-termini interact near the center of the complex, while three of the four C-termini form the distal ends of elongated legs. The end of one leg (leg C) forms a bridge to the non-essential subunits, while that of a second leg (leg B) is essential for function in yeast and humans. For human COG, at least, an additional locus of functionally important interactions is defined by the N-terminal portion of Cog4, predicted to lie along the proximal portion of leg C. Our hypothesis that this region of leg C is a four-helix bundle is consistent with the finding that the COG-interacting portion of Syntaxin 5 is its SNARE motif16, which might displace one of the COG helices or, alternatively, might pack into one of the four surface grooves. Human Sly1 binds to an adjacent region of Cog4; mutations that abolish this interaction compromise retrograde trafficking and the assembly or stability of the relevant SNARE complex37. Taken together, the results reported here represent an important step toward the development of a mechanistic framework for COG function.
Methods
Strain construction and analysis
Haploid strains in both mating types deleted for the following ORFs were obtained from the Yeast Knockout strain collection, available from Open Biosystems: YNL051w, YNL041c, YGL005c, and YML071c. Marker swaps were performed by LiAc transformation of linearized plasmids M4755 and M4758 (ref. 38) to replace the kanMX cassettes for deleted ORFs YGL005c and YML071c with LEU2 and URA3 markers, respectively. The kanMX cassette for deleted ORF YNL041c was swapped out for S. pombe HIS5 using a PCR-amplified His3MX6 cassette from plasmid pFA6a-His3MX6 (ref. 39). Patch matings were performed on YPD plates and diploids were selected on solid synthetic complete dropout media or supplemented synthetic dextrose minimal media. Isolated diploids were grown in large patches at 30°C overnight on solid GNA medium, replicated for overnight growth on fresh GNA medium, then replicated to solid MinSpore medium for sporulation at ambient temperature for 24 hours, followed by 48 hours at 30°C. Tetrads were dissected to isolate double mutant haploids. The resulting strains were then mated, sporulated, and dissected to isolate the haploid quadruple deletion, named JAL017H. The absence of all four COG genes in JAL017H was verified by Southern blotting (Supplementary Fig. 1), as well as by PCR using primers internal to the deleted ORFs (data not shown).
For growth curves, haploid cells were grown in 5 ml of liquid YPD media overnight at 30°C. These cultures were diluted into 5 ml aliquots of YPD at an initial A600 of 0.05. The diluted cultures were incubated with shaking at 23°C, 30°C, and 37°C, respectively. Absorbance was monitored at intervals during subsequent growth. To test for Kar2 secretion, patches of the indicated strains were streaked on solid GNA medium and grown at the indicated temperatures for 2–3 days. Plates were then overlaid with nitrocellulose membrane and incubated at the growth temperature for 30 min. Membranes were lifted from the plates, washed to remove any adhering cells, and immunoblotted using a rabbit polyclonal α-Kar2 primary antibody40 at 1:5000 dilution, followed by HRP-goat α-rabbit IgG secondary antibody (Invitrogen) at 1:3000 dilution. Detection was performed with Lumi-Light Western Blotting Substrate (Roche).
Plasmid construction
The eGFP ORF was cloned into pQLinkN using a PCR product amplified with the following primers to form plasmid pQLinkGN: NT_eGFP_5′_BclI: 5′-GAGCATTGATCAGTGAGC AAGGGCGAGG-3′; NT_eGFP_3′_NdeI_BclI: 5′-ACGTCATGATCACATATGCTTGTACAGCTC GTCCATG-3′ The same ORF was cloned into pQLinkN using a PCR product amplified with the following primers to form plasmid pQLinkGC: CT_eGFP_5′_NotI: GAGCATGCGGCCGCAGTG AGCAAGGGCGAGG; CT_eGFP_3′_EagI: 5′-ACGTCACGGCCGCTACTTGTACAGCTCGTCC ATG-3′. PCR products encoding S. cerevisiae COG subunits were individually cloned into the pQLinkH, pQLinkN, pQLinkGN, and pQLinkGC vectors using BamHI and NotI sites or NdeI and NotI sites. Multi-subunit plasmids were constructed combinatorially from these base clones according to the method of Scheich et al.29.
Protein purification
Multi-subunit constructs were transformed for expression into C43(DE3) cells (Lucigen). Liquid starter cultures were diluted 100-fold into 6 or 12 l of LB with 100 μg ml−1 ampicillin and grown with shaking at 37°C. Protein expression was induced at an OD600 of 0.4 to 0.6 by the addition of 0.5 mM IPTG to the cultures and growth at ambient temperature for 14–18 hours. Cells were harvested by centrifugation, resuspended in lysis buffer (20 mM Tris pH 8.0, 20 mM imidazole, 150 mM NaCl, 1 mM DTT, 0.5% (v/v) protease inhibitor cocktail (Sigma cat. #P8340)), and lysed using an Emulsiflex-C5 cell disruptor (Avestin) at a pressure of ~12,000 psi. Complexes were bound, washed, and eluted from IMAC Sepharose 6 FF resin (GE Healthcare) charged with Ni2+ ions. Eluates were pooled and fractionated using a Source 15Q 10/10 anion exchange column. Complex-containing fractions were pooled, concentrated using a 30 kDa NMWCO Amicon 15 centrifugal ultrafiltration device (Millipore), and loaded on a Superdex 200 16/60 or 10/30 size exclusion column; peak fractions were immediately used for analysis by electron microscopy.
Electron microscopy and image processing
Samples were prepared by conventional negative staining with 0.75% (w/v) uranyl formate as described previously41. Images were collected with a Tecnai T12 electron microscope (FEI, Hillsboro, OR) equipped with a LaB6 filament and operated at an acceleration voltage of 120 kV. Images were recorded on imaging plates at a nominal magnification of 67,000× and a defocus value of −1.5 μm using low dose procedures. Imaging plates were read out with a scanner (DITABIS Digital Biomedical Imaging System AG, Pforzheim, Germany) using a step size of 15 μm, a gain setting of 20,000, and a laser power setting of 30% (ref. 42); 2 × 2 pixels were averaged to yield a pixel size of 4.5 Å on the specimen level.
A total of 11,336 Cog1-4 and 8,271 Cog2-4 particles were interactively selected from the raw images using BOXER, the display program associated with the EMAN software package43. Using the SPIDER software package44, these particles were windowed into 100 × 100 pixel images, rotationally and translationally aligned, and subjected to 10 cycles of multi-reference alignment. Each round of multi-reference alignment was followed by K-means classification specifying 200 output classes. The references used for the first multi-reference alignment were randomly chosen from the particle images. For GFP-labeled samples, a total of 7,904 particles were selected for GFP-Cog1/2/3/4, 8,698 particles for GFP-Cog2/1/3/4, 6,337 particles for GFP-Cog3/1/2/4, 10,014 particles for GFP-Cog4/1/2/3, 6,383 particles for Cog1-GFP/2/3/4, 5,923 particles for Cog2-GFP/1/3/4, 7,827 particles for Cog3-GFP/1/2/4, 7,790 particles for GFP-Cog4(1–553)/1/2/3, and 6,549 particles for GFP-Cog4/1/2/3/Cog8. The particle images were subjected to multi-reference alignment and K-means classification, specifying 200 output classes.
Distance measurements of COG particles and averages were performed using the straight or segmented line distance measurement function in NIH ImageJ software version 1.41o (http://rsbweb.nih.gov/ij/).
Supplementary Material
Acknowledgments
We gratefully acknowledge Mark Rose and Casey Ydenberg (Princeton University) for providing strains, reagents, technical assistance, and valuable advice, as well as Sanford Silverman and Lorraine Schepis (Princeton University) for reagents and technical assistance. We also thank Vladimir Lupashin, Brian Richardson, Daniel Ungar, and Marina Paul for useful discussions. This work was supported by a grant to F.M.H. from the National Institutes of Health (GM071574). C.K.Y. acknowledges fellowships from the Jane Coffin-Childs Memorial Fund and the Canadian Institutes for Health Research. T.W. is an investigator in the Howard Hughes Medical Institute.
Footnotes
Author contributions statement
J.A.L. conducted yeast experiments and prepared samples for EM. C.K.Y. performed EM experiments and analyzed the data. All authors discussed the results and wrote the paper.
References
- 1.Whyte JR, Munro S. Vesicle tethering complexes in membrane traffic. J Cell Sci. 2002;115:2627–2637. doi: 10.1242/jcs.115.13.2627. [DOI] [PubMed] [Google Scholar]
- 2.Cai H, Reinisch K, Ferro-Novick S. Coats, tethers, Rabs, and SNAREs work together to mediate the intracellular destination of a transport vesicle. Dev Cell. 2007;12:671–682. doi: 10.1016/j.devcel.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 3.Sztul E, Lupashin V. Role of vesicle tethering factors in the ER-Golgi membrane traffic. FEBS Lett. 2009;583:3770–83. doi: 10.1016/j.febslet.2009.10.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yu I, Hughson FM. Tethering factors as organizers of intracellular vesicular traffic. Ann Rev Cell Dev Biol. 2010;26 doi: 10.1146/annurev.cellbio.042308.113327. in press. [DOI] [PubMed] [Google Scholar]
- 5.Ungar D, Oka T, Krieger M, Hughson FM. Retrograde transport on the COG railway. Trends Cell Biol. 2006;16:113–120. doi: 10.1016/j.tcb.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 6.Ungar D, et al. Characterization of a mammalian Golgi-localized protein complex, COG, that is required for normal Golgi morphology and function. J Cell Biol. 2002;157:405–415. doi: 10.1083/jcb.200202016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Walter DM, Paul KS, Waters MG. Purification and characterization of a novel 13 S hetero-oligomeric protein complex that stimulates in vitro Golgi transport. J Biol Chem. 1998;273:29565–76. doi: 10.1074/jbc.273.45.29565. [DOI] [PubMed] [Google Scholar]
- 8.Whyte JR, Munro S. The Sec34/35 Golgi transport complex is related to the exocyst, defining a family of complexes involved in multiple steps of membrane traffic. Dev Cell. 2001;1:527–537. doi: 10.1016/s1534-5807(01)00063-6. [DOI] [PubMed] [Google Scholar]
- 9.VanRheenen SM, Cao X, Lupashin VV, Barlowe C, Waters MG. Sec35p, a novel peripheral membrane protein, is required for ER to Golgi vesicle docking. JCB. 1998;141:1107–1119. doi: 10.1083/jcb.141.5.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.VanRheenen SM, et al. Sec34p, a protein required for vesicle tethering to the yeast Golgi apparatus, is in a complex with Sec35p. J Cell Biol. 1999;147:729–742. doi: 10.1083/jcb.147.4.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wuestehube LJ, et al. New mutants of Saccharomyces cerevisiae affected in the transport of proteins from the endoplasmic reticulum to the Golgi complex. Genetics. 1996;142:393–406. doi: 10.1093/genetics/142.2.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ram RJ, Li B, Kaiser CA. Identification of Sec36p, Sec37p, and Sec38p: components of yeast complex that contains Sec34p and Sec35p. Mol Biol Cell. 2002;13:1484–1500. doi: 10.1091/mbc.01-10-0495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Suvorova ES, Duden R, Lupashin VV. The Sec34/Sec35p complex, a Ypt1p effector required for retrograde intra-Golgi trafficking, interacts with Golgi SNAREs and COPI vesicle coat proteins. J Cell Biol. 2002;157:631–43. doi: 10.1083/jcb.200111081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luo Z, Gallwitz D. Biochemical and genetic evidence for the involvement of yeast Ypt6-GTPase in protein retrieval to different Golgi compartments. J Biol Chem. 2003;278:791–9. doi: 10.1074/jbc.M209120200. [DOI] [PubMed] [Google Scholar]
- 15.Schuldiner M, et al. Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell. 2005;123:507–19. doi: 10.1016/j.cell.2005.08.031. [DOI] [PubMed] [Google Scholar]
- 16.Shestakova A, Suvorova E, Pavliv O, Khaidakova G, Lupashin V. Interaction of the conserved oligomeric Golgi complex with t-SNARE Syntaxin5a/Sed5 enhances intra-Golgi SNARE complex stability. J Cell Biol. 2007;179:1179–1192. doi: 10.1083/jcb.200705145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zolov SN, Lupashin VV. Cog3p depletion blocks vesicle-mediated Golgi retrograde trafficking in HeLa cells. J Cell Biol. 2005;168:747–59. doi: 10.1083/jcb.200412003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giaever G, et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–91. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- 19.Deutschbauer AM, et al. Mechanisms of haploinsufficiency revealed by genome-wide profiling in yeast. Genetics. 2005;169:1915–25. doi: 10.1534/genetics.104.036871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fotso P, Koryakina Y, Pavliv O, Tsiomenko AB, Lupashin VV. Cog1p plays a central role in the organization of the yeast conserved oligomeric Golgi complex. J Biol Chem. 2005;280:27613–23. doi: 10.1074/jbc.M504597200. [DOI] [PubMed] [Google Scholar]
- 21.Ungar D, Oka T, Vasile E, Krieger M, Hughson FM. Subunit architecture of the conserved oligomeric Golgi complex. J Biol Chem. 2005;280:32729–35. doi: 10.1074/jbc.M504590200. [DOI] [PubMed] [Google Scholar]
- 22.Ren Y, et al. A structure-based mechanism for vesicle capture by the multisubunit tethering complex Dsl1. Cell. 2009;139:1119–29. doi: 10.1016/j.cell.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tripathi A, Ren Y, Jeffrey PD, Hughson FM. Structural characterization of Tip20p and Dsl1p, subunits of the Dsl1p vesicle tethering complex. Nat Struct Mol Biol. 2009;16:114–123. doi: 10.1038/nsmb.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cavanaugh LF, et al. Structural analysis of conserved oligomeric Golgi complex subunit 2. J Biol Chem. 2007;282:23418–23426. doi: 10.1074/jbc.M703716200. [DOI] [PubMed] [Google Scholar]
- 25.Richardson BC, et al. Structural basis for a human glycosylation disorder caused by mutation of the COG4 gene. Proc Natl Acad Sci U S A. 2009;106:13329–13334. doi: 10.1073/pnas.0901966106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dong G, Hutagalung AH, Fu C, Novick P, Reinisch KM. The structures of exocyst subunit Exo70p and the Exo84p C-terminal domains reveal a common motif. Nat Struct Mol Biol. 2005;12:1094–1100. doi: 10.1038/nsmb1017. [DOI] [PubMed] [Google Scholar]
- 27.Sivaram MV, Furgason ML, Brewer DN, Munson M. The structure of the exocyst subunit Sec6p defines a conserved architecture with diverse roles. Nat Struct Mol Biol. 2006;13:555–556. doi: 10.1038/nsmb1096. [DOI] [PubMed] [Google Scholar]
- 28.Wu S, Mehta SQ, Pichaud F, Bellen HJ, Quiocho FA. Sec15 interacts with Rab11 via a novel domain and affects Rab11 localization in vivo. Nat Struct Mol Biol. 2005;12:879–885. doi: 10.1038/nsmb987. [DOI] [PubMed] [Google Scholar]
- 29.Scheich C, Kummel D, Soumailakakis D, Heinemann U, Bussow K. Vectors for co-expression of an unrestricted number of proteins. Nucleic Acids Res. 2007;35:e43. doi: 10.1093/nar/gkm067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu X, et al. Mutation of the COG complex subunit gene COG7 causes a lethal congenital disorder. Nat Med. 2004;10:518–523. doi: 10.1038/nm1041. [DOI] [PubMed] [Google Scholar]
- 31.Foulquier F, et al. A new inborn error of glycosylation due to a Cog8 deficiency reveals a critical role for the Cog1-Cog8 interaction in COG complex formation. Hum Mol Genet. 2007;16:717–30. doi: 10.1093/hmg/ddl476. [DOI] [PubMed] [Google Scholar]
- 32.Kranz C, et al. COG8 deficiency causes new congenital disorder of glycosylation type IIh. Hum Mol Genet. 2007;16:731–41. doi: 10.1093/hmg/ddm028. [DOI] [PubMed] [Google Scholar]
- 33.Foulquier F, et al. Conserved oligomeric Golgi complex subunit 1 deficiency reveals a previously uncharacterized congenital disorder of glycosylation type II. Proc Natl Acad Sci U S A. 2006;103:3764–9. doi: 10.1073/pnas.0507685103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hsu SC, et al. Subunit composition, protein interactions, and structures of the mammalian brain sec6/8 complex and septin filaments. Neuron. 1998;20:1111–22. doi: 10.1016/s0896-6273(00)80493-6. [DOI] [PubMed] [Google Scholar]
- 35.Sutton RB, Fasshauer D, Jahn R, Brunger AT. Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 A resolution. Nature. 1998;395:347–353. doi: 10.1038/26412. [DOI] [PubMed] [Google Scholar]
- 36.Reynders E, et al. Golgi function and dysfunction in the first COG4-deficient CDG type II patient. Hum Mol Genet. 2009;18:3244–56. doi: 10.1093/hmg/ddp262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Laufman O, Kedan A, Hong W, Lev S. Direct interaction between the COG complex and the SM protein, Sly1, is required for Golgi SNARE pairing. EMBO J. 2009;28:2006–17. doi: 10.1038/emboj.2009.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Voth WP, Jiang YW, Stillman DJ. New ‘marker swap’ plasmids for converting selectable markers on budding yeast gene disruptions and plasmids. Yeast. 2003;20:985–93. doi: 10.1002/yea.1018. [DOI] [PubMed] [Google Scholar]
- 39.Longtine MS, et al. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–61. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 40.Rose MD, Misra LM, Vogel JP. KAR2, a karyogamy gene, is the yeast homolog of the mammalian BiP/GRP78 gene. Cell. 1989;57:1211–21. doi: 10.1016/0092-8674(89)90058-5. [DOI] [PubMed] [Google Scholar]
- 41.Ohi M, Li Y, Cheng Y, Walz T. Negative staining and image classification -powerful tools in modern electron microscopy. Biol Proced Online. 2004;6:23–34. doi: 10.1251/bpo70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li Z, Hite RK, Cheng Y, Walz T. Evaluation of imaging plates as recording medium for images of negatively stained single particles and electron diffraction patterns of two-dimensional crystals. J Electron Microsc (Tokyo) 2010;59:53–63. doi: 10.1093/jmicro/dfp036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 44.Frank J, et al. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 45.Semenza JC, Hardwick KG, Dean N, Pelham HR. ERD2, a yeast gene required for the receptor-mediated retrieval of luminal ER proteins from the secretory pathway. Cell. 1990;61:1349–57. doi: 10.1016/0092-8674(90)90698-e. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.