Abstract
Cysteine is notable among the universal, proteinogenic amino acids for its facile redox chemistry. Cysteine thiolates are readily modified by reactive oxygen species (ROS), reactive electrophilic species (RES), and reactive nitrogen species (RNS). Although thiol switches are commonly triggered by disulfide bond formation, they can also be controlled by S-thiolation, S-alkylation, or modification by RNS. Thiol-based switches are common in both prokaryotic and eukaryotic organisms and activate functions that detoxify reactive species and restore thiol homeostasis while repressing functions that would be deleterious if expressed under oxidizing conditions. Here, we provide an overview of the best-understood examples of thiol-based redox switches that affect gene expression. Intra- or intermolecular disulfide bond formation serves as a direct regulatory switch for several bacterial transcription factors (OxyR, OhrR/2-Cys, Spx, YodB, CrtJ, and CprK) and indirectly regulates others (the RsrA anti-σ factor and RegB sensory histidine kinase). In eukaryotes, thiol-based switches control the yeast Yap1p transcription factor, the Nrf2/Keap1 electrophile and oxidative stress response, and the Chlamydomonas NAB1 translational repressor. Collectively, these regulators reveal a remarkable range of chemical modifications exploited by Cys residues to effect changes in gene expression. Antioxid. Redox Signal. 14, 1049—1063.
Introduction
Cells continually monitor their environment and adapt to their changing surroundings by coordinated changes in gene expression. Environmental sensing is generally the purview of proteins, including, in the simplest cases, proteins that bind DNA and directly control the expression of target genes. Proteins are notable for their propensity to bind many ligands with high affinity and specificity, as evidenced by the remarkable repertoire of enzymes and antibodies, and ligand binding is one important method of signal detection. In addition, many proteins bind cofactors (metal ions, heme, pigments), which significantly expands the range of signals that can be sensed. Finally, some proteins directly react with chemical signals, although this type of signaling is more limited because of the available range of amino acid functional groups. Here, we focus on the ability of sensory proteins to take advantage of the complex redox chemistry of cysteine to sense reactive chemical species and coordinate the expression of appropriate adaptive responses.
Thiol Modifications Caused by ROS, RNS, and RES
In most cells, the cytoplasm is a reducing environment, and protein thiols are maintained in their reduced state. Responsibility for maintaining cytosolic proteins in their reduced form rests with thiol-redox buffers such as glutathione (GSH). GSH is the major thiol reductant in eukaryotic and many prokaryotic cells, and it, in turn, is reduced by glutathione reductase at the expense of NADPH (57). Some bacteria lack GSH and rely instead on alternative low-molecular-weight (LMW) thiol reductants such as mycothiol (MSH) or bacillithiol (BSH), which are widely distributed among the actinobacteria and the Firmicutes, respectively (23, 66, 68).
The ability of the cell to maintain properly the redox state of the cytoplasm can be compromised by reactive oxygen species (ROS), reactive nitrogen species (RNS), and reactive electrophilic species (RES). ROS include hydrogen peroxide and other peroxides, superoxide anion, hypohalous acids, and the highly reactive hydoxyl radical (33). ROS may be generated endogenously from the incomplete reduction of molecular oxygen during respiration and from the autooxidation of reduced molecules in the cell. Exogenous sources of ROS also are widespread and are generated by the oxidative burst of macrophages and as part of plant defense responses (33). RNS include nitric oxide (NO), peroxynitrite (NO3−), and S-nitrosothiols (RSNOs) and lead, directly or indirectly, to protein S-nitrosylation. RES are reactive species that have electron-deficient carbon centers and include quinones, aldehydes, and epoxides. RES react with the nucleophilic cysteine thiol, leading to Cys-S-adducts. RES can be produced during metabolism or as secondary reactive intermediates in response to ROS or RNS. For example, the highly toxic α,β-unsaturated aldehyde methylglyoxal accumulates as a consequence of imbalanced glycolysis.
ROS, RNS, and RES can damage many different cellular components including proteins, nucleic acids, lipids, and metal cofactors. As a result, specific detoxification pathways and defense mechanisms have evolved to destroy the reactive species or to repair the resulting damage (2, 34). These adaptive responses are typically induced by one or more specific stressors, and this regulation is often mediated by redox-sensitive transcription factors. In the majority of those systems in which the response has been defined biochemically, sensing of ROS, RNS, and RES involves redox-active cysteine residues, which are targets for posttranslational modification by the corresponding reactive molecules (Fig. 1) (75). This leads to structural changes and concomitant alteration of transcription factor activity. Sensing is not always mediated by Cys residues: ROS/RNS also can be sensed by metal centers. For example, Bacillus subtilis PerR senses H2O2 by iron-catalyzed histidine oxidation (49), E. coli SoxR senses superoxide anion by oxidation of a [2Fe-2S] center (13), and Escherichia coli NorR senses NO by nitrosylation of a nonheme Fe center (11).
FIG. 1.
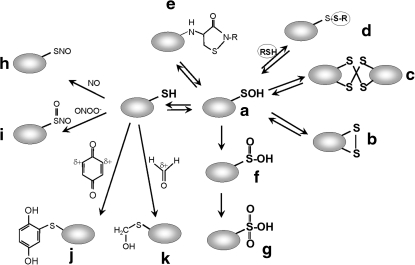
Post-translational cysteine modifications involved in redox-sensing mechanisms. Cysteine can be reversibly oxidized by ROS to the unstable Cys sulfenic acid (R-SOH; a), which can then form an intramolecular disulfide (b), an intermolecular disulfide (c), an S-thiolated adduct with a LMW thiol (RSH) (d), or a cyclic sulfenamide with a polypeptide backbone amide (e). Irreversible Cys modifications include overoxidation to Cys sulfinic (R-SO2H) (f) and sulfonic acids (R-SO3H) (g). RNS, such as nitric oxide (NO) or peroxynitrite (ONOO−), modify Cys by S-nitrosothiol formation (R-SNO) (h) or S-nitrothiol formation (R-SNO2) (i). RES with partial positively charged carbon centers (δ+), such as quinones or aldehydes, lead to thiol-(S)-alkylation of cysteine, resulting in quinone-S-adducts (j) by benzoquinone or S-hydroxymethylthiol modification by formaldehyde (k). This figure is modified from (71).
Transcription factors that sense ROS, RNS, and RES with reactive cysteine residues take full advantage of the complex chemistry of the thiol (RSH) moiety. Although Cys thiol groups often have a pKa near 8, this can be significantly reduced, depending on the protein environment, and many reactive Cys residues are ionized to the more reactive thiolate (RS−) anion at physiologic pH. The thiol modifications known to regulate transcription factor activity can be either reversible (thiol-disulfide switches, S-nitrosylation) or effectively irreversible (overoxidation to the sulfonic acid, thiol-S-alkylation).
Oxidation of protein thiols by ROS leads initially to the cysteine sulfenic acid (R-SOH). Although sulfenic acids may be transiently stabilized in some protein environments (77), they more commonly react rapidly with other thiols or by further oxidation (Fig. 1). Further oxidation leads to the higher and generally irreversible formation of cysteine sulfinic (R-SOOH) or sulfonic acids (R-SO3H). Cys sulfenates are also an important intermediate in the formation of protein–protein disulfides, which may be either intramolecular (disulfide linkage between two Cys residues of the same subunit) or intermolecular (disulfide bond formation between proteins). Alternatively, the sulfenic acid may react with LMW thiols such as GSH, Cys, MSH, or BSH by S-thiolation. S-Thiolation, including the well-characterized S-glutathionylation reaction, is a reversible modification that can protect Cys residues against irreversible overoxidation to the sulfinic and sulfonic acids (31, 57). Although reduction of sulfinic acids within peroxiredoxins may be catalyzed by sulfiredoxins, this repair pathway is not known to operate with thiol-based sensor proteins.
A more recently appreciated product of protein thiol oxidation is the cyclic sulfenamide produced by condensation of the sulfenate with a neighboring backbone amide nitrogen. Although first detected in structural studies of protein tyrosine phosphatases (82, 90), sulfenamide formation has also been observed with B. subtilis OhrR (50).
RNS also lead to reversible thiol-modifications, such as S-nitrosothiol (RS-NO) formation caused by NO, or S-nitrothiol (R-SNO2) formation caused by peroxynitrite (NO3−) (Fig. 1). Formation of S-nitrosothiols in proteins can result from the one-electron oxidation of an initially formed thiol S-nitroxide radical anion (RS-NO−) formed from the thiolate anion and NO. Alternatively, transnitrosation by a LMW S-nitrosothiol (e.g., GSNO) can lead directly to protein S-nitrosothiol formation. A role for a transient (and rapidly reversible) S-nitroxide radical anion was recently proposed for the regulation of soluble guanylate cyclase (in addition to the well-characterized regulation exerted by NO sensing at the heme site) (19).
RES such as aldehydes and quinones react with thiol-containing proteins and LMW thiols largely through thiol-S-alkylation (56) (Fig. 1). In contrast to these alkylating RES species, the electrophilic azocompound diamide leads to “disulfide stress” by catalyzing disulfide-bond formation in thiol-containing proteins. RES can have both direct effects on protein activity and indirect effects mediated by the modification or depletion or both of the LMW thiol pool of the cell.
Cysteine-containing regulators take full advantage of the diverse chemistry of the thiol group to sense ROS, RNS, and RES. We here focus on the best-characterized thiol-containing sensors from both prokaryotic and eukaryotic organisms, with an emphasis on those for which the chemical modifications that elicit the regulatory response are reasonably well known.
Prokaryotic Thiol-Based Regulatory Switches
Thiol-based, redox-responsive switches are common in bacteria and most frequently coordinate the transcription of adaptive responses to chemical stresses that either modify or deplete cellular thiol pools. High-resolution structural information is now available for many of these proteins both in their reduced and modified forms and, in some cases, bound to their DNA target sites.
OxyR: An LysR-family sensor of ROS and RNS
Salmonella typhimurium OxyR was the first-described transcription factor with a dedicated role in sensing ROS. OxyR is a member of the LysR family of DNA-binding proteins and positively regulates a large, peroxide-inducible regulon, including the catalase KatG, the alkylhydroperoxide reductase AhpCF, the DNA-binding ferritin-like protein Dps, glutaredoxin Grx, the glutathione reductase GorA, the ferric homeostasis regulator Fur, the MntH manganese transporter, and the small regulatory RNA OxyS (34). Collectively, the OxyR regulon, as defined in E. coli, mediates adaptation to peroxides by multiple mechanisms. KatG and AhpC degrade H2O2, Grx and GorA help maintain protein thiols in their reduced state, whereas induction of Dps, Fur, and MntH modulates the metal ion environment of the cell to reduce deleterious Fenton chemistry. Fur represses iron import, and Dps functions as a miniferritin to sequester iron. MntH imports Mn(II) to displace adventitiously bound ferrous iron and thereby protects macromolecules from metal-catalyzed oxidation (1).
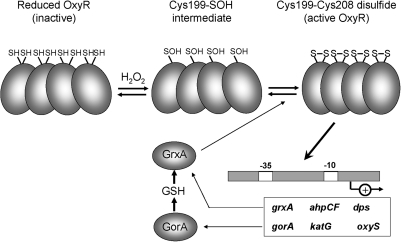
Activation of E. coli OxyR in response to peroxide stress is mediated by disulfide-bond formation. OxyR has six Cys residues, and two of these, C199 and C208, are conserved and essential for redox sensing, as shown by mutational and structural analyses (47). On exposure to ROS, C199 is oxidized to a sulfenic acid intermediate, which then forms an intramolecular disulfide bond with C208 in each subunit of the OxyR tetramer (Fig. 2) (10, 47). The formation of a C199-C208 disulfide on peroxide stress has been verified in vivo (10). OxyR can also be activated under other conditions that deplete the cellular pool of reduced LMW thiols, thereby preventing the efficient reduction of protein thiols: a condition generically referred to as disulfide stress.
FIG. 2.
The disulfide-switch model for transcriptional activation of OxyR in Escherichia coli. OxyR responds to hydrogen peroxide (H2O2) in E. coli and other bacteria. The conserved C199 and C208 residues are essential for redox-sensing activity. C199 is initially oxidized to the sulfenic acid intermediate that rapidly reacts further to form an intramolecular disulfide with Cys208. Oxidized OxyR binds as a tetramer to promoter regions to activate transcription of genes encoding proteins with antioxidant functions, including GrxA, which is involved in the reduction of oxidized OxyR.
Activation of OxyR has been visualized by determination of the crystal structures of the reduced, sulfenate intermediate, and disulfide forms of OxyR (Fig. 9A) (10). In reduced OxyR, C199 is located at the N-terminus of a α-helix and is 17 Å apart from C208. Cys199 is located on an extended loop between the α-helix and the β8 strand. Formation of the C199-C208 disulfide results in unwinding of the α-helix and movement of the α-helix/β8 loop (4). This results in a large structural change in the oligomeric interfaces and relative rotation among the OxyR subunits (10). Oxidation alters the interaction of the OxyR tetramer with DNA sites upstream from OxyR-regulated genes, thereby leading to productive interactions with RNA polymerase (RNAP) and enhanced gene expression.
FIG. 9.
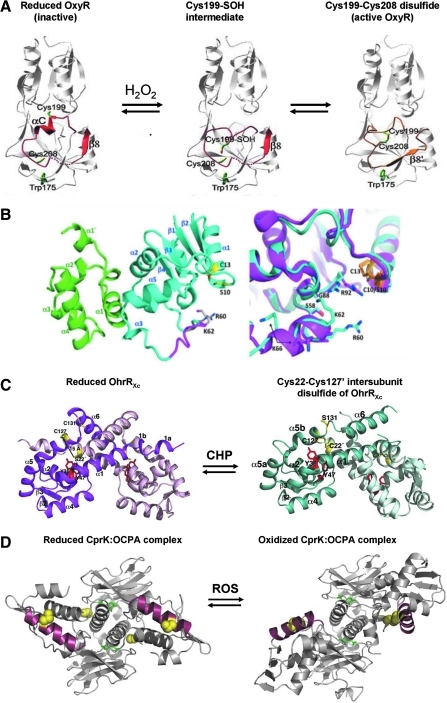
Structures of reduced and oxidized forms of the thiol-based redox sensors OxyR, Spx, OhrR (2-Cys), and CprK. (A) Structures are shown for OxyR in its reduced, Cys199 sulfenic acid intermediate and Cys199-Cys208 intramolecular disulfide conformations. In reduced OxyR, C199 and C208 are separated by 17 Å. The interaction of C199 with the side chains of T100 and R266 stabilizes the C199 thiolate anion. The residues 195 to 223, including αC and β8, are shown in red in reduced and gold in oxidized OxyR and are involved in the structural transition on oxidation. C199 is initially oxidized to the sulfenic acid intermediate that rapidly reacts further to form an intramolecular disulfide with Cys208. During OxyR oxidation, the αC helix melts, and the αC/β8 loop shifts positions. This figure is adapted from (47). (B) Structures are shown for reduced SpxC10S in complex with the αCTD. Left: Spx and αCTD are shown as teal and green ribbons, respectively. Helix α4 is colored magenta in oxidized Spx. Residues S10 and C13 are shown as sticks with carbon and sulfur atoms colored yellow and the γ-oxygen of S10, red. Right: Close-up of the region surrounding helix α4 and residues C10/S10 and C13 after the superposition of the oxidized and reduced αCTD-Spx complex structures. Reduced Spx is shown as a magenta ribbon, and oxidized Spx, as a teal ribbon. The C10-C13 disulfide bond is shown in orange sticks, and S10 and C13 from the reduced structure are shown as yellow sticks. This figure was reproduced from (62). (C) The structures of the 2-Cys OhrR of Xanthomonas campestris OhrRXc in its reduced form (left) and oxidized Cys22-Cys127′ intersubunit disulfide-linked state (right). In reduced OhrR, one subunit is colored magenta, and the other, light pink. In the reduced form, Cys22 and Cys127′ are separated by 15.5 Å. The hydrogen bonds provided by the Y36′ and Y46′ side chains (red) are involved in stabilization of the Cys22 thiolate (yellow). In the oxidized intersubunit disulfide version of the OhrR, one subunit is colored teal, and the other, light blue. On oxidation of OhrR, Cys22 is oxidized to the Cys22-SOH intermediate, disrupting the Y36′-C22-Y47′ hydrogen-bonding network and distorting α5. This allows the movement of Cys127′ by 135-degrees rotation and 8-Å translation, formation of the Cys22-Cys127′ intersubunit disulfide bond, and α6-α6′ helix-swapped reconfiguration, resulting in 28-degree rigid body rotations of the HTH motifs and dissociation from the DNA. This figure is reproduced from (64). (D) The Desulfitobacterium hafniense CprK structure is shown in ligand (OCPA) bound, reduced complex (left) and in the oxidized complex (right) containing intersubunit disulfides between C11 and C200′ (in the DNA-binding HTH motif). The C11 and C200 cysteines are indicated by yellow spheres, the bound OCPA ligand is shown in stick form (green), and the DNA-recognition helix is in red. Note that in the reduced form (left), the two DNA-binding helices are approximately parallel and appropriately spaced for interaction with their cognate DNA operator. Oxidation distorts the protein, disrupting the DNA interaction surface and preventing transcription activation. For further details of the structural changes induced in CprK by ligand binding and protein oxidation, please see (37, 54). This figure is kindly provided by Dr. David Leys, Manchester, UK.
Although OxyR can be activated by intramolecular disulfide-bond formation, the Stamler group has shown that other modifications of Cys199 may also be sufficient to effect regulation, including C199 oxidation to the sulfenate, S-nitrosylation, or S-glutathionylation (44). These observations led to a proposal in which different modified forms of OxyR might have differential abilities to activate gene expression. Whereas reduced and glutathionylated C199 of OxyR result in noncooperative binding, the sulfenate and S-nitrosylated forms bind cooperatively. These results led to the hypothesis that OxyR may experience a diverse set of modifications, and that these may constitute a "code" for coordinating distinct genetic responses. Although this is an attractive idea, the in vivo relevance of these alternative Cys modifications remains to be determined, and subsequent studies of the E. coli stress responses elicited by RNS failed to reveal a significant role for OxyR (61).
OxyR is widely conserved among both Gram-negative and Gram-positive bacteria and numerous othologues have been shown to coordinate the expression of antioxidant functions in response to peroxide stress. Collectively, these studies implicate OxyR orthologues in the regulation of oxidative stress responses, virulence, biofilm formation, fimbrial synthesis, and intestinal or urinary tract colonization in pathogenic bacteria. All characterized OxyR proteins use cysteine residue(s) for sensing, but the details differ between species. Most OxyR orthologues appear to function, like E. coli OxyR, as transcriptional activators in response to oxidative stress. Interestingly, Deinococcus radiodurans OxyR has only one Cys residue (C210) and is oxidized to the Cys sulfenate in vitro (7).
Streptomyces RsrA: An anti-σ factor regulating a disulfide stress response
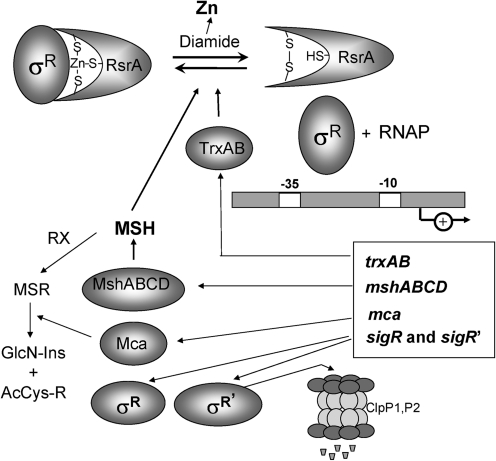
The sensing of ROS by protein thiols typically involves a protein environment that stabilizes a reactive thiolate anion. In some proteins, this is accomplished by coordination to Zn(II), which increases the reactivity of one or more thiolate ligands. One of the best-understood Zn(II)-dependent thiolate switches is the Streptomyces coelicolor RsrA anti-sigma (71). RsrA sequesters the σR transcription factor in an inactive complex (38). However, oxidation of two Cys residues in RsrA to a disulfide destroys the Zn(II)-binding site, leading to metal ion loss, a significant structural change in RsrA, and release of active σR (Fig. 3) (3, 55). The coordination environment of Zn(II) in RsrA has been controversial: most data support a model in which Zn(II) is coordinated to C11, H37, C41, and C44 (94). These four residues are conserved in RsrA orthologues, and substitutions at these sites affect anti-sigma activity. On oxidation, RsrA forms a disulfide bond between C11 and either C41 or C44. The resultant release of σR activates the σR regulon, which includes functions to restore thiol homeostasis.
FIG. 3.
Redox regulation of the Zn-binding antisigma factor RsrA of Streptomyces coelicolor. RsrA is a zinc-binding, redox-sensitive anti-σ factor that sequesters σR under reducing conditions. Diamide treatment causes intramolecular disulfide-bond formation in RsrA, resulting in the release of Zn(II) and active σR. Free σR activates transcription of σR regulon genes, including trxBA and trxC (thioredoxin and thioredoxin reductase), mshA (a glycosyltransferase catalyzing the first step in MSH synthesis), and mca (mycothiol-S-conjugate amidase). Collectively, these thiol-reducing systems function in reduction of oxidized RsrA and restore the thiol redox balance. σR and the unstable σR′protein (with a destabilizing N-terminal extension) are positively autoregulated (see text).
Oxidation of RsrA is elicited in vivo by oxidizing agents that deplete the pool of reduced thiols in the cell (e.g., diamide). Such disulfide stress conditions are physiologically distinct from peroxide-stress and, in S. coelicolor, the disulfide stress response is regulated by RsrA/σR (38). The σR regulon includes genes encoding thioredoxin, thioredoxin reductase, and for the biosynthesis of the low-molecular-weight thiols cysteine and MSH, both of which would be depleted under disulfide stress (67, 74). The σR factor is also positively autoregulated and activates synthesis of an unstable σR' protein that differs from σR by the presence of an N-terminal extension of 55 amino acids (42). The σR regulon is thus controlled by both positive (autoregulation) and negative feedback loops. The major negative feedback loop is the induction of thiol-disulfide reducing systems that reduce oxidized RsrA. The rapid degradation of induced σR` by the stress-induced ClpP1/P2 proteases provides a second negative feedback loop.
Spx: A thiol-based activator and sensor of ROS and RES
Spx is a redox-sensing regulator originally described in B. subtilis, where it controls a large regulon of genes that function in maintenance of the thiol-redox balance of the cell and in the utilization of organosulfur compounds (95). Spx is a structurally atypical transcription activator and is most closely related in sequence to the ArsC (arsenate reductase) family of enzymes. Whereas ArsC proteins couple thiol oxidation to the reduction of arsenate to arsenite, Spx has adopted this same active site to serve as a sensor of disulfide stress by formation of a disulfide bond within a conserved C10XXC13 motif (96).
Exposure to thiol-depleting electrophiles (diamide, quinones, aldehydes) or oxidative-stress conditions (paraquat, peroxides) oxidizes Spx, leading to the activation of Spx regulon genes (63). Target genes for Spx include those encoding the thioredoxin/thioredoxin reductase system (trxA, trxB), a thiol-dependent peroxidase (tpx), FMN-dependent oxidoreductases (nfrA, yugJ), methionine sulfoxide reductase (msrA), and a DJ1/PfpI-family cysteine proteinase (yraA) (63).
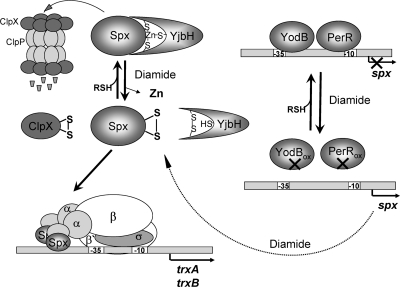
The activation of the Spx regulon in response to oxidants and electrophiles involves complex transcriptional and posttranslational control mechanisms (Fig. 4). Transcription of spx is initiated at four promoters, and involves the σM, σB, and σA containing holoenzymes at sites upstream of the bicistronic yjbC-spx operon and the σA RNAP from a site (spx P3) immediately upstream of spx (96). Induction of spx transcription in response to electrophiles (diamide, quinones) involves inactivation of the peroxide regulon repressor PerR and the MarR-type/DUF24 family repressor YodB that repress the spx P3 promoter (52).
FIG. 4.
Mechanisms regulating Spx levels in response to disulfide stress in Bacillus subtilis. Under nonstress conditions, Spx interacts with the Zn(II)-binding YjbH adaptor protein, which targets Spx to ClpXP proteolytic degradation. YjbH and ClpX both contain Cys-rich Zn-binding sites that are oxidized to an intramolecular disulfide, resulting in the release of Zn(II). The levels of Spx are increased by diamide because of the inactivation of YjbH and ClpX and subsequent decreased proteolysis. The spx gene is regulated by the repressors YodB and PerR, which are both inactivated by diamide stress. Spx has a redox-active CxxC motif that is oxidized by diamide and converts it to a transcriptional activator. Oxidized Spx contacts αCTD of RNAP to activate transcription of trxA and trxB and thereby maintain the thiol-redox state of the cell. (Note: the figure does not specify the precise sites of binding of the regulatory proteins to promoter DNA).
Spx also is a target of post-translational control. Notably, the activity of Spx is directly regulated by thiol oxidation, leading to disulfide bond formation as well as by its interaction with sulfate (46, 65). Oxidized Spx interacts with the α-C-terminal domain (αCTD) of RNAP that binds to DNA sites adjacent to the trxA and trxB promoters (Fig. 4). Unlike most transcription activators, Spx does not interact strongly with specific DNA-binding sites upstream of target genes, although specific binding interactions likely do occur when co-bound with RNA polymerase. Two structures of the Spx/αCTD complex have been resolved, one in the sulfate-bound and one in the sulfate-free conformation, which together reveal the details of protein–protein contact (Fig. 9B) (46, 65).
Spx levels can be elevated under oxidative stress conditions by a reduction in proteolytic turnover. Proteolysis of Spx is normally mediated by the ClpXP protease system. Indeed, spx was originally identified as a suppressor of clpP and clpX mutations, as it accumulates in clpX or clpP mutants to a high level and is responsible for several of the phenotypes associated with loss of ClpXP (96). The proteolytic degradation of Spx is regulated by the ROS-dependent inactivation of two proteins, YjbH and ClpX. The Spx regulon member YjbH contains a Zn-binding His-Cys-rich domain and functions as an adaptor for ClpXP proteolysis of Spx (25). Inactivation of this Zn-binding site by ROS or electrophiles may lead to Zn release, intramolecular disulfide formation, and release of Spx from the adaptor. This would thereby stabilize Spx against proteolysis. Moreover, oxidation of the Cys-rich Zn-finger domain of ClpX in response to diamide stress also leads to Zn release and inactivation of ClpX, which also results in an elevation of Spx levels. Thus, Spx activity may be modulated by as many as three different thiol-disulfide switches including those within Spx itself, the YjbH adaptor protein, and the ClpX unfoldase subunit of the ClpXP protease complex.
Spx plays important roles as both a positive and negative regulator of gene expression. In the absence of RNAP, Spx does not bind directly to its target promoters. However, recent evidence suggests that Spx likely binds to specific sites in the DNA as part of a larger activation complex. It has been shown, for example, that oxidized Spx and the αCTD of RNA polymerase result in a supershift on binding to the trxB promoter, and this complex is disrupted by using DTT (62). The interaction of Spx/αCTD complex with the trxB promoter requires the cis-acting elements −45AGCA-42 and −34AGCG-31, and the first site is also conserved in the nfrA promoter. Thus, the current model suggests that oxidized Spx binds to the αCTD of RNAP and steers the RNAP to transcribe promoters of Spx target genes, such as trxA and trxB (Fig. 4). Negative regulation by Spx appears to occur by a related mechanism in which the binding of Spx to the αCTD occludes the contact surfaces for other activator proteins. This anti-alpha activity likely accounts for the large number of genes that are downregulated in an Spx-dependent manner under oxidative-stress conditions (95, 96).
OhrR: MarR-family sensors of organic hydroperoxides
The OhrR family of regulators sense organic hydroperoxides (OHP) and other ROS by oxidation of a critical, highly conserved Cys residue. In the decade since their first discovery, detailed studies of OhrR family proteins have revealed an intriguing diversity of chemical modifications that are all ultimately initiated by oxidation of the conserved Cys residue near the amino-terminus to a sulfenic acid.
OHPs form as by-products of metabolism and include, for example, lipid hydroperoxides formed from peroxyl radicals and unsaturated fatty acids. The most commonly used OHPs in laboratory studies include the model compounds cumene hydroperoxide (CHP), t-butyl peroxide, and linoleic acid hydroperoxide (LHP). OHPs are detoxified by a variety of thiol-dependent peroxidases including the AhpCF and Ohr systems. The AhpCF alkylhydroperoxide reductase system is composed of a thiol-dependent peroxidase (AhpC) and the FMN-dependent NADPH-oxidoreductase (AhpF) that reduces oxidized AhpC (34). The AhpCF system is an efficient reductant of H2O2 and, consistent with this observation, is often induced as part of peroxide-stress responses. For example, the ahpCF operon is induced by OxyR in enteric bacteria and is regulated by PerR in B. subtilis.
Regulation of OhrA-like peroxidases is mediated by OhrR, a dimeric redox-sensing MarR (multiple antibiotic resistance)-family repressor, as originally defined in B. subtilis (21) and Xanthomonas campestris (88). The ohrR gene is oriented divergently to its ohrA target gene in B. subtilis. A second functionally redundant paralogue, ohrB, is regulated as part of the σB general stress response.
Most MarR-type regulators have a winged helix-turn-helix (HTH) DNA-binding motif and control genes that confer resistance to antibiotics, organic solvents, detergents, ROS, and RES. Several MarR family members are important for the regulation of virulence (16). Other redox-sensing MarR-family regulators include the S. aureus OhrR paralogues MgrA and SarZ, which regulate antibiotic resistance and virulence genes, the multidrug efflux regulator MexR, and Pseudomonas aeruginosa OspR, as reviewed elsewhere in this Forum issue: here we focus on the B. subtilis and Xanthomonas campestris proteins as prototypes for the 1-Cys and 2-Cys subfamilies of OhrR proteins.
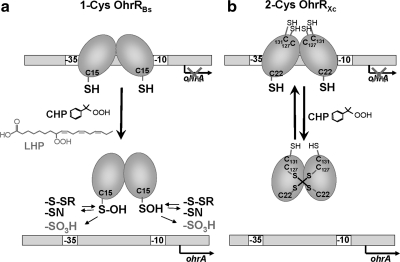
The mechanisms for inactivation of OhrR proteins in response to different OHPs, such as CHP and LHP, have been studied in most detail in B. subtilis and Xanthomonas species. OhrR proteins can be divided into the 2-Cys family, as in X. campestris, which has two redox-active Cys residues (72), and the 1-Cys family with the prototype in B. subtilis (22). For both regulators, the sensing of ROS initiates with the oxidation of a highly conserved peroxidatic Cys residue near the amino-terminus.
The conserved C15 of OhrR of B. subtilis is essential for redox sensing (22). Although the OhrR dimer contains two C15 residues (one in each monomer), intersubunit disulfide bond formation is not observed. Instead, on addition of CHP, OhrR is oxidized to a Cys sulfenic acid in vitro (22). Although this modification was originally thought to inactivate the repressor (32), subsequent studies established that the OhrR C15 sulfenic acid still binds to its operator site, and further modification of the active site is needed to elicit derepression (50).
The fate of the OhrR C15 residue depends both on the nature of the oxidant and the availability of LMW thiols. When isolated from cells treated with CHP, OhrR was present predominantly as the mixed disulfide species with cysteine (S-cysteinylated OhrR) and a 398-Da LMW thiol subsequently defined as bacillithiol (BSH) (50, 68). The S-thiolation of OhrR by Cys and BSH is consistent with the finding that these are the two dominant LMW thiols in B. subtilis: in organisms containing GSH, the protein is instead S-glutathionylated (50). In vitro studies have confirmed that oxidized OhrR dissociates from DNA when it is S-cysteinylated, and this reaction occurs readily even with micromolar concentrations of Cys. Interestingly, OhrR is also inactivated when treated with CHP in vitro in the absence of LMW thiols. Under these conditions, the initially formed sulfenic acid slowly condenses with a backbone amide to generate a cyclic sulfenamide (50). Finally, we noted that in the presence of the strong oxidant LHP, OhrR was irreversibly overoxidized to Cys sulfinic and sulfonic acids (86).
Unlike B. subtilis OhrR, the X. campestris OhrR repressor is inactivated by formation of an intermolecular protein disulfide (73). Phylogenetic analyses indicate that most OhrR-like regulators probably work by a similar mechanism, and these are defined as the 2-Cys subfamily of OhrR proteins because of the presence of both a peroxidatic Cys near the amino-terminus and one or more Cys residues in a carboxyl-terminal domain. The X. campestris C22 residue is positionally and functionally analogous to C15 in B. subtilis OhrR. On treatment with OHP, C22 is oxidized to the sulfenic acid, which then leads to intersubunit disulfide formation between C22 of one subunit and C127′ (where the prime indicates a residue of the opposing subunit in the dimer) (64, 73) (Fig. 5).
FIG. 5.
Redox-sensing mechanisms of the 1-Cys and 2-Cys type OhrR family proteins. OhrR controls the OhrA thiol-dependent peroxidase and contains one conserved reactive Cys15 residue in B. subtilis and three Cys residues (C22, C127, and C131) in X. campestris. (a) The 1-Cys OhrR protein of B. subtilis is initially oxidized by CHP to the Cys sulfenic acid, which further undergoes reversible S-thiolation with Cys and BSH in vivo and may also generate a cyclic sulfenamide when thiols are depleted. In the presence of the strong oxidant LHP, C15 is irreversibly overoxidized to the sulfinic and sulfonic acids. (b) The 2-Cys OhrR protein of X. campestris is regulated by intersubunit disulfide formation between C22 and C127’ of opposing subunits. Both Cys residues are essential for redox sensing in vivo, as shown by mutagenesis (73).
The functional relevance of the evolution of both 1-Cys and 2-Cys mechanisms for OhrR sensors is not yet clear. The introduction of a second cysteine residue in B. subtilis OhrR, at a position equivalent to C127 in the Xanthomonas protein, converts this prototypic 1-Cys protein into a 2-Cys OhrR protein, now regulated by intersubunit disulfide bond formation (85). Thus, no fundamental distinction exists in the reaction mechanism. One consequence of the 1-Cys mechanism, however, is that the protein-inactivation pathway will be very sensitive to the availability of LMW thiols, and if the cell is experiencing both oxidative stress and disulfide stress, the protein is likely to be irreversibly inactivated by overoxidation. This is precisely what was observed when B. subtilis cells pretreated with diamide, to deplete LMW thiol pools, were then treated with CHP (50). In contrast, 2-Cys proteins will be reversibly oxidized to the intermolecular disulfide, regardless of overall cellular thiol status, although one can imagine that reactivation of the repressor by reduction of the protein disulfide might be impaired under disulfide stress conditions.
High-resolution crystal structures are available for B. subtilis 1-Cys OhrR in its free and DNA-bound state (32), for X. campestris OhrR in the reduced and oxidized states (64), and for a homologue from S. aureus (SarZ) in the reduced, sulfenate, and S-thiolated forms (78). These crystal structures support a model in which modification of the initially formed Cys sulfenate by S-thiolation leads to an allosteric change in the DNA-binding domains and release of the repressor from its operator (Fig. 9C). For example, in the reduced X. campestris protein, the hydrogen bonds of the Y36' and Y46' side chains stabilize the C22 thiolate located 15.5 Å from its redox partner C127′. Oxidation of C22 to sulfenic acid by OHP disrupts this hydrogen-bonding network and results in a rotation of C127′ by 135 degrees accompanied by an 8-Å translation, thereby allowing intersubunit disulfide formation between C22 and C127′. The resulting 28-degree rigid body rotation of the domains containing the HTH motifs results in dissociation of OhrR from the operator (64). Although the structural studies to date have only characterized uniformly oxidized OhrR proteins (with both active sites in each dimer oxidized), studies with a single-chain OhrR (scOhrR) variant suggest that oxidation of only one active-site cysteine (of the two symmetric cysteines in the dimer) is likely sufficient for inactivation of the repressor (15). In principle, this may lead to a more sensitive induction of the Ohr peroxidase when cells are exposed to relatively low levels of OHPs.
YodB as MarR/DUF24-family sensor for quinones and diamide
In addition to OhrR, B. subtilis contains several other MarR-family redox sensors belonging to the MarR/DUF24 subfamily (51). In contrast to OhrR, DUF24 family regulators respond specifically to RES (e.g., diamide, quinones, and aldehydes) but not to ROS (2). The azocompound diamide causes disulfide stress by the direct catalysis of disulfide bond formation including S-thiolations of cytoplasmic proteins (31, 81). Quinones have two distinct mechanisms of action, an oxidative and an electrophilic mode. Quinones can be reduced incompletely to reactive semiquinone anions that in turn reduce molecular oxygen to ROS. As electrophiles, quinones form S-adducts with cellular thiols via the irreversible thiol-(S)-alkylation reaction. Protein–protein crosslinking can lead to aggregation of many thiol-containing cytoplasmic proteins (56). As consequence of RES stress, the thiol redox buffer of the cell is depleted (56, 70).
The MarR-type transcription factors YodB, MhqR, and YvaP control specific detoxification pathways that confer resistance to diamide and quinones (2). Together these sensors control paralogous azoreductases (AzoR1 and AzoR2), nitroreductases (YodC and MhqN), and thiol-dependent dioxygenases (CatE, MhqA, MhqE, and MhqO) that are involved in the reduction or ring-cleavage of the toxic compounds (51, 56). The MarR/DUF24-family YodB-repressor is highly conserved among low-GC Gram-positive bacteria, including Bacillus, Listeria, Staphylococcus, Enterococcus, and Clostridia species. YodB has three Cys residues (C6, C101, and C108), and C6 is conserved among MarR/DUF24-family proteins. Mutational analyses indicate that the C6 residue is important for DNA binding and also contributes to redox sensing in vivo (8, 51). Current evidence supports a model in which YodB resembles the 2-Cys type OhrR regulators and is inactivated by formation of a C6-C101′ intersubunit disulfide in response to treatment with diamide and quinones both in vitro and in vivo (8).
In addition to YodB, B. subtilis encodes MarR/DUF24-family regulators YybR, YdeP, YkvN, YtcD, YdzF, HxlR, and YvaP of mostly unknown function. HxlR is an activator of the formaldehyde-responsive hxlAB operon encoding the genes specifying the ribulose monophosphate pathway that functions in formaldehyde detoxification (93). Because the N-terminal Cys residue is also conserved in HxlR, it seems likely that this serves as a sensing residue for aldehydes, presumably via post-translational Cys modification.
Another 1-Cys MarR/DUF24-family regulator, CgR1435 (QorR), was recently characterized as a redox-sensing repressor of a quinone oxidoreductase QorA in Corynebacterium glutamicum (14). The DNA-binding activity of QorR was inhibited by diamide and H2O2 in vitro. In this case, the redox-sensing mechanism apparently involves intersubunit disulfide formation between the two C17 residues of QorR, although this has yet to be confirmed in vivo.
MerR/NmlR-like sensors for RES and RNS
MerR is the prototype for the eponymous family of regulatory proteins that most commonly control metal ion resistance determinants and efflux pumps. MerR (mercury resistance regulator) is a well-characterized representative that senses Hg(II) by coordination to three Cys residues, which then converts MerR from a repressor to a transcription activator. Several other MerR family members also sense metals by coordination to cysteine. One important redox-sensing regulator of the MerR family is SoxR, which contains a redox-active [2Fe-2S] center that is oxidized by superoxide anions or nitrosylated by RNS, leading to activation of soxS expression.
Recently, a subfamily of redox-sensing MerR regulators (the NmlR subfamily) has been identified in Neisseria species, Haemophilus influenza, and Streptococcus pneumoniae. NmlR regulators respond to RES and NO stress, regulate genes that protect cells against RNS (nitrosative stress), and thereby contribute to virulence (40, 87). Neisseria gonorrhoea NmlR was first characterized as redox-controlled regulator that positively controls the class III alcohol dehydrogenase AdhC (known to also possess S-nitrosoglutathione reductase activity), a Cpx-type ATPase, and a thioredoxin reductase. The NmlR regulon protects against both NO and disulfide stress (41). Mutational analyses reveal that four Cys residues (C40, 54, 71, and 95) are important for NmlR function, and a model has been proposed in which these cysteines contribute to a Zn(II) binding site that stabilizes a repressor form of the protein (41). In response to oxidative stress, NmlR is modified, possibly by disulfide bond formation, leading to loss of repression or activation or both of target promoters.
NmlR homologues have a common function in the regulation of divergently transcribed adh-like genes that are known or likely to help protect against RNS. Mutations in the nmlR and adhC genes of Neisseria species, H. influenzae, and S. pneumoniae rendered cells sensitive to GSNO stress, because of the lower GSNO reductase activity. AdhR, an NmlR homologue from B. subtilis, positively regulates the adhA-yraA operon (encoding a thiol-dependent formaldehyde reductase and DJ-1/PfpI-family cysteine proteinase YraA) and yraC (encoding a γ-carboxymuconolactone decarboxylase) (70). In contrast to other NmlR homologues, AdhR of B. subtilis responds specifically to aldehydes (formaldehyde and methylglyoxal), but not to NO. Mutation of C52 in AdhR abolished transcriptional activation of the adhA-yraA operon by formaldehyde in vivo. Thus, AdhR is hypothesized to be redox-regulated via thiol-S-alkylation by aldehydes at C52 (70).
Redox regulation of photosynthesis: The RegBA two-component system and the PpsR/CrtJ repressor
Rhodobacter spp. serve as model systems for the regulation of gene expression in facultative, photosynthetic bacteria. Rhodobacter spp. perform aerobic respiration under high oxygen tension and anoxygenic photosynthesis under low oxygen tension. Consequently, the genes encoding the photosynthesis apparatus and pigment-binding proteins are repressed by oxygen. These include the photosynthetic reaction center (RC) and transmembrane light-harvesting complexes 1 and 2 (LH1 and LH2) encoded by the puc (LH2) and puf operons (RC and LH1).
Photosynthesis genes are globally regulated by the redox-sensing two-component system RegBA in R. capsulatus (also known as PrrBA in R. sphaeroides) (18). Orthologous two-component systems are found in a wide variety of alpha- and gamma-proteobacteria. The RegB histidine kinase has a conserved redox-active Cys residue that is oxidized to an intermolecular disulfide, thereby inactivating the RegB kinase when oxygen levels are high (18). The redox signal detected by RegB is the ubiquinone pool in the respiratory chain (89). RegB, in turn, phosphorylates the RegA response regulator, a DNA-binding transcription factor that regulates, both positively and negatively, a large regulon of target genes.
Redox-sensing repressors (CrtJ of R. capsulatus, PpsR of R. sphaeroides, and their orthologues) also control photosynthesis gene expression in response to oxygen and light (18, 59). PpsR and CrtJ are homotetrameric repressors with two central PAS domains (17). Under oxidizing conditions, CrtJ and PpsR bind to conserved operators in the promoters of the bacteriochlorophyll (bch), carotenoid (crt), and LH2 (puc) operons and the heme biosynthesis genes (17). The CrtJ and PpsR repressors undergo intramolecular disulfide formation between the two conserved redox-sensing residues (Cys251 and C424 in PpsR; C249 and C420 in CrtJ) on oxidation (59). Mutational analyses showed that both CrtJ Cys residues are important for function: C249A and C420A mutants are impaired in DNA-binding in vivo. A similar thiol-disulfide switch has been shown for PpsR only in vitro: the redox state of PspR is not affected under oxidizing and light conditions in vivo (9).
Comparison of the redox properties of the dithiol/disulfide couple for CrtJ and PpsR revealed interesting differences. The redox potential (Em value) of PpsR was significantly more negative (-320 mV) than that of CrtJ (-180 mV) (43). CrtJ is only oxidized under aerobic conditions, even though oxygen has no effect on the redox potential in the cytoplasm. Thus, CrtJ is not always in redox equilibrium with the reducing cytoplasm (-222 to −224 mV) (59). Two PpsR orthologues have also been characterized from Bradyrhizobium. Interestingly, PpsR1 contains only a single Cys residue, whereas PpsR2 lacks Cys residues (36). Thus, considerable diversity in the redox-sensing mechanisms may exist in this family of repressor proteins.
In addition to redox sensing through a thiol-disulfide switch, PpsR repressors are also regulated in response to light. In R. sphaeroides, regulation of PpsR activity in response to oxygen and blue light involves the AppA antirepressor. AppA binds PspR in response to low oxygen levels, resulting in derepression of photosynthesis genes (58). Modulation of AppA activity is mediated by both an N-terminal photoreceptor BLUF domain [sensor of Blue Light Uses FAD] and a C-terminal Cys-rich heme-binding domain (26). The C-terminal domain transmits the redox signals, but not the light signal, to affect PpsR activity (28).
CprK as a redox-regulated, CRP-FNR family regulator of halorespiration
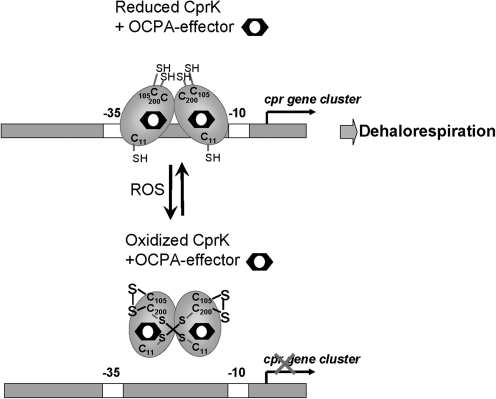
Desulfitobacterium dehalogenans is an anaerobic Gram-positive bacterium that is able to conserve energy through halorespiration, the reductive dehalogenation of haloorganic compounds, such as chloroalkenes and chlorophenols, which serve as terminal electron acceptors. The genes for o-chlorophenolacetic acid (OCPA) reductive dehalogenase (cprAB) are encoded by the cpr cluster in Desulfitobacterium spp. The cpr gene cluster is controlled by CprK that belongs to the CRP-FNR-(cAMP receptor protein-fumarate nitrate respiration regulator)-family of transcription factors (45). CprK is a homodimeric DNA-binding protein with a C-terminal winged HTH domain and an N-terminal sensor domain, which binds OCPA. Binding of the effector OCPA causes conformational changes that enable binding to “haloboxes” in the cpr gene promoter (24, 79, 80). The effector OCPA can bind to reduced or oxidized CprK, but only reduced CprK can activate transcription (37, 60). Thus, activation of CprK is controlled by both effector binding and the redox state of the cell.
Redox control of CprK involves a thiol-disulfide switch that inactivates CprK under aerobic conditions in D. halogenans and D. hafniense. CprK from D. halogenans has five Cys residues (C11, C105, C111, C161, and C200). On oxidation, CprK loses DNA-binding activity (80) because of the formation of a C11-C200′ intermolecular disulfide or an intramolecular disulfide bond between C105 and C111 (79) or both. In contrast, the C11 and C200 CprK mutant proteins of D. hafniense retained redox-sensitivity under aerobic conditions when expressed in E. coli (24). Recent studies confirmed the C11-C200′ intermolecular disulfide in CprK of D. halogenans by H2O2 and diamide treatment after expression in E. coli cells (27). The loss of DNA-binding activity and redox-control of CprK C11 mutant was further confirmed in vitro and in vivo. The two modes of dehalorespiration control by CprK are shown in Fig. 6, involving redox and effector-binding switches.
FIG. 6.
CprK regulation in Desulfitobacterium dehalogenans. CprK is present in a reduced state in the cell under anaerobic conditions. In response to oxidative stress, CprK forms a Cys11-Cys200′ intersubunit disulfide bond and a Cys105-Cys200 intramolecular disulfide, which inhibit CprK DNA-binding activity. Oxidized and reduced CprK bind the effector OCPA with similar affinity. Reduced, ligand-bound CprK binds DNA and activates transcription of the cpr gene cluster for halorespiration.
Crystal structures of the allosteric states of CprK from D. hafniense were resolved in the ligand-free reduced (2.0 Å), ligand-free oxidized (3.2 Å), ligand-bound reduced (1.8 Å), and DNA-bound (1.8 Å) states, and together suggest a model for the CprK-activation mechanism (Fig. 9D) (37, 54). In the absence of ligands, CprK is flexible with the C11 and C200′ residues of apposed monomers in proximity, explaining the susceptibility of CprK to intersubunit disulfide formation. Oxidation of CprK constrains the DNA-binding domains, reduces flexibility, and disrupts the interdomain network that establishes the DNA-binding surface. Ligand binding to reduced CprK shifts the DNA-binding domains into positions that allow operator binding and transcriptional activation. Activation of CprK is an example of allosteric regulation: the binding of the ligand to reduced CprK first induces changes in the N-terminal effector-interaction domain that in turn cause structural changes in the DNA-binding domain required for specific DNA contacts. Amino acid residues that play key roles in the allosteric activation mechanism of CprK are not conserved among CRP-FNR–type regulators, suggesting that distinct allosteric mechanisms are used to control different metabolic processes (54).
Eukaryotic Thiol Switches
Saccharomyces cerevisiae Yap1p
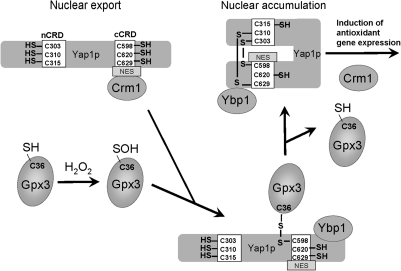
The S. cerevisiae Yap1p is a model peroxide-sensing transcription factor regulated by cysteine oxidation (5, 29). Yap1p is a basic leucine zipper (bZIP)-class transcription factor that responds to ROS by activating the expression of ∼100 or more proteins, including several with obvious roles in thiol homeostasis, such as thioredoxin, GSH biosynthetic enzymes, and glutathione reductase (48). Orthologous regulators include Pap1 in Schizosaccharomyces pombe and Cap1 in Candida albicans. Yap1p contains two redox-active, cysteine-rich domains (CRDs) important for signaling. The mechanism of regulation of Yap1p been recently reviewed (5), and is here briefly summarized.
Yap1p is regulated by subcellular localization with redox-changes in the accessibility of a C-terminal nuclear export signal playing a key role (Fig. 7) (92). Peroxides oxidize Yap1p, leading ultimately to the formation of one or more intramolecular disulfide bonds bridging the two CRDs. Oxidation of Yap1p is indirectly mediated by glutathione peroxidase Gpx3 (12). Oxidation of Gpx3 by substrate peroxides leads to a C36 sulfenic acid, which reacts with Yap1p C598 (in the C-terminal CRD). This initial covalent Yap1p-Gpx3 complex is resolved by thiol-disulfide exchange, leading to reduced Gpx3 and Yap1p containing a C303-C598 disulfide. Similar Gpx3-mediated processes are thought to generate additional disulfide bonds linking C310-C629 and C315-C629, which further activates Yap1p.
FIG. 7.
Regulation of yeast transcription factor Yap1p in response to peroxide stress. The activity of Yap1p is regulated by nuclear localization. Yap1p contains two Cys-rich domains (C-terminal and N-terminal CRD) that harbor redox-sensitive Cys residues that are reduced under nonstress conditions. The nuclear export sequence (NES) can bind to the nuclear export receptor (Crm1), and Yap1p is exported to the cytosol, preventing its accumulation in the nucleus. On peroxide stress, C36 of the glutathione peroxidase (Gpx3) is oxidized to sulfenate and interacts with C598 of Yap1p through intermolecular disulfide formation to a Yap1-Gpx3 intermediate that binds the Yap1-binding protein 1 (Ybp1). Thiol-disulfide exchange reactions lead subsequently to C303-C598 disulfide formation and other intramolecular disulfide bonds (e.g., C310-C629) in Yap1p, accompanied by conformational changes of Yap1p and the release of Gpx3. The NES export signal is not accessible for Crm1 interaction, which leads to accumulation of Yap1p in the nucleus. This in turn leads to activation of transcription of Yap1p antioxidant target genes (5).
Although disulfide-bond formation between the two CRDs is likely the major pathway activating Yap1p in response to ROS, other modifications elicited by RES and metalloids can also stimulate Yap1p activity. Activation of Yap1p-dependent genes by the thiol oxidant diamide, electrophiles and alkylating agents, and heavy metals likely involves intramolecular domain disulfide-bond formation or direct thiol modifications or both. A similar mechanism regulates the S. pombe Yap1p orthologue Pap1, in which cysteine(s) are directly modified by diethylmaleate (6).
Keap1/Nrf2
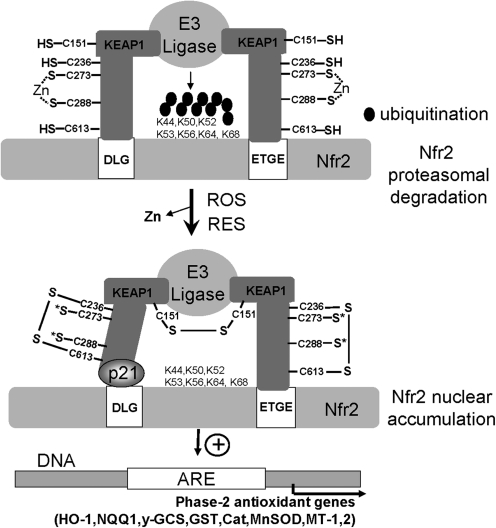
Humans also have a robust cellular response to oxidative stress and electrophiles. The Nrf2 transcription factor plays a key role in the activation of genes preceded by antioxidant response elements (AREs) by formation of heterodimeric complexes with other transcription factors. Altogether, at least 200 genes can be induced in response to ROS and RES. Nrf2 activity is normally limited by its association with its cysteine-rich, redox-sensing partner protein, Keap1. The Keap1 protein associates with Nrf2 and targets it for ubiquitin-dependent proteosomal degradation. Oxidation or S-alkylation of Keap1 leads to release of Nrf2, which was proposed to result in nuclear translocation (35). However, recent results suggest instead that Keap1 may be shuttling between the cytoplasm and the nucleus, and Nrf2 may be nucleus localized (39, 69) (Fig. 8).
FIG. 8.
Redox regulation of the KEAP1/Nfr2 system in response to ROS and RES. The Keap1 sensor protein contains more than 25 Cys residues, including the redox-sensitive C151 and the C273 and C288 residues that coordinate Zn. Under nonstress conditions, Keap1 binds to the DLG and ETGE sites in the Nfr2 transcription factor, positioning the lysine residues of Nfr2 optimal for ubiquitination by the E3 ligase and subsequent degradation of Nfr2. On exposure to ROS, the C151 residues form an intersubunit disulfide bond in Keap1, and intramolecular disulfide bonds between C236-C613 have also been detected. Oxidation of C273 and C288 leads to Zn release. S* indicates unknown oxoforms (75). RES leads to alkylation of many different Cys residues, including C151, C273, and C288 and others. Disulfide bonds or S-alkylation of Keap1 decreases binding to the DLG site of Nfr2. Oxidation of Keap1 also masks the NES, leading to nuclear accumulation of Nfr2, which is further stabilized by binding to p21. This leads to transcriptional activation of Nfr2 that binds to antioxidant response elements (AREs) in the promoters of phase-2 genes encoding heme oxygenase (HO-1), quinone oxidoreductase (NQQ1), γ-glutamylcysteine synthetase (γ-GCS), glutathione S-transferase (GST), catalase (Cat), superoxide dismutase (MnSod), or metallothioneins (MT-1,2) and other antioxidant-function genes. This figure is adapted from (75).
The chemical correlates of Keap1 oxidation have been well studied and include a variety of possible modifications (39, 69). Keap1 proteins typically have 25 or more Cys residues, which presents a formidable challenge in attempting to define the relevant thiol modifications. Three of these (C151, C273, and C288) appear to be central to the signaling mechanism. At least two (C273 and C288) contribute to a Zn(II)-binding site, which presumably contributes to the ionization, and consequent high reactivity, of the coordinating cysteine thiolates at physiologic pH (76). Reaction of these Cys thiolates leads to Zn(II) release and disulfide-bond formation. Recently, two important disulfide linkages have been determined that occur on ROS exposure in Keap1, the C151 intermolecular disulfide bridge between two subunits and the intramolecular long-range C236-C613 disulfide that result in conformational changes in Keap1(20). As a result, oxidized Keap1 protein dissociates from Nrf2, which can then activate transcription of phase-2 genes with antioxidant and thiol-protecting functions.
Emerging systems: Other cysteine-dependent eukaryotic signaling systems
Cysteines, and their ability to be reversibly oxidized and covalently modified, play roles in a wide variety of other eukaryotic signaling systems (4, 5, 75, 77). In many cases, the molecular details have yet to emerge, but it can be anticipated that in most cases, these new systems will represent variations on the themes noted earlier.
We noted that anoxygenic, phototrophic bacteria repress the expression of photosynthetic reaction complexes in the presence of oxygen. However, photosynthetic reaction complexes are also regulated by ROS in oxygenic phototrophs. Photosynthetic eukaryotes coordinate the synthesis of light-harvesting complexes in response to changes in the cytosolic redox state by using cysteine-containing, redox-active translational repressors. The emerging model system for this type of regulation is the NAB1 repressor from Chlamydomonas reinhardtii (91). NAB1 contains two cysteine residues (C181 and C226) within its C-terminal RNA recognition domain that are implicated in regulation. Mutant analyses indicate that the single and double Cys mutants have distinct phenotypes, which suggests that regulation may be more complex than a simple thiol-disulfide switch (91).
A second example of a novel and unexpected role for cysteine modification in eukaryotic signaling is provided by the soluble guanylate cyclase (sGC) enzyme, which mediates the many physiologic effects of nitric oxide in mammals. In response to NO, sGC synthesizes cGMP, which triggers smooth muscle relaxation, resulting in vasodilation. The ability of sGC to sense and respond to NO has long been ascribed to its bound heme cofactor, but recent results indicate that the NO–heme interaction only partially activates sGC: full activation requires an additional NO-dependent interaction with one or more cysteine thiolates. Because this second stage of activation is rapidly reversible and does not require an oxidant, formation of a stable S-nitrosothiol adduct is seemingly excluded (or at least not required) for this activation. Marletta and co-workers (19) suggest that the relevant species may be a thionitroxide (RSNO−) formed from the thiolate anion (RS−) and NO (19). If verified, this further expands the range of thiol modifications implicated in regulating protein activity.
Concluding Remarks
The critical role of protein disulfides in stabilization of secreted proteins has been appreciated for 60 years, since the earliest days of protein chemistry. Indeed, the determination of the first complete protein sequence, of bovine pancreatic insulin, was complicated by the presence of intermolecular disulfide bonds linking the two chains (83). More recently, the central role of the thiol sulfenate in mediating oxidative modifications has been appreciated (77), and the burgeoning field of redox proteomics has allowed a global perspective on the extent of protein oxidation in both normal and genetically or chemically stressed cells (30, 53, 84). The correlates between protein oxidation and human disease are widely appreciated, although deconvoluting cause and effect remains a challenge (84). Despite more than 60 years of work, the full chemical versatility of the cysteine residue, and its ability to regulate protein function, still holds surprises. The most recent additions to the panoply of cysteine-based regulatory modifications include the cyclic sulfenamide (PTPase 1b, OhrR) and the possible transient of formation thionitroxide anions (sGC) (19, 50, 82, 90).
Abbreviations Used
- αCTD
C-terminal domain of the alpha subunit of bacterial RNA polymerase
- ARE
antioxidant response element
- BSH
bacillithiol
- CHP
cumene hydroperoxide
- CRD
cysteine-rich domain
- Cys
cysteine
- GSH
glutathione
- GSNO
S-nitrosoglutathione
- HTH
helix-turn-helix
- LMW
low molecular weight
- MSH
mycothiol
- NO
nitric oxide
- OCPA
o-chlorophenolacetic acid
- RES
reactive electrophilic species
- RNAP
RNA polymerase
- RNS
reactive nitrogen species
- ROS
reactive oxygen species
- RSNO
S-nitrosothiol
Acknowledgments
We thank Dr. David Leys (Manchester, UK) for providing the Figure shown in 9D. This work was supported by grants from the National Science Foundation (MCB0640616 to JDH), the National Institutes of Health (GM059323 to JDH), and the Deutsche Forschungsgemeinschaft (AN746/2-1 to HA).
References
- 1.Anjem A. Varghese S. Imlay JA. Manganese import is a key element of the OxyR response to hydrogen peroxide in Escherichia coli. Mol Microbiol. 2009;72:844–858. doi: 10.1111/j.1365-2958.2009.06699.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antelmann H. Hecker M. Zuber P. Proteomic signatures uncover thiol-specific electrophile resistance mechanisms in Bacillus subtilis. Expert Rev Proteom. 2008;5:77–90. doi: 10.1586/14789450.5.1.77. [DOI] [PubMed] [Google Scholar]
- 3.Bae JB. Park JH. Hahn MY. Kim MS. Roe JH. Redox-dependent changes in RsrA, an anti-sigma factor in Streptomyces coelicolor: zinc release and disulfide bond formation. J Mol Biol. 2004;335:425–435. doi: 10.1016/j.jmb.2003.10.065. [DOI] [PubMed] [Google Scholar]
- 4.Barford D. The role of cysteine residues as redox-sensitive regulatory switches. Curr Opin Struct Biol. 2004;14:679–686. doi: 10.1016/j.sbi.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 5.Brandes N. Schmitt S. Jakob U. Thiol-based redox switches in eukaryotic proteins. Antioxid Redox Signal. 2009;11:997–1014. doi: 10.1089/ars.2008.2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Castillo EA. Ayte J. Chiva C. Moldon A. Carrascal M. Abian J. Jones N. Hidalgo E. Diethylmaleate activates the transcription factor Pap1 by covalent modification of critical cysteine residues. Mol Microbiol. 2002;45:243–254. doi: 10.1046/j.1365-2958.2002.03020.x. [DOI] [PubMed] [Google Scholar]
- 7.Chen H. Xu G. Zhao Y. Tian B. Lu H. Yu X. Xu Z. Ying N. Hu S. Hua Y. A novel OxyR sensor and regulator of hydrogen peroxide stress with one cysteine residue in Deinococcus radiodurans. PLoS One. 2008;3:e1602. doi: 10.1371/journal.pone.0001602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chi BK. Albrecht D. Gronau K. Becher D. Hecker M. Antelmann H. The redox-sensing regulator YodB senses quinones and diamide via a thiol-disulfide switch in Bacillus subtilis. Proteomics. 2010;10:3155–3164. doi: 10.1002/pmic.201000230. [DOI] [PubMed] [Google Scholar]
- 9.Cho SH. Youn SH. Lee SR. Yim HS. Kang SO. Redox property and regulation of PpsR, a transcriptional repressor of photosystem gene expression in Rhodobacter sphaeroides. Microbiology. 2004;150:697–706. doi: 10.1099/mic.0.26777-0. [DOI] [PubMed] [Google Scholar]
- 10.Choi H. Kim S. Mukhopadhyay P. Cho S. Woo J. Storz G. Ryu SE. Structural basis of the redox switch in the OxyR transcription factor. Cell. 2001;105:103–113. doi: 10.1016/s0092-8674(01)00300-2. [DOI] [PubMed] [Google Scholar]
- 11.D'Autreaux B. Tucker NP. Dixon R. Spiro S. A non-haem iron centre in the transcription factor NorR senses nitric oxide. Nature. 2005;437:769–772. doi: 10.1038/nature03953. [DOI] [PubMed] [Google Scholar]
- 12.Delaunay A. Pflieger D. Barrault MB. Vinh J. Toledano MB. A thiol peroxidase is an H2O2 receptor and redox-transducer in gene activation. Cell. 2002;111:471–481. doi: 10.1016/s0092-8674(02)01048-6. [DOI] [PubMed] [Google Scholar]
- 13.Ding H. Demple B. In vivo kinetics of a redox-regulated transcriptional switch. Proc Natl Acad Sci U S A. 1997;94:8445–8449. doi: 10.1073/pnas.94.16.8445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ehira S. Ogino H. Teramoto H. Inui M. Yukawa H. Regulation of quinone oxidoreductase by the redox-sensing transcriptional regulator QorR in Corynebacterium glutamicum. J Biol Chem. 2009;284:16736–16742. doi: 10.1074/jbc.M109.009027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eiamphungporn W. Soonsanga S. Lee JW. Helmann JD. Oxidation of a single active site suffices for the functional inactivation of the dimeric Bacillus subtilis OhrR repressor in vitro. Nucleic Acids Res. 2009;37:1174–1181. doi: 10.1093/nar/gkn1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ellison DW. Miller VL. Regulation of virulence by members of the MarR/SlyA family. Curr Opin Microbiol. 2006;9:153–159. doi: 10.1016/j.mib.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 17.Elsen S. Jaubert M. Pignol D. Giraud E. PpsR: a multifaceted regulator of photosynthesis gene expression in purple bacteria. Mol Microbiol. 2005;57:17–26. doi: 10.1111/j.1365-2958.2005.04655.x. [DOI] [PubMed] [Google Scholar]
- 18.Elsen S. Swem LR. Swem DL. Bauer CE. RegB/RegA, a highly conserved redox-responding global two-component regulatory system. Microbiol Mol Biol Rev. 2004;68:263–279. doi: 10.1128/MMBR.68.2.263-279.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fernhoff NB. Derbyshire ER. Marletta MA. A nitric oxide/cysteine interaction mediates the activation of soluble guanylate cyclase. Proc Natl Acad Sci U S A. 2009;106:21602–21607. doi: 10.1073/pnas.0911083106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fourquet S. Guerois R. Biard D. Toledano MB. Activation of NRF2 by nitrosative agents and H2O2 involves KEAP1 disulfide formation. J Biol Chem. 2010;285:8463–8471. doi: 10.1074/jbc.M109.051714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fuangthong M. Atichartpongkul S. Mongkolsuk S. Helmann JD. OhrR is a repressor of ohrA, a key organic hydroperoxide resistance determinant in Bacillus subtilis. J Bacteriol. 2001;183:4134–4141. doi: 10.1128/JB.183.14.4134-4141.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fuangthong M. Helmann JD. The OhrR repressor senses organic hydroperoxides by reversible formation of a cysteine-sulfenic acid derivative. Proc Natl Acad Sci U S A. 2002;99:6690–6695. doi: 10.1073/pnas.102483199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gaballa A. Newton GL. Antelmann H. Parsonage D. Upton H. Rawat M. Claiborne A. Fahey RC. Helmann JD. Biosynthesis and functions of bacillithiol, a major low-molecular-weight thiol in Bacilli. Proc Natl Acad Sci U S A. 2010;107:6482–6486. doi: 10.1073/pnas.1000928107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gabor K. Verissimo CS. Cyran BC. Ter Horst P. Meijer NP. Smidt H. de Vos WM. van der Oost J. Characterization of CprK1, a CRP/FNR-type transcriptional regulator of halorespiration from Desulfitobacterium hafniense. J Bacteriol. 2006;188:2604–2613. doi: 10.1128/JB.188.7.2604-2613.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garg SK. Kommineni S. Henslee L. Zhang Y. Zuber P. The YjbH protein of Bacillus subtilis enhances ClpXP-catalyzed proteolysis of Spx. J Bacteriol. 2009;191:1268–1277. doi: 10.1128/JB.01289-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gomelsky M. Klug G. BLUF: a novel FAD-binding domain involved in sensory transduction in microorganisms. Trends Biochem Sci. 2002;27:497–500. doi: 10.1016/s0968-0004(02)02181-3. [DOI] [PubMed] [Google Scholar]
- 27.Gupta N. Ragsdale SW. Dual roles of an essential cysteine residue in activity of a redox-regulated bacterial transcriptional activator. J Biol Chem. 2008;283:28721–28728. doi: 10.1074/jbc.M800630200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Han Y. Braatsch S. Osterloh L. Klug G. A eukaryotic BLUF domain mediates light-dependent gene expression in the purple bacterium Rhodobacter sphaeroides 2.4.1. Proc Natl Acad Sci U S A. 2004;101:12306–12311. doi: 10.1073/pnas.0403547101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Herrero E. Ros J. Belli G. Cabiscol E. Redox control and oxidative stress in yeast cells. Biochim Biophys Acta. 2008;1780:1217–1235. doi: 10.1016/j.bbagen.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 30.Hochgrafe F. Mostertz J. Albrecht D. Hecker M. Fluorescence thiol modification assay: oxidatively modified proteins in Bacillus subtilis. Mol Microbiol. 2005;58:409–425. doi: 10.1111/j.1365-2958.2005.04845.x. [DOI] [PubMed] [Google Scholar]
- 31.Hochgrafe F. Mostertz J. Pother DC. Becher D. Helmann JD. Hecker M. S-cysteinylation is a general mechanism for thiol protection of Bacillus subtilis proteins after oxidative stress. J Biol Chem. 2007;282:25981–25985. doi: 10.1074/jbc.C700105200. [DOI] [PubMed] [Google Scholar]
- 32.Hong M. Fuangthong M. Helmann JD. Brennan RG. Structure of an OhrR-ohrA operator complex reveals the DNA binding mechanism of the MarR family. Mol Cell. 2005;20:131–141. doi: 10.1016/j.molcel.2005.09.013. [DOI] [PubMed] [Google Scholar]
- 33.Imlay JA. Pathways of oxidative damage. Annu Rev Microbiol. 2003;57:395–418. doi: 10.1146/annurev.micro.57.030502.090938. [DOI] [PubMed] [Google Scholar]
- 34.Imlay JA. Cellular defenses against superoxide and hydrogen peroxide. Annu Rev Biochem. 2008;77:755–776. doi: 10.1146/annurev.biochem.77.061606.161055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Itoh K. Wakabayashi N. Katoh Y. Ishii T. O'Connor T. Yamamoto M. Keap1 regulates both cytoplasmic-nuclear shuttling and degradation of Nrf2 in response to electrophiles. Genes Cells. 2003;8:379–391. doi: 10.1046/j.1365-2443.2003.00640.x. [DOI] [PubMed] [Google Scholar]
- 36.Jaubert M. Zappa S. Fardoux J. Adriano JM. Hannibal L. Elsen S. Lavergne J. Vermeglio A. Giraud E. Pignol D. Light and redox control of photosynthesis gene expression in Bradyrhizobium: dual roles of two PpsR. J Biol Chem. 2004;279:44407–44416. doi: 10.1074/jbc.M408039200. [DOI] [PubMed] [Google Scholar]
- 37.Joyce MG. Levy C. Gabor K. Pop SM. Biehl BD. Doukov TI. Ryter JM. Mazon H. Smidt H. van den Heuvel RH. Ragsdale SW. van der Oost J. Leys D. CprK crystal structures reveal mechanism for transcriptional control of halorespiration. J Biol Chem. 2006;281:28318–28325. doi: 10.1074/jbc.M602654200. [DOI] [PubMed] [Google Scholar]
- 38.Kang JG. Paget MS. Seok YJ. Hahn MY. Bae JB. Hahn JS. Kleanthous C. Buttner MJ. Roe JH. RsrA, an anti-sigma factor regulated by redox change. EMBO J. 1999;18:4292–4298. doi: 10.1093/emboj/18.15.4292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kaspar JW. Niture SK. Jaiswal AK. Nrf2:INrf2 (Keap1) signaling in oxidative stress. Free Radic Biol Med. 2009;47:1304–1309. doi: 10.1016/j.freeradbiomed.2009.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kidd SP. Jiang D. Jennings MP. McEwan AG. Glutathione-dependent alcohol dehydrogenase AdhC is required for defense against nitrosative stress in Haemophilus influenzae. Infect Immun. 2007;75:4506–4513. doi: 10.1128/IAI.00487-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kidd SP. Potter AJ. Apicella MA. Jennings MP. McEwan AG. NmlR of Neisseria gonorrhoeae: a novel redox responsive transcription factor from the MerR family. Mol Microbiol. 2005;57:1676–1689. doi: 10.1111/j.1365-2958.2005.04773.x. [DOI] [PubMed] [Google Scholar]
- 42.Kim MS. Hahn MY. Cho Y. Cho SN. Roe JH. Positive and negative feedback regulatory loops of thiol-oxidative stress response mediated by an unstable isoform of sigmaR in actinomycetes. Mol Microbiol. 2009;73:815–825. doi: 10.1111/j.1365-2958.2009.06824.x. [DOI] [PubMed] [Google Scholar]
- 43.Kim SK. Mason JT. Knaff DB. Bauer CE. Setterdahl AT. Redox properties of the Rhodobacter sphaeroides transcriptional regulatory proteins PpsR and AppA. Photosynth Res. 2006;89:89–98. doi: 10.1007/s11120-006-9086-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim SO. Merchant K. Nudelman R. Beyer WF., Jr Keng T. DeAngelo J. Hausladen A. Stamler JS. OxyR: a molecular code for redox-related signaling. Cell. 2002;109:383–396. doi: 10.1016/s0092-8674(02)00723-7. [DOI] [PubMed] [Google Scholar]
- 45.Korner H. Sofia HJ. Zumft WG. Phylogeny of the bacterial superfamily of Crp-Fnr transcription regulators: exploiting the metabolic spectrum by controlling alternative gene programs. FEMS Microbiol Rev. 2003;27:559–592. doi: 10.1016/S0168-6445(03)00066-4. [DOI] [PubMed] [Google Scholar]
- 46.Lamour V. Westblade LF. Campbell EA. Darst SA. Crystal structure of the in vivo-assembled Bacillus subtilis Spx/RNA polymerase alpha subunit C-terminal domain complex. J Struct Biol. 2009;168:352–356. doi: 10.1016/j.jsb.2009.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee C. Lee SM. Mukhopadhyay P. Kim SJ. Lee SC. Ahn WS. Yu MH. Storz G. Ryu SE. Redox regulation of OxyR requires specific disulfide bond formation involving a rapid kinetic reaction path. Nat Struct Mol Biol. 2004;11:1179–1185. doi: 10.1038/nsmb856. [DOI] [PubMed] [Google Scholar]
- 48.Lee J. Godon C. Lagniel G. Spector D. Garin J. Labarre J. Toledano MB. Yap1 and Skn7 control two specialized oxidative stress response regulons in yeast. J Biol Chem. 1999;274:16040–16046. doi: 10.1074/jbc.274.23.16040. [DOI] [PubMed] [Google Scholar]
- 49.Lee JW. Helmann JD. The PerR transcription factor senses H2O2 by metal-catalysed histidine oxidation. Nature. 2006;440:363–367. doi: 10.1038/nature04537. [DOI] [PubMed] [Google Scholar]
- 50.Lee JW. Soonsanga S. Helmann JD. A complex thiolate switch regulates the Bacillus subtilis organic peroxide sensor OhrR. Proc Natl Acad Sci U S A. 2007;104:8743–8748. doi: 10.1073/pnas.0702081104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Leelakriangsak M. Huyen NT. Towe S. van Duy N. Becher D. Hecker M. Antelmann H. Zuber P. Regulation of quinone detoxification by the thiol stress sensing DUF24/MarR-like repressor, YodB in Bacillus subtilis. Mol Microbiol. 2008;67:1108–1124. doi: 10.1111/j.1365-2958.2008.06110.x. [DOI] [PubMed] [Google Scholar]
- 52.Leelakriangsak M. Kobayashi K. Zuber P. Dual negative control of spx transcription initiation from the P3 promoter by repressors PerR and YodB in Bacillus subtilis. J Bacteriol. 2007;189:1736–1744. doi: 10.1128/JB.01520-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Leichert LI. Gehrke F. Gudiseva HV. Blackwell T. Ilbert M. Walker AK. Strahler JR. Andrews PC. Jakob U. Quantifying changes in the thiol redox proteome upon oxidative stress in vivo. Proc Natl Acad Sci U S A. 2008;105:8197–8202. doi: 10.1073/pnas.0707723105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Levy C. Pike K. Heyes DJ. Joyce MG. Gabor K. Smidt H. van der Oost J. Leys D. Molecular basis of halorespiration control by CprK, a CRP-FNR type transcriptional regulator. Mol Microbiol. 2008;70:151–167. doi: 10.1111/j.1365-2958.2008.06399.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li W. Bottrill AR. Bibb MJ. Buttner MJ. Paget MS. Kleanthous C. The role of zinc in the disulphide stress-regulated anti-sigma factor RsrA from Streptomyces coelicolor. J Mol Biol. 2003;333:461–472. doi: 10.1016/j.jmb.2003.08.038. [DOI] [PubMed] [Google Scholar]
- 56.Liebeke M. Pother DC. van Duy N. Albrecht D. Becher D. Hochgrafe F. Lalk M. Hecker M. Antelmann H. Depletion of thiol-containing proteins in response to quinones in Bacillus subtilis. Mol Microbiol. 2008;69:1513–1529. doi: 10.1111/j.1365-2958.2008.06382.x. [DOI] [PubMed] [Google Scholar]
- 57.Masip L. Veeravalli K. Georgiou G. The many faces of glutathione in bacteria. Antioxid Redox Signal. 2006;8:753–762. doi: 10.1089/ars.2006.8.753. [DOI] [PubMed] [Google Scholar]
- 58.Masuda S. Bauer CE. AppA is a blue light photoreceptor that antirepresses photosynthesis gene expression in Rhodobacter sphaeroides. Cell. 2002;110:613–623. doi: 10.1016/s0092-8674(02)00876-0. [DOI] [PubMed] [Google Scholar]
- 59.Masuda S. Dong C. Swem D. Setterdahl AT. Knaff DB. Bauer CE. Repression of photosynthesis gene expression by formation of a disulfide bond in CrtJ. Proc Natl Acad Sci U S A. 2002;99:7078–7083. doi: 10.1073/pnas.102013099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mazon H. Gabor K. Leys D. Heck AJ. van der Oost J. van den Heuvel RH. Transcriptional activation by CprK1 is regulated by protein structural changes induced by effector binding and redox state. J Biol Chem. 2007;282:11281–11290. doi: 10.1074/jbc.M611177200. [DOI] [PubMed] [Google Scholar]
- 61.Mukhopadhyay P. Zheng M. Bedzyk LA. LaRossa RA. Storz G. Prominent roles of the NorR and Fur regulators in the Escherichia coli transcriptional response to reactive nitrogen species. Proc Natl Acad Sci U S A. 2004;101:745–750. doi: 10.1073/pnas.0307741100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nakano MM. Lin A. Zuber CS. Newberry KJ. Brennan RG. Zuber P. Promoter recognition by a complex of Spx and the C-terminal domain of the RNA polymerase alpha subunit. PLoS One. 2010;5:e8664. doi: 10.1371/journal.pone.0008664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nakano S. Kuster-Schock E. Grossman AD. Zuber P. Spx-dependent global transcriptional control is induced by thiol-specific oxidative stress in Bacillus subtilis. Proc Natl Acad Sci U S A. 2003;100:13603–13608. doi: 10.1073/pnas.2235180100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Newberry KJ. Fuangthong M. Panmanee W. Mongkolsuk S. Brennan RG. Structural mechanism of organic hydroperoxide induction of the transcription regulator OhrR. Mol Cell. 2007;28:652–664. doi: 10.1016/j.molcel.2007.09.016. [DOI] [PubMed] [Google Scholar]
- 65.Newberry KJ. Nakano S. Zuber P. Brennan RG. Crystal structure of the Bacillus subtilis anti-alpha, global transcriptional regulator, Spx, in complex with the alpha C-terminal domain of RNA polymerase. Proc Natl Acad Sci U S A. 2005;102:15839–15844. doi: 10.1073/pnas.0506592102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Newton GL. Buchmeier N. Fahey RC. Biosynthesis and functions of mycothiol, the unique protective thiol of Actinobacteria. Microbiol Mol Biol Rev. 2008;72:471–494. doi: 10.1128/MMBR.00008-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Newton GL. Fahey RC. Regulation of mycothiol metabolism by sigma(R) and the thiol redox sensor anti-sigma factor RsrA. Mol Microbiol. 2008;68:805–809. doi: 10.1111/j.1365-2958.2008.06222.x. [DOI] [PubMed] [Google Scholar]
- 68.Newton GL. Rawat M. La Clair JJ. Jothivasan VK. Budiarto T. Hamilton CJ. Claiborne A. Helmann JD. Fahey RC. Bacillithiol is an antioxidant thiol produced in Bacilli. Nat Chem Biol. 2009;5:625–627. doi: 10.1038/nchembio.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nguyen T. Nioi P. Pickett CB. The Nrf2-antioxidant response element signaling pathway and its activation by oxidative stress. J Biol Chem. 2009;284:13291–13295. doi: 10.1074/jbc.R900010200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nguyen TT. Eiamphungporn W. Mader U. Liebeke M. Lalk M. Hecker M. Helmann JD. Antelmann H. Genome-wide responses to carbonyl electrophiles in Bacillus subtilis: control of the thiol-dependent formaldehyde dehydrogenase AdhA and cysteine proteinase YraA by the MerR-family regulator YraB (AdhR) Mol Microbiol. 2009;71:876–894. doi: 10.1111/j.1365-2958.2008.06568.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Paget MS. Buttner MJ. Thiol-based regulatory switches. Annu Rev Genet. 2003;37:91–121. doi: 10.1146/annurev.genet.37.110801.142538. [DOI] [PubMed] [Google Scholar]
- 72.Panmanee W. Vattanaviboon P. Eiamphungporn W. Whangsuk W. Sallabhan R. Mongkolsuk S. OhrR, a transcription repressor that senses and responds to changes in organic peroxide levels in Xanthomonas campestris pv. phaseoli. Mol Microbiol. 2002;45:1647–1654. doi: 10.1046/j.1365-2958.2002.03116.x. [DOI] [PubMed] [Google Scholar]
- 73.Panmanee W. Vattanaviboon P. Poole LB. Mongkolsuk S. Novel organic hydroperoxide-sensing and responding mechanisms for OhrR, a major bacterial sensor and regulator of organic hydroperoxide stress. J Bacteriol. 2006;188:1389–1395. doi: 10.1128/JB.188.4.1389-1395.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Park JH. Roe JH. Mycothiol regulates and is regulated by a thiol-specific antisigma factor RsrA and sigma(R) in Streptomyces coelicolor. Mol Microbiol. 2008;68:861–870. doi: 10.1111/j.1365-2958.2008.06191.x. [DOI] [PubMed] [Google Scholar]
- 75.Paulsen CE. Carroll KS. Orchestrating redox signaling networks through regulatory cysteine switches. ACS Chem Biol. 2010;5:47–62. doi: 10.1021/cb900258z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Penner-Hahn J. Zinc-promoted alkyl transfer: a new role for zinc. Curr Opin Chem Biol. 2007;11:166–171. doi: 10.1016/j.cbpa.2007.02.038. [DOI] [PubMed] [Google Scholar]
- 77.Poole LB. Karplus PA. Claiborne A. Protein sulfenic acids in redox signaling. Annu Rev Pharmacol Toxicol. 2004;44:325–347. doi: 10.1146/annurev.pharmtox.44.101802.121735. [DOI] [PubMed] [Google Scholar]
- 78.Poor CB. Chen PR. Duguid E. Rice PA. He C. Crystal structures of the reduced, sulfenic acid, and mixed disulfide forms of SarZ, a redox active global regulator in Staphylococcus aureus. J Biol Chem. 2009;284:23517–23524. doi: 10.1074/jbc.M109.015826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pop SM. Gupta N. Raza AS. Ragsdale SW. Transcriptional activation of dehalorespiration: identification of redox-active cysteines regulating dimerization and DNA binding. J Biol Chem. 2006;281:26382–26390. doi: 10.1074/jbc.M602158200. [DOI] [PubMed] [Google Scholar]
- 80.Pop SM. Kolarik RJ. Ragsdale SW. Regulation of anaerobic dehalorespiration by the transcriptional activator CprK. J Biol Chem. 2004;279:49910–49918. doi: 10.1074/jbc.M409435200. [DOI] [PubMed] [Google Scholar]
- 81.Pother DC. Liebeke M. Hochgrafe F. Antelmann H. Becher D. Lalk M. Lindequist U. Borovok I. Cohen G. Aharonowitz Y. Hecker M. Diamide triggers mainly S thiolations in the cytoplasmic proteomes of Bacillus subtilis and Staphylococcus aureus. J Bacteriol. 2009;191:7520–7530. doi: 10.1128/JB.00937-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Salmeen A. Andersen JN. Myers MP. Meng TC. Hinks JA. Tonks NK. Barford D. Redox regulation of protein tyrosine phosphatase 1B involves a sulphenyl-amide intermediate. Nature. 2003;423:769–773. doi: 10.1038/nature01680. [DOI] [PubMed] [Google Scholar]
- 83.Sanger F. Sequences, sequences, and sequences. Annu Rev Biochem. 1988;57:1–28. doi: 10.1146/annurev.bi.57.070188.000245. [DOI] [PubMed] [Google Scholar]
- 84.Seo YH. Carroll KS. Profiling protein thiol oxidation in tumor cells using sulfenic acid-specific antibodies. Proc Natl Acad Sci U S A. 2009;106:16163–16168. doi: 10.1073/pnas.0903015106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Soonsanga S. Lee JW. Helmann JD. Conversion of Bacillus subtilis OhrR from a 1-Cys to a 2-Cys peroxide sensor. J Bacteriol. 2008;190:5738–5745. doi: 10.1128/JB.00576-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Soonsanga S. Lee JW. Helmann JD. Oxidant-dependent switching between reversible and sacrificial oxidation pathways for Bacillus subtilis OhrR. Mol Microbiol. 2008;68:978–986. doi: 10.1111/j.1365-2958.2008.06200.x. [DOI] [PubMed] [Google Scholar]
- 87.Stroeher UH. Kidd SP. Stafford SL. Jennings MP. Paton JC. McEwan AG. A pneumococcal MerR-like regulator and S-nitrosoglutathione reductase are required for systemic virulence. J Infect Dis. 2007;196:1820–1826. doi: 10.1086/523107. [DOI] [PubMed] [Google Scholar]
- 88.Sukchawalit R. Loprasert S. Atichartpongkul S. Mongkolsuk S. Complex regulation of the organic hydroperoxide resistance gene (ohr) from Xanthomonas involves OhrR, a novel organic peroxide-inducible negative regulator, and posttranscriptional modifications. J Bacteriol. 2001;183:4405–4412. doi: 10.1128/JB.183.15.4405-4412.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Swem LR. Gong X. Yu CA. Bauer CE. Identification of a ubiquinone-binding site that affects autophosphorylation of the sensor kinase RegB. J Biol Chem. 2006;281:6768–6775. doi: 10.1074/jbc.M509687200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.van Montfort RL. Congreve M. Tisi D. Carr R. Jhoti H. Oxidation state of the active-site cysteine in protein tyrosine phosphatase 1B. Nature. 2003;423:773–777. doi: 10.1038/nature01681. [DOI] [PubMed] [Google Scholar]
- 91.Wobbe L. Blifernez O. Schwarz C. Mussgnug JH. Nickelsen J. Kruse O. Cysteine modification of a specific repressor protein controls the translational status of nucleus-encoded LHCII mRNAs in Chlamydomonas. Proc Natl Acad Sci U S A. 2009;106:13290–13295. doi: 10.1073/pnas.0900670106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wood MJ. Storz G. Tjandra N. Structural basis for redox regulation of Yap1 transcription factor localization. Nature. 2004;430:917–921. doi: 10.1038/nature02790. [DOI] [PubMed] [Google Scholar]
- 93.Yurimoto H. Hirai R. Matsuno N. Yasueda H. Kato N. Sakai Y. HxlR, a member of the DUF24 protein family, is a DNA-binding protein that acts as a positive regulator of the formaldehyde-inducible hxlAB operon in Bacillus subtilis. Mol Microbiol. 2005;57:511–519. doi: 10.1111/j.1365-2958.2005.04702.x. [DOI] [PubMed] [Google Scholar]
- 94.Zdanowski K. Doughty P. Jakimowicz P. O'Hara L. Buttner MJ. Paget MS. Kleanthous C. Assignment of the zinc ligands in RsrA, a redox-sensing ZAS protein from Streptomyces coelicolor. Biochemistry. 2006;45:8294–8300. doi: 10.1021/bi060711v. [DOI] [PubMed] [Google Scholar]
- 95.Zuber P. Spx-RNA polymerase interaction and global transcriptional control during oxidative stress. J Bacteriol. 2004;186:1911–1918. doi: 10.1128/JB.186.7.1911-1918.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zuber P. Management of oxidative stress in Bacillus. Annu Rev Microbiol. 2009;63:575–597. doi: 10.1146/annurev.micro.091208.073241. [DOI] [PubMed] [Google Scholar]