Abstract
Proteins play major roles in most biological processes; as a consequence, protein expression levels are highly regulated. While extensive post-transcriptional, translational and protein degradation control clearly influence protein concentration and functionality, it is often thought that protein abundances are primarily determined by the abundances of the corresponding mRNAs. Hence surprisingly, a recent study showed that abundances of orthologous nematode and fly proteins correlate better than their corresponding mRNA abundances. We tested if this phenomenon is general by collecting and testing matching large-scale protein and mRNA expression datasets from seven different species: two bacteria, yeast, nematode, fly, human, and plant. We find that steady-state abundances of proteins show significantly higher correlation across these diverse phylogenetic taxa than the abundances of their corresponding mRNAs (p=0.0008, paired Wilcoxon). These data support the presence of strong selective pressure to maintain protein abundances during evolution, even when mRNA abundances diverge.
Proteins play major roles in most biological processes, ranging from central metabolism to cell structure, maintenance, and replication. Consequently, protein expression levels are subject to diverse and complex control. Due to extensive post-transcriptional, translation and stability regulation, protein abundance is only partly determined by accumulation and degradation of the corresponding mRNAs (e.g., as in references [1–3]), with perhaps 20-60% of the variation in steady-state protein abundances attributable to mRNA levels, depending upon organism and conditions [4]. A recent study of the nematode and fly proteomes made the remarkable observation that the abundances of orthologous nematode and fly proteins correlated better than their corresponding mRNA abundances [3]. The difficulty in making such measurements on a proteome scale has until recently held back such comparisons, and it is unknown whether this observation is generally true. We asked if this phenomenon is indeed general by collecting and testing matching large-scale protein and mRNA expression datasets from seven different species. We find that steady-state abundances of proteins show significantly higher correlation across diverse phylogenetic taxa than the abundances of their corresponding mRNAs (p=0.0008, paired Wilcoxon). These data support the presence of strong selective pressure to maintain protein abundances during evolution. A necessary consequence is that protein stability and post-transcriptional regulatory schemes must compensate for divergent mRNA levels to maintain protein levels at evolutionarily optimized levels.
Specifically, we assembled large-scale quantitative protein expression datasets and measured protein abundances from bacteria (E. coli, P. aeruginosa), fungi (Baker’s yeast, S. cerevisiae), plants (the leaf proteome of rice, O. sativa), insects (fruit fly, D. melanogaster), nematodes (C. elegans), and humans, as described in the online supplement. For each species, we identified or collected mRNA expression datasets from matching strain and growth conditions. We limited datasets to those from similar measurement platforms. For mRNA, we compiled data from single channel DNA microarrays and counting methods if available (Table S1). For proteins, we used mass spectrometry based shotgun proteomics, measuring absolute abundances with a label-free weighted spectral counting approach [2]. We then computed orthologous genes between each pair of species using InParanoid [5]. (Alternate choices of measurement platforms, quantitation, and calculation of orthology, described below, all give similar results.)
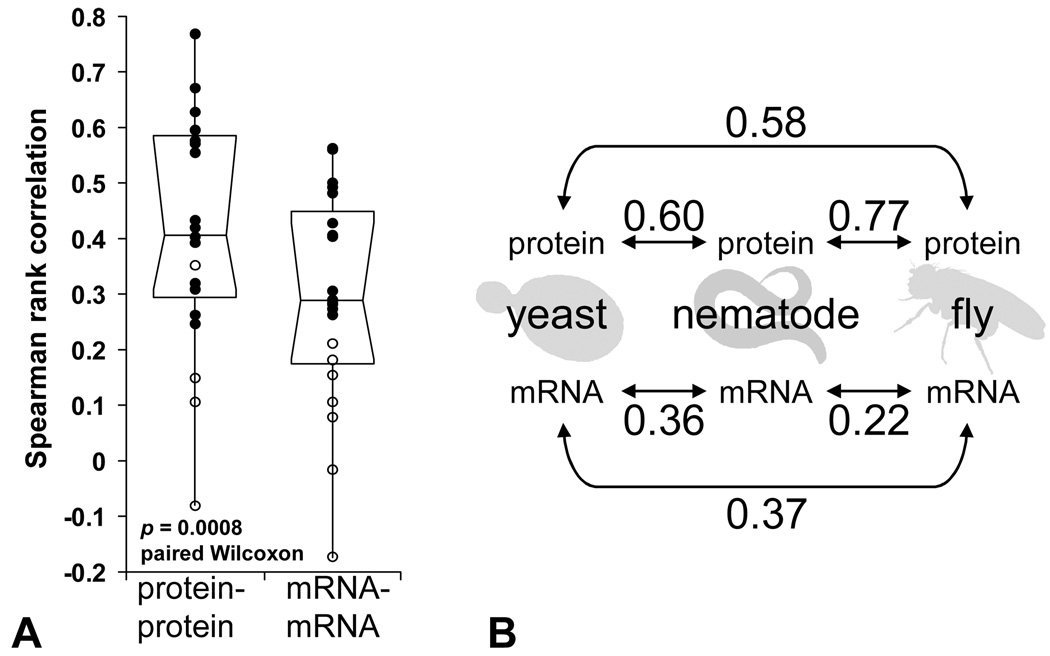
We then determined the extent to which steady-state protein concentrations were conserved between each pair of organisms by calculating the rank correlation of the protein abundances originating from orthologous genes, as shown for human and yeast in Figure 1A. Similarly, we measured the rank correlation in the abundances of the corresponding mRNAs. Importantly, we limited all comparisons to only those genes for which we had both protein and mRNA measurements, thereby controlling for possible sources of bias related to selection of genes, including technology-specific abundance biases (for example, the tendency for mass spectrometry to selectively sample abundant proteins). The relative conservation of protein and mRNA abundances could then be estimated by comparing the resulting rank correlations, listed in full in Figure 1B. Of the 21 organism pairs considered, the correlation in protein abundances was greater than that of mRNAs in 17 cases, and less than that in only four. The trend can be clearly seen in the distributions of protein-protein and mRNA-mRNA correlations (Figure 2A), supporting a significantly greater conservation of protein abundances than for the abundances of the corresponding mRNAs (p = 0.0008, paired Wilcoxon).
Figure 1.
(A) General scheme for collecting, organizing, and analyzing protein and mRNA expression datasets involved in the study. For each organism, expression datasets were either assembled or measured in-house, with protein and mRNA abundances estimated by mass spectrometry and single-channel microarrays, respectively. For the genes orthologous between each pair of organisms, we calculated the Spearman rank correlation between their corresponding protein levels and between their corresponding mRNA levels, as reported in (B). Blue and red represent protein-protein and mRNA-mRNA correlations, respectively, with darker boxes indicating those correlations with p-value < 0.01. White boxes down the diagonal are the protein-mRNA correlations within each species.
Figure 2.
Protein abundances are better conserved across seven taxa than mRNA abundances. (A) Summary of the measured distributions of protein-protein correlations and mRNA-mRNA correlations as outlier box-plots indicating the median +/− 1 quartile, with whiskers indicating ±1.5 interquartile ranges. Individual observations with p-value < 0.01 are plotted as filled circles, and observations with p-value ≥ 0.01 as open circles. Additional statistical tests are in the online Supplement. (B) Correlations using only SAGE or RNAseq transcript abundance measurements for organisms for which those data were available. Protein abundance correlations are substantially larger than mRNA abundance correlations in all three available cases (n = 700, 774, and 2680 for yeast-nematode, yeast-fly, and fly-nematode comparisons, respectively).
We attempted to rule out the possibility of either technical artifacts or conflating trends giving rise to our observations as follows: the trend was also observed when we considered mRNA measurements based only on sequencing (SAGE and RNA-seq) rather than DNA microarrays (Figure 2B; only 3 such comparisons available) and was highly statistically significant when we considered average mRNA abundance measurements obtained by multiple techniques (i.e., mixing microarrays and SAGE or RNA-seq; p < 0.0001, Table S4), and when we omitted any one organism (all p < 0.01). To control for errors in assigning orthology, we considered an alternate method of calculating orthologs (p = 0.025, Table S5); both cases behaved similarly and showed a similarly significant trend. Finally, both mRNA and protein abundances are known to be inversely correlated to gene length [6]. To eliminate the possibility that our observation is due to correlations to a third variable, gene length, we measured the partial correlations for either protein or mRNA levels given gene lengths; again, protein levels were significantly better conserved than mRNA levels even after correcting for gene length (p = 0.018, Table S6). Also, protein abundance correlations were significantly higher than mRNA abundance correlations (p < 0.05, paired Wilcoxon) regardless of whether all observations were considered or whether only correlations with significant p-values were considered, for all comparisons described above (Tables S4 to S7).
To investigate if the differences in correlations are due to differences in the underlying measurement errors, we assessed (for a subset of the data) measurement reliability through correlation analysis of technical replicates. Measurements of mRNA concentrations tend to have higher reproducibilities than measurements of protein concentrations (Rs=0.99 and 0.80, respectively, Figure S1), arguing against general measurement errors as an explanation of the lower mRNA-mRNA correlations. We occasionally observed a contribution from expression level, e.g., for the fly-nematode comparison: the observed difference in correlation coefficient is most pronounced for the least abundant mRNAs and proteins; conversely, highly expressed proteins and mRNAs are similarly conserved in their abundance across the two organisms. However, this trend did not hold for all organism pairs (data not shown).
Higher conservation of protein abundances suggests that abundances of proteins are to some degree optimized and that evolutionary pressure helps to maintain these levels despite changing mRNA levels, as also exhibited by only partially correlated mRNA and protein levels within a species. Extensive regulation of protein abundances must therefore compensate for divergent mRNA expression levels to maintain proteins at favored levels. It remains to be seen if evolutionary or molecular signatures of such compensatory regulation can be detected. For example, it has been speculated that transcriptional bursts, observed to increase variance in mRNA abundances, may be buffered by long protein half-lives [7]. Furthermore, divergence of mRNA expression levels is an evolutionarily well known process [8], and a remarkable conservation of protein expression levels across organisms has been observed recently [9]. Within a population of organisms of the same species, variation in mRNA abundances may be a mechanism to increase molecular diversity so as to improve chances of survival under stress conditions. Under normal conditions, less varied protein expression levels are presumably needed for proper cellular function, with variation of mRNA expression buffered by mechanisms that are yet to be defined. Finally, these data also suggest that for conserved genes, direct assessment of protein levels may often be more informative of the cellular state than analysis of mRNA levels, despite the widespread use of mRNA expression levels as proxy measurements for protein expression levels.
Supplementary Material
Acknowledgements
We thank Sabine Schrimpf and colleagues for providing data from their publication. This work was supported by grants from the N.S.F., N.I.H., and Welch (F1515) and Packard Foundations to E.M.M, NIH grant #GM55962 to PCR, and NIH grant # 5R01AI075068 to MW. MW is a Burroughs Wellcome Investigator in the Pathogenesis of Infectious Disease.
References
- 1.Anderson L, Seilhamer J. A comparison of selected mRNA and protein abundances in human liver. Electrophoresis. 1997;18:533–537. doi: 10.1002/elps.1150180333. [DOI] [PubMed] [Google Scholar]
- 2.Lu P, Vogel C, Wang R, Yao X, Marcotte EM. Absolute protein expression profiling estimates the relative contributions of transcriptional and translational regulation. Nat Biotechnol. 2007;25:117–124. doi: 10.1038/nbt1270. [DOI] [PubMed] [Google Scholar]
- 3.Schrimpf SP, Weiss M, Reiter L, Ahrens CH, et al. Comparative functional analysis of the Caenorhabditis elegans and Drosophila melanogaster proteomes. PLoS Biol. 2009;7:e48. doi: 10.1371/journal.pbio.1000048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Sousa Abreu R, Penalva LO, Marcotte EM, Vogel C. Global signatures of protein and mRNA expression levels. Mol Biosyst. 2009;5:1512–1526. doi: 10.1039/b908315d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Remm M, Storm CE, Sonnhammer EL. Automatic clustering of orthologs and in-paralogs from pairwise species comparisons. J Mol Biol. 2001;314:1041–1052. doi: 10.1006/jmbi.2000.5197. [DOI] [PubMed] [Google Scholar]
- 6.Nie L, Wu G, Zhang W. Correlation of mRNA expression and protein abundance affected by multiple sequence features related to translational efficiency in Desulfovibrio vulgaris: a quantitative analysis. Genetics. 2006;174:2229–2243. doi: 10.1534/genetics.106.065862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Raj A, Peskin CS, Tranchina D, Vargas DY, Tyagi S. Stochastic mRNA synthesis in mammalian cells. PLoS Biol. 2006;4:e309. doi: 10.1371/journal.pbio.0040309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Khaitovich P, Enard W, Lachmann M, Paabo S. Evolution of primate gene expression. Nat Rev Genet. 2006;7:693–702. doi: 10.1038/nrg1940. [DOI] [PubMed] [Google Scholar]
- 9.Weiss M, Schrimpf S, Hengartner MO, Lercher MJ, von Mering C. Shotgun proteomics data from multiple organisms reveals remarkable quantitative conservation of the eukaryotic core proteome. Proteomics. 10:1297–1306. doi: 10.1002/pmic.200900414. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.